Abstract
Human neuroserpin (hNS) is a serine protease inhibitor that belongs to the serpin superfamily and is expressed in nervous tissues. The serpin fold is generally characterized by a long exposed loop, termed the reactive center loop, that acts as bait for the target protease. Intramolecular insertion of the reactive center loop into the main serpin β-sheet leads to the serpin latent form. As with other known serpins, hNS pathological mutants have been shown to accumulate as polymers composed of quasi-native protein molecules. Although hNS polymerization has been intensely studied, a general agreement about serpin polymer organization is still lacking. Here we report a biophysical characterization of native hNS that is shown to undergo two distinct conformational transitions, at 55°C and 85°C, both leading to distinct latent and polymeric species. The latent and polymer hNS forms obtained at 45°C and 85°C differ in their chemical and thermal stabilities; furthermore, the hNS polymers also differ in size and morphology. Finally, the 85°C polymer shows a higher content of intermolecular β-sheet interactions than the 45°C polymer. Together, these results suggest a more complex conformational scenario than was previously envisioned, and, in a general context, may help reconcile the current contrasting views on serpin polymerization.
Introduction
Human neuroserpin (hNS) belongs to the serpin serine protease inhibitor superfamily. It specifically inhibits tissue plasminogen activator (tPA) in nervous tissues, where it is mainly expressed (1,2). hNS is held to play an important, yet poorly understood, role in learning, memory, and synaptic plasticity (3,4). So far, five pathological hNS mutations have been characterized as being responsible for an autosomally dominant disease known as familial encephalopathy with neuroserpin inclusion bodies (FENIB). This disease results in early-onset dementia and epilepsy, with each mutated variant giving rise to different degrees of severity (1,5). As reported for other serpins, in vivo the pathological mutations lead to the formation of latent and polymeric forms that accumulate intra/extracellularly (6). The biophysical properties of two known hNS pathological mutants, Ser49Pro and Ser52Arg, have been reported (7–9). In particular, it was shown that Ser49Pro is thermally less stable than the WT protein (7,9), whereas the Ser52Arg variant is as stable as wild-type (WT) hNS but is a very poor tPA inhibitor (8).
All serpins (composed of 350–400 amino acids) are globular proteins typically characterized by a long exposed polypeptide loop, termed the reactive center loop (RCL), that carries a recognition sequence for the target protease. After protease binding and cleavage of the RCL, the serpin molecule undergoes a large conformational change whereby the cleaved RCL is inserted (intramolecularly) as the fourth β-strand (called s4A) of the adjacent A-sheet (10). During this conformational change, the protease remains covalently bound to the RCL and is dragged to the opposite pole of the serpin molecule (∼70 Å away). Additionally, the bound protease active site is sterically distorted, preventing hydrolysis of the acyl-enzyme complex almost indefinitely (weeks) (11). In this respect hNS is an exception, since the hNS/tPA complex is stable for less than an hour (7,12,13). During the conformational change from the native to the RCL-cleaved form, a consistent stabilization of the serpin molecule takes place, yielding a cleaved form that is hyperstable (14). In contrast, the native uncleaved serpin is considered to represent a metastable conformation (2).
Uncleaved serpins can display a second conformation, known as the latent form, that can be produced in vitro from the native serpin molecule by heating, chemical treatment, or pH change (2). The latent and cleaved serpin forms share the same gross structural organization, whereby the RCL (intact in the latent form) is inserted as strand s4A in the A-sheet, and fold stability (2). Different levels of latentization have been reported. Usually, in the fully achieved latent form, the RCL is totally inserted into the A-sheet and the nearby short s1C strand unfolds (15) (Fig. 1). However, cases in which the RCL is only partly inserted into the A-sheet have been reported (15–17). Furthermore, the structure of an α1-antichymotrypsin mutant has been shown to adopt an aberrant conformation (known as the δ conformation) whereby the RCL is partly inserted and the loop between helix hF and s3A fills the lower part of the A-sheet (18). Since in the latent form the RCL is not accessible to proteases, latent serpins are inactive as inhibitors.
Figure 1.
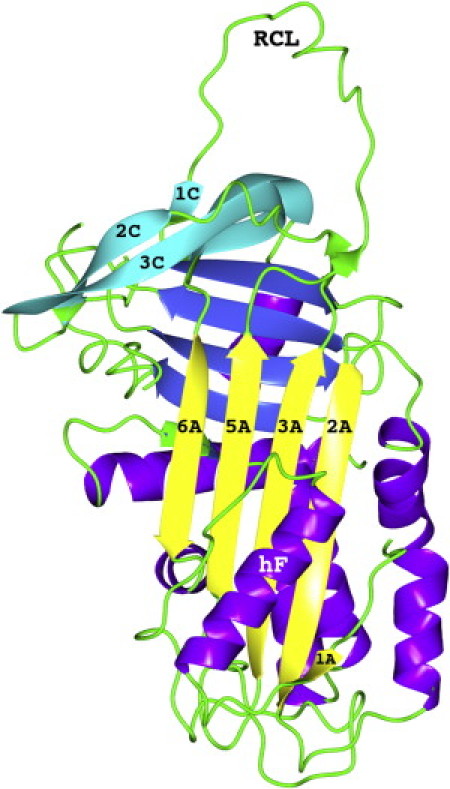
Cartoon representation of native hNS (pdb code 3F5N). β-Strands belonging to the A- and C-sheets, RCL, and helix F are labeled according to the accepted serpin nomenclature. (A-sheet is shown in yellow, B-sheet in blue, and C-sheet in cyan; helices are purple, and coils, including RCL, are shown in green online).
Although the serpin monomeric forms have been structurally well characterized through crystallographic analyses (19), the assembly principles for serpin polymerization are still a matter of debate. In all current polymer models the RCL is mainly held responsible for bridging neighboring serpin molecules, yielding long strings of linked protein molecules (beads-on-a-string). More specifically, several pathological serpin polymers are thought to be formed in vivo via the insertion of the RCL, as strand s4A, into the A-sheet of a neighboring serpin molecule (20). Other models have also been proposed; for example, it was recently suggested that the RCL and the following strand s5A are both involved in the intermolecular swap mechanism (21) (in the primary neuroserpin sequence, strand 5A starts right after the end of the RCL (which in cleaved and latent forms becomes strand 4A). Nevertheless, despite the relevance of serpinopathies for human health, the fine details of polymerization and the precise identification of the serpin regions that are actually swapped in the polymers are still debated (6).
Here we present a biophysical characterization of four molecular species of native (uncleaved) hNS that were isolated under different experimental conditions. Based on hNS thermal and chemical melting experiments, dynamic light scattering (DLS), and Fourier transform infrared spectroscopy (FTIR), our results highlight an ensemble composed of two distinct hNS latent forms and two biophysically and morphologically different hNS polymeric species that, to our knowledge, have not previously been reported for any known serpin. These observations have implications for the poorly understood serpin polymerization mechanisms and may reconcile, at least partly, the currently contrasting models for pathological serpin polymerization.
Materials and Methods
hNS expression and purification
For this study, hNS was purified to homogeneity according to a previously published procedure (13).
Production of different hNS forms
Latent-45 and polymer-45 were produced by incubating overnight hNS (0.05 or 0.5 mg/mL) at 45°C in 10 mM Tris, 50 mM KCl, pH 8.0. The two species were then separated by size exclusion chromatography (SEC, Superdex 200 10/300 GL, GE Healthcare, Little Chalfont, UK). Similarly, latent-85 and polymer-85 were produced by incubating hNS at 85°C for 2 h.
Circular dichroism
Circular dichroism (CD) spectra were recorded on a J-108 spectropolarimeter (Jasco, Tokyo, Japan) equipped with a Peltier-type temperature-control system. Quartz cuvettes of 0.1 cm path length and a protein concentration of 0.25 mg mL−1 were used. Typically, scans were acquired in 0.1 nm steps over a 205–260 nm wavelength range with a time constant of 0.5 s, a 2 nm bandwidth, a scan rate of 10 nm min−1, and baseline-corrected by subtracting a buffer spectrum. Molar ellipticity per mean residue, [θ] in deg.cm2 dmol−1, was calculated from the equation [θ] = [θ]obs mrw/10lC, where [θ]obs is the ellipticity measured in degrees, mrw is the mean residue molecular mass (113.7 Da), C is the protein concentration in gl−1, and l is the optical path length of the cell (in centimeters). Thermal unfolding curves were recorded in temperature mode at 220 nm and 216 nm, from 25°C to 100°C, with a scan rate of 1.0 K min−1. Melting temperature values were obtained by fitting the data to a single titration curve using GraFit 5 (Erithacus Software, London, UK). Protein denaturation by guanidinium hydrochloride (GdHCl) for the monomeric forms was obtained after 30 min incubation at room temperature in the presence of different amounts of the denaturant. The following denaturing conditions were used: 0–6 M GdHCl, 10 mM TRIS/HCl (pH 7.40), 50 mM KCl, and enzyme protein concentration of 0.25 mg/mL. The samples were subsequently analyzed by SEC in the presence of different concentrations of GdHCl to assess whether suboptimal concentrations of GdHCl would trigger the formation of hNS oligomers. As a result, only monomeric species were observed (data not shown). (GdHCl)-induced denaturation was monitored by CD, and [GdHCl]1/2 values were obtained by fitting the data to a single titration curve using GraFit 5.
Native polyacrylamide gel electrophoresis
Monomeric and polymeric samples were loaded onto native polyacrylamide gel electrophoresis (PAGE) slabs consisting of a 7.5% acrylamide resolving gel and a 5% stacking gel. The native gels were run at 90V in standard sample and running buffers on ice to prevent sample denaturation and polymer dissociation. The gel was then stained with Coomassie brilliant blue R250.
Characterization of polymeric species
Details on the GdHCl-induced denaturation, transmission electron microscopy (TEM), DLS, and FTIR spectroscopy performed in this work are available in the Supporting Material.
Results
Native neuroserpin
The biophysical characterization of native hNS was initially performed by means of far-UV CD analysis. First, the variation of molar ellipticity at 216 nm was monitored during a temperature ramp (20–95°C), following the approach of Belorgey et al. (7). The recorded CD signal highlighted two consecutive transitions with a contained change of molar ellipticity (data not shown).
To gain better insight into the nature of hNS behavior upon an increase in temperature, we performed a more complex temperature ramp (Fig. 2 a). A native hNS sample was first heated to 70°C and then cooled down to the starting temperature of 20°C. The same hNS sample was then heated up to 95°C and then cooled down to 20°C. After each step of the temperature ramp, the samples were cooled and a far-UV CD spectrum was recorded, and a native PAGE was run in parallel (note that only the monomeric fraction is shown in Fig. 2 b, because the polymeric species did not enter in the gel). As shown in Fig. 2 a (upper trace, green online), the CD data highlight a transition at T1/2 = 56.4 ± 0.2°C (T1 in Fig. 2 a), matching the Tm = 56.6 ± 0.3°C reported by Belorgey et al. (7). A second, smaller change in molar ellipticity was observed at T1/2 = 87.0 ± 0.9°C (T2 in Fig. 2 a) when the same sample, which was first cooled down to 20°C, was subsequently heated to 95°C (Fig. 2 a, middle trace, blue online). Of interest, a similar transition (at 89.4 ± 0.4°C) was observed by Onda et al. when they performed temperature ramp studies on the hNS latent form (9).
Figure 2.
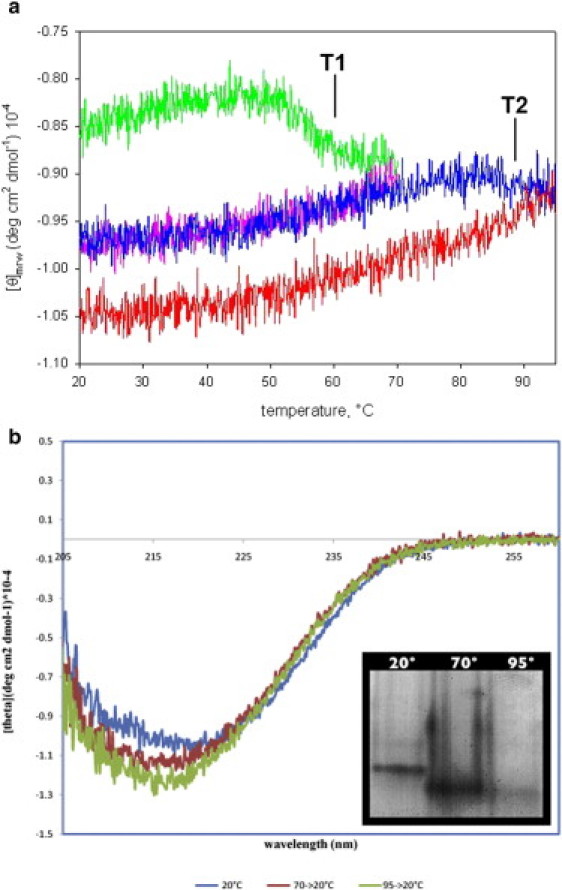
(a) Temperature ramp of native hNS followed through the far-UV CD signal at 216 nm. In the first segment hNS was heated from 20°C to 70°C (upper trace, green online) and then cooled down to 20°C (middle, purple online). In the third segment hNS was heated to 95°C (middle, blue online) and again cooled down to 20°C (lower trace, red online). The two transitions are highlighted and labeled T1 and T2, respectively. (b) The CD spectra recorded after each step of the temperature ramp are shown at 20°C (upper spectrum, blue online) after being heated at 70°C (middle, red online) and 95°C (lower, green online). (Inset) The same samples analyzed by native PAGE.
Contrary to what was previously hypothesized (7), the CD spectra indicate that hNS is folded at each temperature, and that only minor structural changes take place (Fig. 2 b). Moreover, the observed molar ellipticity changes (Fig. 2 a) indicate that at 56.6°C and 87.0°C hNS undergoes (contained) structural transitions that increase the overall secondary structure content. It is worth noting that both transitions are irreversible: when hNS is cooled down, either from 70°C or from 95°C, the observed molar ellipticity does not match the signal recorded before the thermal ramps (Fig. 2 a, middle and lower traces, purple and red online). Finally, in the native PAGE, shown in Fig. 2 b, the three samples show the typical behavior for native and latent serpin in the sample at 20°C and samples cooled down from 70°C and 95°C, respectively.
Because the stability of native hNS cannot be assessed by temperature ramp experiments, due to the irreversible interconversion events discussed above, we tested the stability of protein against chemical denaturation. Fig. 3 a shows the profile of native hNS unfolding in the presence of GdHCl, resulting in Cm = 2.9 ± 0.1 M. An independent chemical unfolding experiment (see Fig. S2) performed at 222 nm shows that native hNS unfolding is a two-step process (Cm = 3.0 ± 0.1 M), as previously shown for hNS and other serpins (8,22). Finally, at subdenaturating concentrations of the denaturant (1 M GdHCl), hNS loses its native fold in a single CD transition (at T1/2 = 50.5 ± 0.1°C).
Figure 3.
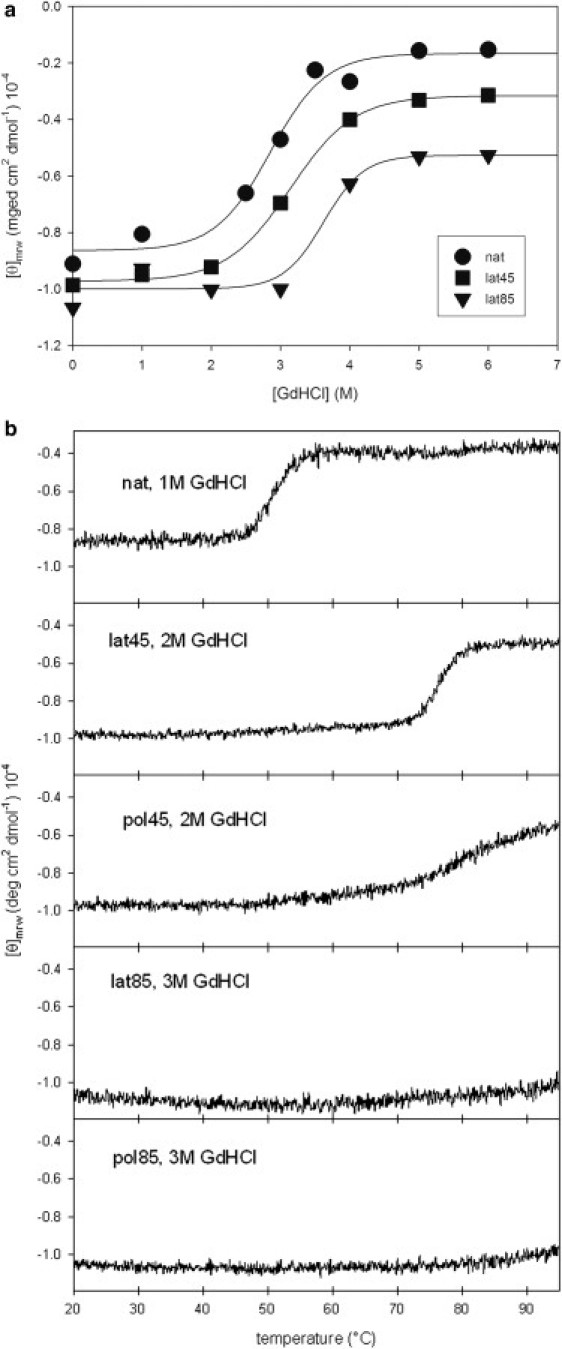
(a) Chemical unfolding of native hNS (circles), latent-45 (squares), and latent-85 (triangles), followed through far-UV CD at 216 nm by increasing concentrations of GdHCl (for a more thorough unfolding curve of native hNS and partial unfolding curves for latent forms followed at 222 nm, see Fig. S2). (b) Thermal unfolding (monitored through far-UV CD at 216 nm) of native hNS with 1 M GdHCl, latent-45 with 2 M GdHCl, latent-85 with 3 M GdHCl, polymer-45 with 2 M GdHCl, and polymer-85 with 3 M GdHCl.
To investigate how the described transitions might be related to hNS latent and polymeric forms, and to trigger their formation, hNS was incubated for 12 h at 45°C in agreement with previously reported protocols (7). Subsequently, and in consideration of the observed conformational transition at 85°C, a native hNS sample was independently incubated at 85°C for 2 h. As a result, both incubation protocols highlighted the appearance of one monomeric and one polymeric species. The two species resulting from each incubation protocol were separated by SEC, which showed that the monomeric species eluted at a column volume comparable to that of native hNS, whereas the polymeric forms eluted in the dead volume of the column, thus displaying a size of ≥600 kDa. Since both monomeric forms isolated (at 45°C and 85°C, respectively) proved inactive as tPA inhibitors, were uncleaved (data not shown), and showed a mobility in native PAGE typical of latent forms as compared with a sample of native hNS (Fig. 4 c, inset), they were classified as latent hNS (hereafter called latent-45 and latent-85, respectively; analogously, the two polymeric forms isolated from the two protocols are referred to as polymer-45 and polymer-85). A time-course analysis by native PAGE of hNS incubated at 45°C and 85°C showed that although the hNS polymeric fractions initially behaved similarly to previously described hNS polymers (Fig. S3 a), at the end of the two incubations the polymers were too large to enter the native PAGE run. The incubation times (12 h at 45°C and 2 h at 85°C, respectively) ensured that the polymerization process could be completed without affecting the overall hNS fold (see Fig. 4 b and 6 b). Because all four species were stable under the conditions employed here, we were able to prepare/isolate and further analyze them by far-UV CD and SEC as detailed below. Moreover, to investigate the nature of the polymer-45 and -85 species, we also employed DLS, FTIR, and TEM approaches.
Figure 4.
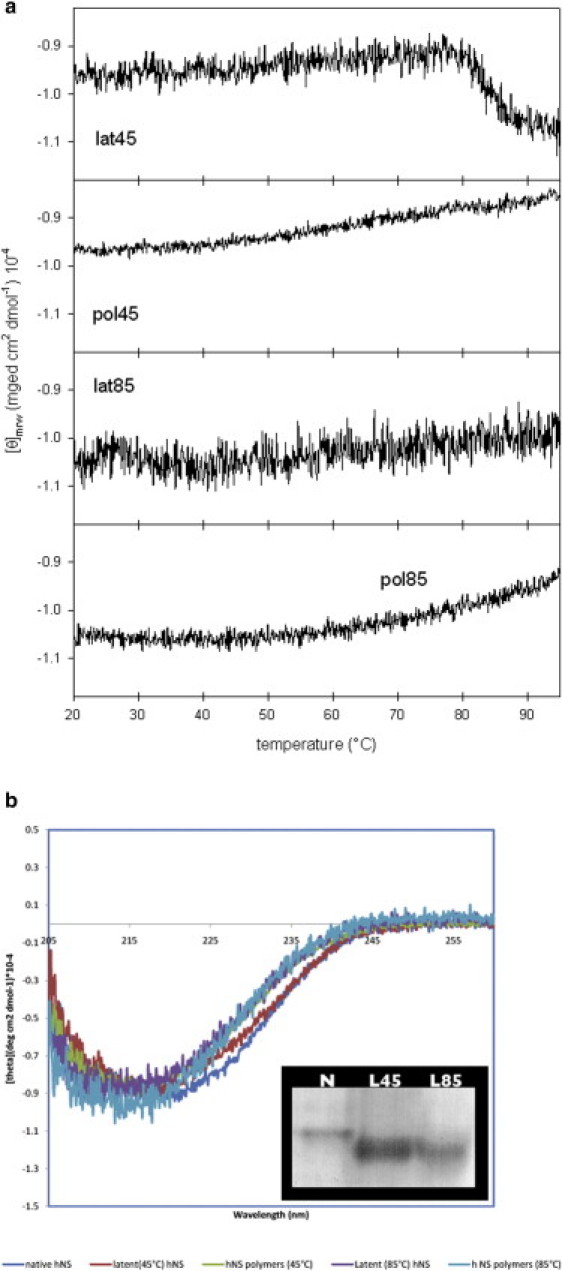
(a) Temperature ramps of hNS latent-45, latent-85, polymer-45, and polymer-85 in the 20–95°C temperature range, monitored through far-UV CD at 216 nm. (b) Far-UV CD spectra of native hNS, latent-45, polymer-45, latent-85, and polymer-85 are superimposed (online: native hNS is blue, latent-45 red, polymer-45 green, latent-85 purple, and polymer-85 cyan). (Inset) Native PAGE of native hNS (N), latent-45 (L45), and latent-85 (L85).
Figure 6.
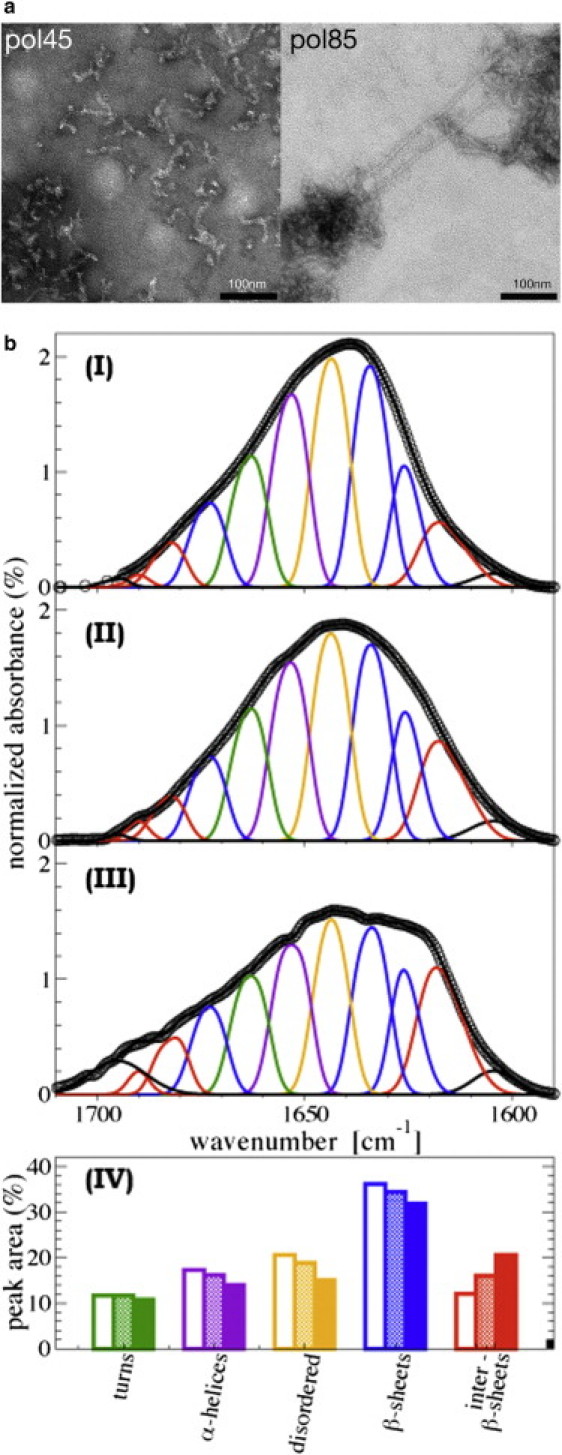
(a) TEM pictures of polymer-45 and polymer-85. (b) FTIR spectra of monomeric native hNS at 25°C (panel I), hNS polymer-45 (panel II), and hNS polymer-85 (panel III). The amide I band areas (1600–1700 cm−1) are normalized to 100 (open circles). Solid lines are the best fit to the data, obtained as the sum of different spectral components (colored curves online). (Panel IV) Percentage of each amide I band spectral component (color-coded online) as for the spectra deconvolution in panels I–III above. Native hNS, open bars; polymer-45, dotted bars; polymer-85, solid bars.
Latent-45
A thermal ramp on a latent-45 sample showed a change in molar ellipticity at T1/2 = 83.9 ± 0.1°C, close to previously reported values for latent hNS (9) (Fig. 4 a). The far-UV CD spectra of this species at 20°C and 95°C are very similar (Fig. S1), indicating that latent-45 retains the hNS overall fold. Moreover, the change in ellipticity observed at 83.9°C for latent-45 seems to correspond to the one observed at a 89.4°C by Onda et al. (9) in the thermal ramp of latent hNS; however, in contrast to native hNS, latent-45 does not display any CD signal change in the 50–60°C temperature range.
Latent-45 was later chemically unfolded by GdHCl: it unfolds at higher GdHCl concentration compared with native hNS (Cm = 3.5 ± 0.3 M; Fig. 3 a). Such different behavior between native and latent hNS is in keeping with what was previously observed through transverse urea gradient PAGE (9), and with the general behavior of latent serpins that are definitely more stable than their native counterparts (for a review see (2)). Combining GdHCl with a temperature ramp, latent-45 unfolds with a Tm of 66.9 ± 0.1°C in the presence of 2M GdHCl, showing once more a distinctly higher stability than native hNS (Fig. 3 b).
Latent-85
The thermal and chemical stability of latent-85 were assessed as described above for latent-45. In contrast to what was observed for native hNS and latent-45, the latent-85 thermal ramp (20°C to 95°C) did not lead to any observable change in CD signal (Fig. 4 a), suggesting that through the incubation at 85°C hNS reached its ultimate stable conformation. The latent-85 chemical stability in GdHCl is slightly higher than that of latent-45, with a Cm = 3.6 ± 0.1 M (Fig. 3 a). It is noteworthy that the three monomeric hNS species did not reach the same level of unfolding: latent-45 and (even more so) latent-85, retained a residual structure in 6 M GdHCl. Such a resistance to chemical denaturation was previously observed (23). Although unfolding of native hNS is in fact a two-step process, the first minor unfolding step is not observable for either of the two latent species (Fig. S2). Finally, a mixed thermal/chemical approach did not result in unfolding of latent-85. A 20–95°C temperature ramp run in the presence of 3 M GdHCl did not lead to any unfolding signal, highlighting qualitatively the remarkable stability of this hNS species (Fig. 3 b).
Once latent-45 and latent-85 were completely unfolded and then refolded, they displayed a mobility in native PAGE typical of native hNS (Fig. S3 b).
Polymer-45
As mentioned above, incubation of native hNS at 45°C also resulted in the formation of a polymeric species, in keeping with previous reports (9). Both polymer-45 and -85 were biophysically characterized following the approaches employed for the latent forms. A thermal ramp monitored by far-UV CD of polymer-45, from 20°C to 95°C, did not suggest the occurrence of major conformational readjustments (Fig. 4 a), in agreement with findings by Onda et al. (9), and the polymer was not dissociated into its monomeric components (data not shown). We then used SEC to follow the chemical depolymerization/unfolding of polymer-45 induced by GdHCl. A series of polymer-45 samples were incubated with increasing amounts of GdHCl for 30 min at room temperature before they were individually loaded onto the column. Fig. 5 shows the disappearance of polymer-45 (three independent experiments of depolymerization were carried out, and the standard deviations are shown as error bars). The GdHCl concentration at which 50% of polymer-45 (C50%pol) was dissociated was estimated as 2.7 ± 0.1 M. The SEC runs at increasing GdHCl concentrations show that the two main species present during the depolymerization were the monomer and the polymer; however, a nonnegligible shoulder accounting for the presence of species with higher molecular weight is evident (Fig. S4 a). Remarkably, although some intermediate species between the polymeric and monomeric forms are visible, the depolymerization process mainly shows a cooperative behavior in which the polymer and monomer are the most represented forms, with very few intermediate oligomers (Fig. S4 a).
Figure 5.
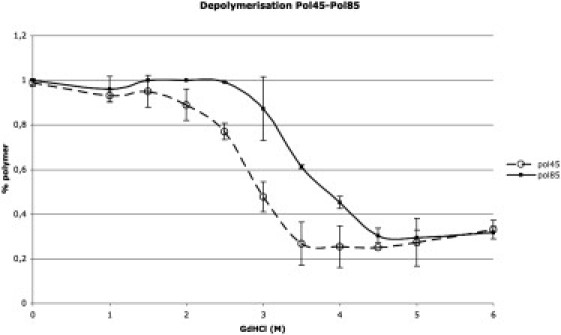
Chemical depolymerization of polymer-45 and polymer-85 by increasing concentrations of GdHCl. The black line with triangles and the dashed line with open circles show the polymer-85 and polymer-45 dissolution, respectively. Standard deviations are shown as error bars.
The combined effect of GdHCl and temperature was also analyzed. Polymer-45 unfolds with a Tm = 77.0 ± 0.1°C in the presence of 2 M GdHCl. The melting curve is reminiscent of what observed for latent-45, which showed a comparable Tm at the same GdHCl concentration (Fig. 3 b).
Polymer-85
The experiments performed on polymer-45 were repeated on polymer-85. Upon heating to 95°C, polymer-85 did not show any change in the far-UV signal (Fig. 4 a), and no depolymerization was observed after the temperature ramp (data not shown). The chemical stability of polymer-85 was assessed via the same protocol used for polymer-45. Surprisingly, polymer-85 showed a much higher chemical stability (versus GdHCl), with a C50%pol of 3.5 ± 0.1 M (Fig. 5 b). As for polymer-45, the two main species observed in SEC were the polymer and the monomer, suggesting that polymer-85 depolymerization is a cooperative process (Fig. S4 b). Finally, the combination of GdHCl and temperature highlighted an extraordinary stability of polymer-85. A thermal ramp to 95°C in 3 M GdHCl did not lead to unfolding, in similarity to the above findings for latent-85 (Fig. 3 b).
To assess whether the samples of polymer-45 and polymer-85 incubated with GdHCl for 30 min had reached thermodynamic equilibrium, samples of polymers were incubated with GdHCl for 15, 30, and 60 min. The levels of depolymerization after 30 and 60 min were identical, suggesting that the 30 min incubation allowed attainment of equilibrium (data not shown).
DLS and TEM experiments
The size and morphology of the neuroserpin polymers were assessed by TEM (Fig. 6 a) and DLS. In particular, whereas polymer-45 appears to be similar in size and shape to previously reported TEM pictures of serpin polymers (24), polymer-85 appears to be longer, thicker, and more rigid than polymer-45 (Fig. 6 a). Consistent with TEM observations, DLS experiments revealed that polymers that form after 12 h of incubation at 45°C have a size (i.e., hydrodynamic radius (Rh)) not exceeding a few tens of nanometers (an average of 42 nm with a polydispersity of 25%); the same size is reached upon overnight incubation at intermediate temperatures, below 60°C (V. Martorana and R. Noto, Istituto di Biofisica, Consiglio Nazionale delle Ricerche, CNR, Palermo Italy, personal communication, 2010). On the other hand, after only 5 min of incubation at 85°C, the size of aggregates reaches the value of several hundred nanometers (an average of 475 nm with a polydispersity of 25%). Given the appearance of polymer-85 as long fibers (Fig. 6 b and Fig. S5 a), we performed a Thioflavin fluorescence test to determine whether the polymeric species that forms at 85°C has an amyloid nature. Thioflavin fluorescence was measured for all five of the hNS species discussed here, and the results showed no differences between monomeric and polymeric species (data not shown).
FTIR characterization of the polymeric hNS species
We explored key structural features of hNS polymer-45 and polymer-85 at the molecular scale by FTIR spectroscopy. In particular, we focused on the Amide I′ band, located between 1600 and 1700 cm−1, which is mainly due to stretching of the backbone carbonyl group and thus is extremely sensitive to secondary structure details (25,26). The FTIR spectra of polymer-45 and polymer-85 samples were measured along with those of native hNS (Fig. 6 b, panels I–III). Inspection of the recorded native hNS FTIR spectra showed a highly structured amide I′ band that was deconvoluted into different component bands associated with specific secondary structure features of the protein (as reported in Materials and Methods). The percentage of the different components is displayed in Fig. 6 b, panel IV.
The FTIR spectra for native, polymer-45, and polymer-85 hNS exhibit appreciable differences in the relative amounts of disordered structures, α-helices, and turns (measured from the bands centered at 1644, 1653, and 1663 cm−1; shown in Fig. 6 b online in yellow, purple, and green, respectively). Such differences would be in keeping with a certain degree of restructuring of the native hNS fold in the polymerized species. However, the key difference emerging from analysis of the FTIR spectra concerns the amount of β-sheet components. Whereas the bands typically associated with parallel or antiparallel β-sheet are centered at 1634 and 1626 cm−1 (blue in Fig. 6 b online), the component at lower wave numbers (centered at 1617 cm−1; red in Fig. 6 b online) can be ascribed to antiparallel β-sheet structure displaying a variable number of strands (27) or, equivalently, to a variable amount of interstrand interactions (28). In our spectra the amount of this component (labeled as inter-β-sheet in Fig. 6 b, panel IV) increases in the spectra of the polymerized species compared to that of native hNS, along with a corresponding decrease of the bands at higher wave numbers. An analogous shift of the bands related to β-sheets has been observed and associated with the formation of amyloid fibrils, which are characterized by the stacking of several β-strands (28–31). In our experiments, the increase in the 1617 cm−1 band can be related to the formation of multistranded structures, and is compatible with the intermolecular insertion of the RCL as additional strand in hNS A-sheet. Of interest, we note that the amount of multistranded (intermolecular) β-sheets (band at 1617 cm−1) is higher in polymer-85 than in polymer-45. Taken together, the FTIR results show that the two isolated polymers differ in their structural assemblies, and particularly that polymer-85 displays a more extensive set of intermolecular β-sheet-like interactions than polymer-45. Of note, these observations are in agreement with the polymer-45/polymer-85 stability results described above.
Discussion
This work provides a biophysical characterization of WT hNS in terms of its conformational, polymerization, and stability trends. After observing two distinct and consecutive hNS conformational changes induced by increasing temperatures, we set up two specific incubation protocols in an attempt to isolate the conformationally distinct species. Each protocol yielded a mixture of polymeric and monomeric hNS (termed latent-45/polymer-45 and latent-85/polymer-85, respectively) according to the temperature at which they were isolated. These species were then biophysically characterized, and the results showed that the hNS conformational landscape displays two additional levels of complexity relative to previous findings in serpins. Based on their overall properties, we conclude that latent-45 and polymer-45 match the latent and polymeric hNS species previously characterized (7,9,32). Indeed, the protocol we adopted to produce latent-45 is very similar to those previously used to obtain latent hNS, and, of significance, the temperature ramp we devised for the production of latent-45 has a profile identical to that adopted for latent hNS by Onda et al. (9). However, whereas those authors interpreted the observed drop in CD signal at 89.4°C as being due to the melting of latent hNS, our data suggest that such a change (molar ellipticity drops to more negative values) should instead underlie further secondary structure regularization, as indicated by the far-UV CD spectra of latent-45 at 20°C and 95°C (Fig. S1).
An analysis of the data reported here suggests that the hNS species obtained by heating the latent-45 form is identical to the isolated and previously unreported latent-85 species, which is hyperstable, likely representing an energy minimum (i.e., highest stability) for hNS in the monomeric form. One can best appreciate the different stabilities of hNS latent-45 and latent-85 by comparing the results of the mixed chemical/thermal unfolding experiments, where latent-45 displays a Tm = 66.9°C in 2 M GdHCl, and latent-85 does not unfold in 3 M GdHCl up to 95°C. Two alternative models may explain the presence of two hNS latent forms. Upon heating of native hNS, the change in CD signal observed at 56.4°C most likely reflects the insertion of the RCL into the A-sheet. The nature of the conformational change that occurs at 87.0°C is less easily explained. The CD signal change is definitely smaller than the one observed at 56.4°C, accounting for a further, albeit minor, gain in secondary structure (Fig. 2 a). On one hand, latent-45 may reflect an incomplete insertion of the RCL into the A-sheet, in similarity to what has been observed for WT PAI-1, a different serpin that targets tPA (15). On the other hand, the latent-45/-85 pair may recall what was previously observed in the structures of Δ42 and Δ51 latent tengpin (16,17). Different truncations of tengpin N-terminus modulate protein stability and conformation (16,17), and in particular the Δ42 truncation leads to a superlatent species with a hyperinserted RCL. Therefore, the additional (although contained) structural regularization observed for latent-85, relative to latent-45, may be explained by a more extended integration of strand s4A into the A-sheet and/or by the regularization of the s1C strand, which may hydrogen-bond to the edge strand of the A-sheet. A third possibility is that latent-45 is in fact an inactive but aberrant conformer, as in the δ conformation observed for a mutant of α1-antichymotryspin (18). All such models for the latent-45 → latent-85 transition would explain the CD signal changes and the higher chemical stability achieved. A high-resolution structural approach, such as x-ray crystallography, is needed to explore the fine details of latent-45/-85.
Concerning the polymeric species, the data reported here suggest that hNS polymerizes differently at 45°C and 85°C. Polymer-45 was obtained under conditions very similar to those adopted for the hNS polymer produced and characterized in previous studies (7–9,32). Such polymers are believed to be formed by swapping of the RCL region between adjacent hNS molecules, with insertion of (a part of) the RCL into the A-sheet of the neighboring serpin molecule (7,9,32).
We have shown that one can produce a second kind of polymer by incubating native hNS at 85°C for 2 h. The different properties of polymer-45 and polymer-85 are evident in their chemical stability against depolymerization and against mixed chemical/thermal unfolding. Both polymers display a strong chemical stability, in keeping with previous dissociation work performed on α1-antitrypsin polymers that showed a high chemical stability of the polymeric forms (33). Polymer-85 requires a concentration of GdHCl 0.8 M higher than polymer-45 to be disassembled into monomers (Fig. 5), and does not unfold upon heating in 3 M GdHCl up to 95°C (Fig. 3 b) (polymer-45 unfolds at 75°C in 2 M GdHCl). Furthermore, the DLS, TEM, and FTIR results suggest that the two polymers differ in size, morphology, and β-sheet components. An ω-loop, a unique feature of hNS among serpins, was recently identified in a crystal structure of hNS (34). This structural feature is known to support protein-protein interaction, and may partly explain the stability displayed by polymer-85 against depolymerization.
A most intriguing question concerns the structural organization of polymer-85, particularly if it is assumed that polymer-45 is based on an s4A/RCL insertion mechanism, since this would rule out the possibility that polymer-85 is formed according to the very same mechanism. Serpin (reversible) polymerization based on the s7A/RCL mechanism was previously reported for PAI-1 and MENT; however, this type of polymer has limited intermolecular interactions (35–37) and would hardly explain the outstanding stability of polymer-85. It has been reported that the latent form of the S49P hNS mutant (but not the WT) can form polymers (9), and such polymers have been proposed to be formed by a C-sheet mechanism. According to this model, if strand 1C is partially or completely displaced, the RCL of an adjacent serpin molecule can interact with strand s2C, yielding polymers (38). However, a distinctive feature of C-sheet polymerization is that RCL-cleaved serpin molecules can be incorporated into the polymer, whereas this is not possible for the s4A/RCL polymer model (33). Indeed, when a mixture of native and cleaved hNS species are incubated at 45°C or 85°C, the cleaved hNS is found in the monomeric fraction only, indicating that neither polymer-45 nor -85 is a C-sheet polymer (Fig. S6).
An alternative mechanism of A-sheet polymerization was recently proposed (21). Based on the crystal structure of a domain-swapped dimer of antithrombin, where the RCL and the following s5A (in the primary neuroserpin sequence, strand 5A starts right after the end of the RCL (which in cleaved and latent forms becomes strand 4A) are the swapped region, it has been suggested that the formation of serpin polymers by intermolecular RCL insertion would also entail the swap of s5A (21). Such a model is compatible with the biophysical data reported here for hNS polymer-85. In particular, the high thermal and chemical stability observed imply a tight association between the polymer components, and the 1617 cm−1 FTIR band suggests an increased amount of inter-β-sheet hydrogen bonding in polymer-85. These observations would therefore indicate that the fine structural organizations of (hNS) polymer-45 and polymer-85 are not the same. However, the data reported here are compatible with other possible schemes, such as 1), if polymer-45 is formed by partial RCL intermolecular insertion into the A-sheet, then in polymer-85 the RCL may be fully inserted; or 2), if in polymer-45 the RCL is completely inserted into the A-sheet, then polymer-85 may be built by an as yet undisclosed interaction or domain swap.
Conclusion
Our results indicate that one given serpin may form more than one polymer type. Although the physicochemical conditions we employed to produce polymer-85 are extreme, it should be considered that serpin stability and the energy required for specific conformational changes may vary markedly under different conditions, as previously noted (39). Thus, a polymer that for hNS (here in 10 mM Tris, 50 mM KCl, pH 7.4) can be formed only at high temperature might become accessible under different conditions and/or by other serpins. Furthermore, pathological mutations that lead to highly unstable serpin variants may dramatically alter the protein energy landscape and trigger formation of the most stable polymer under much milder physicochemical conditions. Therefore, in principle, some mutations may lower the energetic barrier for the formation of polymer-85 to a level that allows its formation in vivo. Accordingly, the hNS mutant S49P forms latent and polymeric species at a temperature 7° lower than the WT protein (7).
The data reported here for hNS polymer-45/polymer-85, together with previous in vivo evidence indicating differential polymer/antibody recognition (40), suggest that one unique model of serpin polymerization is probably an oversimplified view, and that conformational plasticity may still be an underestimated molecular property within the serpin superfamily. Thus, serpinopathies should be considered as an ensemble of conformational diseases whose molecular bases may not necessarily adhere to just one common serpin polymerization model. Furthermore, even in the context of one specific serpinopathy, different mutations within the same serpin molecule might lead to differently assembled polymers.
Acknowledgments
We thank Prof. G. Merlini (IRCCS Policlinico San Matteo, and University of Pavia, Pavia, Italy) for helpful discussions and continuous support. We also thank Ms. Nadia Santo (Centro Interdipartimentale di Microscopia Avanzata, University of Milano, Milan, Italy) for technical support; V. Martorana and R. Noto for close collaboration and access to unpublished data; M. Mangione, M. D'Amico, A. Cupane, A. Emanuele, and P.L. San Biagio for stimulating discussions; and Elena Miranda for suggestions during the re-editing of this manuscript.
This work was supported by the Fondazione Cariplo (N.O.B.E.L. project Transcriptomics and Proteomics Approaches to Diseases of High Sociomedical Impact: a Technology Integrated Network).
Supporting Material
References
- 1.Miranda E., Lomas D.A. Neuroserpin: a serpin to think about. Cell. Mol. Life Sci. 2006;63:709–722. doi: 10.1007/s00018-005-5077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whisstock J.C., Bottomley S.P. Molecular gymnastics: serpin structure, folding and misfolding. Curr. Opin. Struct. Biol. 2006;16:761–768. doi: 10.1016/j.sbi.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Hastings G.A., Coleman T.A., Lawrence D.A. Neuroserpin, a brain-associated inhibitor of tissue plasminogen activator is localized primarily in neurons. Implications for the regulation of motor learning and neuronal survival. J. Biol. Chem. 1997;272:33062–33067. doi: 10.1074/jbc.272.52.33062. [DOI] [PubMed] [Google Scholar]
- 4.Yepes M., Lawrence D.A. Tissue-type plasminogen activator and neuroserpin: a well-balanced act in the nervous system? Trends Cardiovasc. Med. 2004;14:173–180. doi: 10.1016/j.tcm.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Coutelier M., Andries S., Godfraind C. Neuroserpin mutation causes electrical status epilepticus of slow-wave sleep. Neurology. 2008;71:64–66. doi: 10.1212/01.wnl.0000316306.08751.28. [DOI] [PubMed] [Google Scholar]
- 6.Gooptu B., Lomas D.A. Conformational pathology of the serpins: themes, variations, and therapeutic strategies. Annu. Rev. Biochem. 2009;78:147–176. doi: 10.1146/annurev.biochem.78.082107.133320. [DOI] [PubMed] [Google Scholar]
- 7.Belorgey D., Crowther D.C., Lomas D.A. Mutant neuroserpin (S49P) that causes familial encephalopathy with neuroserpin inclusion bodies is a poor proteinase inhibitor and readily forms polymers in vitro. J. Biol. Chem. 2002;277:17367–17373. doi: 10.1074/jbc.M200680200. [DOI] [PubMed] [Google Scholar]
- 8.Belorgey D., Sharp L.K., Lomas D.A. Neuroserpin Portland (Ser52Arg) is trapped as an inactive intermediate that rapidly forms polymers: implications for the epilepsy seen in the dementia FENIB. Eur. J. Biochem. 2004;271:3360–3367. doi: 10.1111/j.1432-1033.2004.04270.x. [DOI] [PubMed] [Google Scholar]
- 9.Onda M., Belorgey D., Lomas D.A. Latent S49P neuroserpin forms polymers in the dementia familial encephalopathy with neuroserpin inclusion bodies. J. Biol. Chem. 2005;280:13735–13741. doi: 10.1074/jbc.M413282200. [DOI] [PubMed] [Google Scholar]
- 10.Carrell R.W., Lomas D.A. α1-Antitrypsin deficiency—a model for conformational diseases. N. Engl. J. Med. 2002;346:45–53. doi: 10.1056/NEJMra010772. [DOI] [PubMed] [Google Scholar]
- 11.Huntington J.A., Read R.J., Carrell R.W. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 12.Barker-Carlson K., Lawrence D.A., Schwartz B.S. Acyl-enzyme complexes between tissue-type plasminogen activator and neuroserpin are short-lived in vitro. J. Biol. Chem. 2002;277:46852–46857. doi: 10.1074/jbc.M207740200. [DOI] [PubMed] [Google Scholar]
- 13.Ricagno S., Caccia S., Bolognesi M. Human neuroserpin: structure and time-dependent inhibition. J. Mol. Biol. 2009;388:109–121. doi: 10.1016/j.jmb.2009.02.056. [DOI] [PubMed] [Google Scholar]
- 14.Kaslik G., Kardos J., Gráf L. Effects of serpin binding on the target proteinase: global stabilization, localized increased structural flexibility, and conserved hydrogen bonding at the active site. Biochemistry. 1997;36:5455–5464. doi: 10.1021/bi962931m. [DOI] [PubMed] [Google Scholar]
- 15.Mottonen J., Strand A., Goldsmith E.J. Structural basis of latency in plasminogen activator inhibitor-1. Nature. 1992;355:270–273. doi: 10.1038/355270a0. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Q., Buckle A.M., Whisstock J.C. The N terminus of the serpin, tengpin, functions to trap the metastable native state. EMBO Rep. 2007;8:658–663. doi: 10.1038/sj.embor.7400986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q., Law R.H., Buckle A.M. A structural basis for loop C-sheet polymerization in serpins. J. Mol. Biol. 2008;376:1348–1359. doi: 10.1016/j.jmb.2007.12.050. [DOI] [PubMed] [Google Scholar]
- 18.Gooptu B., Hazes B., Lomas D.A. Inactive conformation of the serpin α(1)-antichymotrypsin indicates two-stage insertion of the reactive loop: implications for inhibitory function and conformational disease. Proc. Natl. Acad. Sci. USA. 2000;97:67–72. doi: 10.1073/pnas.97.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marszal E., Shrake A. Serpin crystal structure and serpin polymer structure. Arch. Biochem. Biophys. 2006;453:123–129. doi: 10.1016/j.abb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Lomas D.A., Evans D.L., Carrell R.W. The mechanism of Z α1-antitrypsin accumulation in the liver. Nature. 1992;357:605–607. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
- 21.Yamasaki M., Li W., Huntington J.A. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature. 2008;455:1255–1258. doi: 10.1038/nature07394. [DOI] [PubMed] [Google Scholar]
- 22.James E.L., Whisstock J.C., Bottomley S.P. Probing the unfolding pathway of α1-antitrypsin. J. Biol. Chem. 1999;274:9482–9488. doi: 10.1074/jbc.274.14.9482. [DOI] [PubMed] [Google Scholar]
- 23.Shortle D., Ackerman M.S. Persistence of native-like topology in a denatured protein in 8 M urea. Science. 2001;293:487–489. doi: 10.1126/science.1060438. [DOI] [PubMed] [Google Scholar]
- 24.Davis R.L., Shrimpton A.E., Lomas D.A. Familial dementia caused by polymerization of mutant neuroserpin. Nature. 1999;401:376–379. doi: 10.1038/43894. [DOI] [PubMed] [Google Scholar]
- 25.Barth A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta. 2007;1767:1073–1101. doi: 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Krimm S., Bandekar J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv. Protein Chem. 1986;38:181–364. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
- 27.Kubelka J., Keiderling T.A. Differentiation of β-sheet-forming structures: ab initio-based simulations of IR absorption and vibrational CD for model peptide and protein β-sheets. J. Am. Chem. Soc. 2001;123:12048–12058. doi: 10.1021/ja0116627. [DOI] [PubMed] [Google Scholar]
- 28.Zandomeneghi G., Krebs M.R., Fändrich M. FTIR reveals structural differences between native β-sheet proteins and amyloid fibrils. Protein Sci. 2004;13:3314–3321. doi: 10.1110/ps.041024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dzwolak W., Ravindra R., Winter R. Aggregation of bovine insulin probed by DSC/PPC calorimetry and FTIR spectroscopy. Biochemistry. 2003;42:11347–11355. doi: 10.1021/bi034879h. [DOI] [PubMed] [Google Scholar]
- 30.Frare E., Mossuto M.F., Fontana A. Characterization of oligomeric species on the aggregation pathway of human lysozyme. J. Mol. Biol. 2009;387:17–27. doi: 10.1016/j.jmb.2009.01.049. [DOI] [PubMed] [Google Scholar]
- 31.Frare E., Mossuto M.F., Fontana A. Identification of the core structure of lysozyme amyloid fibrils by proteolysis. J. Mol. Biol. 2006;361:551–561. doi: 10.1016/j.jmb.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 32.Chiou A., Hägglöf P., Klenerman D. Probing neuroserpin polymerization and interaction with amyloid-β peptides using single molecule fluorescence. Biophys. J. 2009;97:2306–2315. doi: 10.1016/j.bpj.2009.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lomas D.A., Elliott P.R., Carrell R.W. α1-Antitrypsin Mmalton (Phe52-deleted) forms loop-sheet polymers in vivo. Evidence for the C sheet mechanism of polymerization. J. Biol. Chem. 1995;270:16864–16870. doi: 10.1074/jbc.270.28.16864. [DOI] [PubMed] [Google Scholar]
- 34.Takehara S., Onda M., Lomas D.A. The 2.1-A crystal structure of native neuroserpin reveals unique structural elements that contribute to conformational instability. J. Mol. Biol. 2009;388:11–20. doi: 10.1016/j.jmb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 35.McGowan S., Buckle A.M., Whisstock J.C. X-ray crystal structure of MENT: evidence for functional loop-sheet polymers in chromatin condensation. EMBO J. 2006;25:3144–3155. doi: 10.1038/sj.emboj.7601201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nar H., Bauer M., Declerck P.J. Plasminogen activator inhibitor 1. Structure of the native serpin, comparison to its other conformers and implications for serpin inactivation. J. Mol. Biol. 2000;297:683–695. doi: 10.1006/jmbi.2000.3604. [DOI] [PubMed] [Google Scholar]
- 37.Sharp A.M., Stein P.E., Read R.J. The active conformation of plasminogen activator inhibitor 1, a target for drugs to control fibrinolysis and cell adhesion. Structure. 1999;7:111–118. doi: 10.1016/S0969-2126(99)80018-5. [DOI] [PubMed] [Google Scholar]
- 38.Carrell R.W., Stein P.E., Wardell M.R. Biological implications of a 3 A structure of dimeric antithrombin. Structure. 1994;2:257–270. doi: 10.1016/s0969-2126(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 39.Belorgey D., Hagglof P., Onda M., Lomas D.A. pH-Dependent stability of neuroserpin is mediated by histidines 119 and 138; implications for the control of β-sheet A and polymerization. Protein Sci. 2009;19:220–228. doi: 10.1002/pro.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janciauskiene S., Eriksson S., Lomas D.A. Differential detection of PAS-positive inclusions formed by the Z, Siiyama, and Mmalton variants of α1-antitrypsin. Hepatology. 2004;40:1203–1210. doi: 10.1002/hep.20451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


