Abstract
As public availability of gene expression profiling data increases, it is natural to ask how these data can be used by neuroscientists. Here we review the public availability of high-throughput expression data in neuroscience and how it has been re-used, and tools that have been developed to facilitate re-use. There is increasing interest in making expression data re-use a routine part of the neuroscience tool-kit, but there are a number of challenges. Data must become more readily available in public databases; efforts to encourage investigators to make data available are important, as is education on the benefits of public data release. Once released, data must be better-annotated. Techniques and tools for data re-use are also in need of improvement. Integration of expression profiling data with neuroscience-specific resources such as anatomical atlases will further increase the value of expression data.
Keywords: Microarray, gene expression analysis, meta-analysis
Introduction
Science as a human endeavor is founded on the principle of sharing experimental findings. The concept of sharing the raw data underlying the findings is less familiar. Most researchers are content to publish and examine the “exemplar” photographs, the summary graphs and the tables that make up the typical results section of a journal article. The desire and need to release or look at the primary data are more rarely expressed. Publishing raw data requires a mechanism to do so; utilizing the raw data requires sufficient motivation. Modern biology and communications now provide both the means (the Internet and powerful computers to enable re-analysis of large amounts of data) and the motivation (too much complex data to exhaustively analyze in a single study). There are massive challenges, both technical and cultural, to making data sharing (and therefore re-use) wide-spread in neuroscience but also great potential benefit (Eckersley et al., 2003; Koslow, 2000).
High-throughput gene expression data (the topic of this review) is frequently put forward as an example motivating the need to improve data sharing, but there are more mature success stories in other areas of genomics. The wide-spread availability of nucleotide and protein sequence data, which started decades ago, has reaped enormous benefits, many of which would not have been realized if the community had not decided that sequence submission was a requirement for publication, and that supporting sequence databases was worthwhile. The publishing of biomolecule structure coordinates in PDB starting in the early 1970s has been essential to making the structures useful (Berman et al., 2007). The essential difference between these types of data and expression data is that a gene sequence or a protein structure is “canonical” – it is not tied to a specific individual (an important exception being sequence polymorphisms, which are submitted to distinct databases such as dbSNP). In a sense that is not always true of other primary data types, sequence or structure data can be the primary “finding” of a study without further processing or interpretation. Sequence and structure data are also relatively (an important qualifier!) simple, with a small, well-defined alphabet of entities to store (nucleotides or amino acids in the case of primary sequence data; 3-D coordinates of atoms in the case of crystal structures).
In contrast to sequences and structures, most primary data in neuroscience are both individual (involving the analysis of a specific person or biological sample in a specific laboratory, which can never be exactly reproduced) and complex (difficult to describe fully in a readily-transmissible manner). Sharing primary data on a large scale requires data that are less individual and/or less complex. Along the data complexity axis, expression data stands out as, in practice, as being tractable. It is stored digitally from the outset, and standards have been developed for transmission (The Microarray Gene Expression Markup Language, MAGE-ML, for example) and description (Minimum information about a microarray experiment, or MIAME) (Brazma et al., 2001; Spellman et al., 2002). Many journal editors have embraced the concept that primary expression data should be made available on publication (see http://www.mged.org).
Expression data also has the advantage of one type of reduced individuality: many of the same genes are assayed in multiple studies. This means that for a given gene, it is possible to find relevant expression data; in this way, microarray data are like sequence data. The task of interpreting the data remains and is no less difficult than for other complex data (e.g., determining the relation of the expression patterns to some experimental parameter).
The efforts that have gone into making expression data an easily-sharable commodity have paid off, and there are thousands of studies available on the Internet (Barrett et al., 2007; Parkinson et al., 2007). The rest of this review focuses on the re-use of expression data in the neurosciences, both in practice and in principle, and uses some of our experiences in building expression data re-analysis tools as a backdrop to a discussion of existing challenges and opportunities. Table 1 summarizes web-accessible resources relevant to the sharing and comparing of gene expression data in neuroscience. To illustrate the application of some of the ideas presented in this review, a data re-use case study is presented as a supplement (http://www.bioinformatics.ubc.ca/pavlidis/lab/reuse).
Table 1.
Resources for gene expression data re-use
| Expression data repositories | ||
| Gene Expression Omnibus (GEO) |
www.ncbi.nlm.nih.gov/geo | Major repository of expression data, based at the NCBI |
| NIH Microarray Consortium | http://arrayconsortium.tgen.org/ | Assists NIH-funded neuroscience labs with expression studies and makes data public |
| ArrayExpress | www.ebi.ac.uk/arrayexpress | Major repository of expression data, based at the EBI. |
| Neuroscience-focused data re-use resources | ||
| GeneNetwork | www.genenetwork.org | Database linking behavioral and gene expression traits with genetic maps |
| Gemma | www.bioinformatics.ubc.ca/Gemma | Meta-analysis resource for expression data |
| Other comparison facilities | ||
| Bioconductor | www.bioconductor.org | Contains several packages implementing meta-analysis algorithms |
| Oncomine | www.oncomine.org | Cancer profiling database |
| CleanEx | www.cleanex.isb-sib.ch | Provides public human and mouse data with identifiers suitable for cross-experiment comparison |
| Amazonia | http://amazonia.montp.inserm.fr/ | Meta-analysis focused on stem cells |
| Gene Aging Nexus | gan.usc.edu | Database of gene expression studies of aging |
| Annotation resources | ||
| MGED Ontology | mged.sourceforge.net/ontologies | Ontology specific for microarray experiment description |
| Open Biomedical Ontologies | obofoundry.org | Gathering of ontologies for biomedical research |
| Data transport and description standards | ||
| MIAME | www.mged.org/Workgroups/MIAME/miame.html | Standard for information required in microarray experiment descriptions |
| MAGE-OM and MAGE-ML | www.mged.org/Workgroups/MAGE/mage.html | Object model and markup language for describing expression experiments |
| Functional Genomics Experiment (FuGE) |
fuge.sourceforge.net | Object model for describing functional genomics experiments |
| SOFT | www.ncbi.nlm.nih.gov/projects/geo/info/soft2.html | Primary format used by GEO |
|
Other brain gene expression resources |
||
| Allen Brain Atlas | www.brain-map.org | Expression patterns of 25,000 genes in adult mouse brain |
| GENSAT | www.gensat.org | Expression patterns of fluorescent protein reporters in mouse brain |
| Mouse Atlas | www.mouseatlas.org/data/mouse | SAGE data from 198 tissues including many brain regions at different developmental stages |
Modes of re-use
There are a number of ways that published expression data can be used by others. At the most basic level, the results can be referred to or compared to new results (not necessarily from microarray data) in an anecdotal fashion. If detailed analysis results are available, it is possible to compare published results systematically. At the next level of complexity, the raw data can be accessed and re-analyzed, either to revisit the original conclusions of the data generators, or to ask new or modified questions of the data. These relatively straightforward types of re-use are bound to be common when researchers want to compare their own data to existing results in more detail than is afforded simply by reading the paper.
The “comparison” approach to re-use can be facilitated by providing tools that make the data available in specialized ways. Integration with other types of data can be an important way to increase the value and interpretability of expression profiling data, and this is likely to be critical for re-use in neuroscience. For example, differences in expression patterns between brain regions and types of neurons are a potentially rich source of information about function and its modulation, but because expression profiling studies tend to have low spatial resolution (and often low cell-type resolution), the clues obtained from profiling have to be bolstered with data taking the precise site of expression into account. The approach of “genetical genomics” is a way of connecting genetic variation and phenotypes at the organismal level (e.g., behavior) to molecular networks (Schadt et al., 2005; Williams, 2006). In addition, detailed databases of neuronal function in terms of electrophysiology and pharmacology (Crasto et al., 2007) and connectivity (Bota et al., 2005) might offer further opportunities to help bridge the gaps between molecular biology and behavior. As discussed in more detail below, and summarized in Table 1, there are several tools and databases that offer ways of accessing and using expression data over the web in ways that is increasingly integrative.
A special type of data re-use, of particular interest to us, is meta-analysis. Meta-analysis is usually defined as the “analysis of analyses”, or the combination of independent analysis results, but for our purposes it is useful to expand this definition to include the re-analysis of raw data from multiple studies for the purpose of combining the results, or even the combining of data sets into “mega-datasets”. The goal of a meta-analysis is often to attempt to provide a more powerful test of a hypothesis than is provided by any individual study, thereby making better use of studies which, taken alone, yielded results that were not considered statistically significant. Another use of meta-analysis is to compare or contrast studies which differ in some systematic way, to identify commonalities or differences. Multiple data sets can also be used to identify novel patterns that were not sought by the original data producers. All of these types of meta-analysis are relevant to expression studies. Meta-analysis is described in more detail in the next section.
A final way data can be re-used is by researchers in statistics or computer science who are trying to hone algorithms for expression analysis. In this case the data are generally used in a cross-validation setting, often pitting one algorithm against another with relatively objective measures of performance. Some data sets have become part of a standard repertoire that computational biologists test their algorithms on, such as the “Golub Leukemia” data set (Golub et al., 1999). While this use might seem of limited direct interest to biologists, it is worthwhile to consider that if one wants a good algorithm for a particular type of data set, there is hardly a better way to see that happen than to let the data loose on the hungry community of algorithm researchers.
Meta analysis
Performing a traditional meta-analysis is challenging, with numerous pitfalls (Cooper and Hedges, 1994; Hunter and Schmidt, 1990). Relevant studies must be identified, the data or results must be extracted into a comparable form, and appropriate methods must be used to compare or combine the results. The problem of varying study quality is important and difficult to address, as is the problem of “missing studies” which are never published due to negative findings (the “desk-drawer effect”), leading to over-optimistic meta-analysis results. Statisticians have developed strategies for dealing with these problems, but it is often difficult to get past the first step of identifying studies that are sufficiently comparable. Even if the same hypothesis is being tested, the methodology often varies sufficiently to make comparison challenging. In basic biological science identifying multiple comparable studies is particularly problematic as laboratories tend not to be interested in exactly retesting a hypothesis that has already been tested and published by someone else. In contrast, in clinical studies it is relatively common, and even necessary, to ask the same (or nearly the same) question of multiple independent samples.
For expression studies, there are some interesting ways around the problem of finding comparable data sets. Most trivially, because quite a few microarray studies have a clinical focus, there are indeed multiple studies which compare, for example, similar tumor types. This fact has been exploited by a number of researchers (for reviews see (Larsson et al., 2006; Moreau et al., 2003; Rhodes and Chinnaiyan, 2004)). Another way to approach the problem is to recognize that related biological phenomena might yield related results. This encourages the comparison of the same phenomena across species (such as aging (McCarroll et al., 2004)). Biologists are often interested in finding the commonalities in these cases to identify basic principles. One simply has to define a level of commonality that is relevant to a particular question. In some cases this could involve any data from any organism, where one is interested in identifying the most widespread, generic aspects of gene expression. In general we expect that researchers have a “middle ground” level of data set commonality that addresses a specific area of interest without being so restricted as to exclude all data from consideration (Figure 1). It helps that expression data is often treated as “hypothesis-generating” or “exploratory” in the sense that patterns that are found are likely to be used as the grist for new experiments, rather than treated as an end in itself. However, even the analysis of data sets without considering their experimental design can yield insights, though the patterns that emerge are more likely to be related to functions that are not tissue or cell-type specific (Lee et al., 2004; Stuart et al., 2003).
Figure 1.
Conceptualization of data selection for re-use. Criteria that are too stringent or too lax make comparisons difficult.
Once it has been determined that a meta-analysis involving the combination of multiple studies is desirable, there are several methodological routes to take. Because the raw data are often available, it makes sense in some cases to simply combine the data sets together into a “mega-dataset”. To do this, the data must be carefully renormalized, especially if different types of platforms are being combined. Forming mega-datasets seems more commonly performed when the data were in fact collected by a single lab (Eisen et al., 1998) or at least from the same platform. Depending on the aims of the study, it might be better to use a statistical model to account for differences between studies, for example treating the experiment source as a random effect in a mixed-effects model.
If forming a mega-dataset proves problematic, the meta-analysis literature offers up a menu of options. First, one must determine what value will be used to represent the “result” of each study. The choices generally break down into using p-values from a hypothesis test, or effect sizes. P-values can be combined using a number of methods, of which the best known is Fisher’s, which uses the fact that p-values are uniformly distributed under the null hypothesis (Cooper and Hedges, 1994; Rhodes et al., 2002). Effects sizes (e.g., fold-changes or correlations) can be combined under a number of different models which can be used to perform hypothesis tests. In essence effects size techniques form a weighted average of the size of the effect in each study and an estimate of the variance of the effects across the studies to form a new test statistic. This statistic can be compared to a theoretical distribution to generate a new p-value (Choi et al., 2003). A simpler but less sensitive option is to use a “vote-counting” approach where each data set simply casts a vote as to whether an effect is observed, typically based on significance tests on each data set. Vote counting has the advantage that is it not necessary to assume that all the data sets should show an effect, which is an underlying assumption of the effect size technique in the simple cases; alternatively one has explicitly choose a model to account for differences between studies, which may not be justifiable. The main difficulty with vote counting is determining how many votes are needed before significance is achieved. In a simple meta-analysis, the number of positive votes under the null hypothesis will follow a binomial distribution. However, gene expression studies are much more complex, because many genes are tested and the same genes are not present in all studies. Therefore studies using vote-counting have to use other methods such as resampling to estimate the significance of findings repeated across studies (Lee et al., 2004; Stuart et al., 2003).
Trends in public availability of expression data
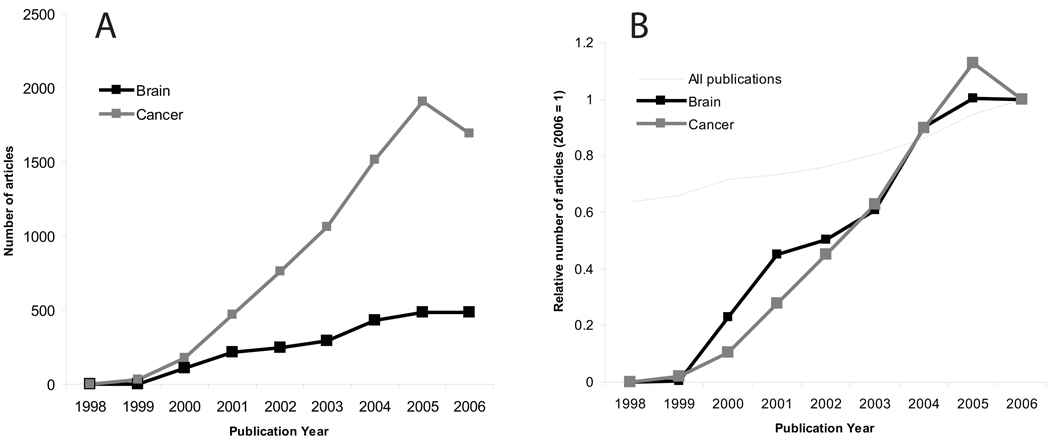
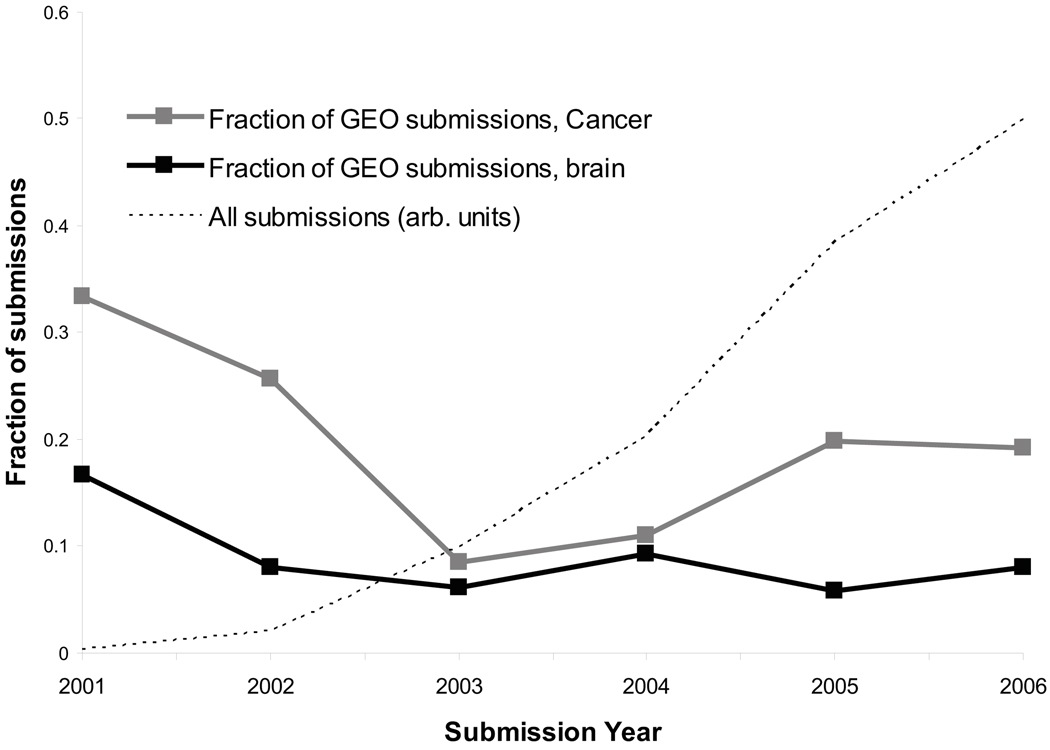
To date published re-analyses of expression data overwhelmingly involve human tumors, and examples of re-analysis in neuroscience are few (see discussion below). This suggests that either neuroscience-related expression data is less available, or there are fewer researchers interested in re-analyzing the available data. To gain some insight into this issue we examined the rate of publications and data submissions to public databases, using admittedly crude methodology. As shown in Figure 2, publications on expression profiling are far more numerous for cancer than for brain, but the rate of growth is similar. These publications are of limited use for re-analysis if the raw data are not available, so we searched a major repository of expression data, the Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geo/), for datasets that met the same criteria as our publication search (Figure 3). We found that cancer-related submissions were again more numerous than brain-related submissions. A comparison of Figure 2 and Figure 3 suggests that, if anything, brain-related submissions to GEO are in fact more frequent given the number of publications. This analysis is subject to a number of caveats, including the ability of the searches to identify the relevant publications or data (though variant searches gave similar results) and that the data in Figure 2 may include re-analyses as well as new data papers. There are also studies that appear in both groups (brain tumor studies). Finally, data from a single publication can sometimes be split into more than one GEO series (by the submitter), yielding an over-count of expression data sets.
Figure 2.
Trends in publications on expression profiling. We searched PubMed for entries using the search criteria “Gene Expression Profiling $M $Y[publication date]”, where $M was either “cancer” or “brain” and $Y was a year (1998–2006); or the total number of PubMed entries by year. A. Raw numbers showing that profiling papers accessible with the keyword “cancer” were consistently much more numerous than for “brain”. B. Data normalized by the number of publications in 2006, showing the similarity of the growth curves. Data for all PubMed entries are shown for comparison: submissions about profiling outpace the growth of PubMed by a wide margin.
Figure 3.
Trends in submissions to GEO. We used the GEO web interface to identify experiment series submissions in each year, using the same keywords that were used for the PubMed analysis in Figure 3 (e.g., “GSE[Entry Type] AND 2002[Publication Date] AND cancer”). Values are expressed as the fraction of all GEO series submissions. The growth curve of GEO overall is shown in arbitrary units for comparison. Submissions with the keyword “brain” follow a similar trend to “cancer” but with consistently smaller numbers of submissions.
We tentatively conclude that there is little evidence to suggest a general unwillingness of neuroscientists to make their data publicly available, and indeed most journals now require public submission of expression data on publication. However, there are some important gaps in the availability of neuroscience-related expression data. Data from studies using human brain samples from studies examining experimental factors other than cancer are relatively rare in public databases. This is despite many publications about expression patterns in schizophrenia, bipolar disorder and other neuropsychiatric disorders (see (Mirnics and Pevsner, 2004) for review). In GEO we identified 22 series that use human non-tumor brain samples, but these actually correspond to only 15 different studies because some studies were broken up into multiple series accessions. This includes three studies of Alzheimer’s disease but only one on schizophrenia. In another major expression data resource, the European Bioinformatics Institute (EBI) ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) we identified only one human brain study relating to abnormal brain function or disease (E-AFMX-6, a study of Huntington’s disease).
There are several factors likely to account for the under-representation of human brain expression data in public databases. First, a number of published studies of schizophrenia or bipolar disorder (Iwamoto et al., 2004b; Jurata et al., 2004; Polesskaya et al., 2003; Sokolov et al., 2003; Tkachev et al., 2003) used samples which were obtained under license from the Stanley Medical Research Institute; the terms of the license do not permit release of the raw data to third parties (http://www.stanleyresearch.org/programs/brain_collection.asp). Second, as it is considerably more difficult to get human brain samples (almost always obtained post-mortem) than tumors (which are routinely biopsied), we find that investigators are sometimes less willing to share their hard-won data. Finally, not all journals require data submission (for example, Molecular Psychiatry (http://www.nature.com/mp/for_authors.html)), removing an incentive.
Using reasonably stringent criteria, at this writing we identify 334 GEO data sets that appear to be brain-related (including competitive genome hybridization, studies with multiple tissues types, and tumor studies) in mouse, human or rat, associated with 170 PubMed citations (the list is available as a supplement). In ArrayExpress, 67 data sets (from a total of 1683) were returned in response to the query “brain”, of which most (50) involve human, mouse or rat samples. Aarnio et al. (2005) performed a remarkably thorough survey of the state of neuroscience microarray publications as of June 2004, and identified 448 papers describing microarray studies. Unfortunately, only 56 were reported as having full data available (Aarnio et al., 2005). Some of the available data sets they identified are available through author or publisher web sites rather than the public repositories. The spreadsheet giving the full details of Aarnio et al.’s literature search is available at http://www.uku.fi/aivi/neuro/genomics/supplement.xls.
It is worth mentioning some other data sources which sometimes have data that are not in the main public repositories. The NIH Microarray Consortium (http://arrayconsortium.tgen.org/) is an expression analysis service for neuroscientists holding NIH grants. The Consortium provides data 6 months after data collection. While these data sets are all put in GEO after publication of a paper, at this writing there are about a dozen mouse and rat data sets which appear to be public only though the Consortium web site. The Stanford Microarray Database (SMD, http://smd.stanford.edu/) data is submitted to GEO and ArrayExpress, but offers a variety of analysis tools and supplementary information. Most of the studies in SMD are not neuroscience-related, except for a few brain tumor studies. Similarly, the National Cancer Institute caArrayDB (https://caarraydb.nci.nih.gov/) contains data for around 60 studies, though most are probably of lesser interest to neuroscientists. The caArrayDB data sets do not appear to be mirrored in GEO.
How can investigators be encouraged to release expression data (or indeed any type of data)? One strategy, which we refer to as “the stick”, is to require release as a condition for funding or publication. As mentioned above, many but not all journals require microarray data release. The NIH guidelines on data release (http://grants.nih.gov/grants/policy/data_sharing/) are a step in the direction of requiring greater openness. The other strategy is “the carrot”, in which investigators recognize the benefits of releasing their data. The growing popularity of “open” software and journal publishing models raises some hope that a cultural shift is taking place, in which the benefits of a “share and share alike” attitude are increasingly being taken up by scientists (Brown et al., 2003; Watson, 2007).
Gene expression data re-use in neuroscience
Expression data can be re-used either by re-analyzing the original data, or by re-using the summarized results in some way. The latter case is not always easily distinguished from the act of simply citing a brief comparison to earlier work (The 170 papers mentioned above are cited over 800 times in papers in PubMed Central journals alone (http://www.pubmedcentral.nih.gov/)). There are some cases we can identify where re-use of published results was substantial without requiring access to the raw data. Iwamoto et al. (2004a) made use of a list of genes differentially expressed in schizophrenics published by Hakak et al. (2001) to show that the same genes appear to be differentially expressed between schizophrenics and controls in their own data set (Hakak et al., 2001; Iwamoto et al., 2004a). In a similar re-use of processed data, Sibille et al. (2007) showed that the effects of aging on gene expression are similar in the mouse and previously-published data on human brain (Erraji-Benchekroun et al., 2005) and also made use of direct use of published results on a BDNF knockout mouse (Glorioso et al., 2006).
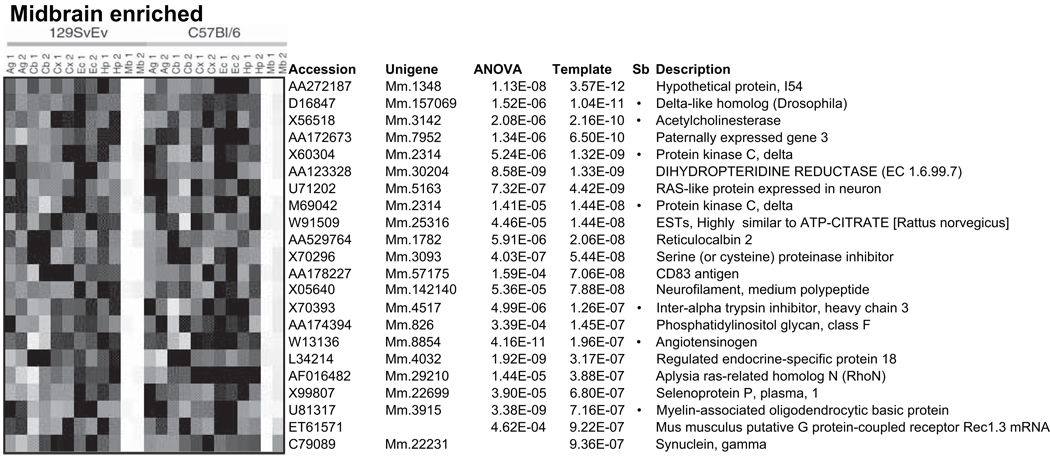
There are only a few cases we have identified where gene expression data from neuroscience studies was actually re-analyzed, as opposed to simply cited or used in summary form. An early case of re-analysis comes from work undertaken by one of us (P.P.) to re-analyze a previously published mouse brain data set from Sandberg et al. (Sandberg et al., 2000). The focus of the re-analysis was to compare the power of different methodologies for identifying differentially-expressed genes (Pavlidis and Noble, 2001), rather than asking new biological questions or performing a meta-analysis (Figure 4). More recently, two groups (Gu and Gu, 2003, 2004; Hsieh et al., 2003) independently re-analyzed a data set examining the divergence of human, orangutan and chimpanzee brain expression patterns (Enard et al., 2002), again with a focus on using different methods to answer the same question. In a case of a focused analysis of a previously-available data set, Perkins et al. (2007) studied microRNA-containing transcripts in human brain samples from the Harvard Brain Tissue Resource, and included in their analysis an Affymetrix data set that has been available to registered investigators on the HBTR web site for several years (http://www.brainbank.mclean.org/). Finally, data from (Sugino et al., 2006), comprising expression patterns for purified GFP-labeled neurons of 11 distinct classes (available from GEO as GSE2882 or from http://mouse.bio.brandeis.edu/), were reanalyzed to identify genes with expression correlated with Egr1 (Ponomarev et al., 2006).
Figure 4.
Re-analysis of mouse brain data from Sandberg et al. (2000). An example of how re-analysis of existing data can uncover previously unrecognized patterns. Pavlidis et al. (2001) identified genes showing brain-regionalization of expression using analysis of variance, in this case in the midbrain compared to five other regions, in two mouse strains. A comparison to the existing analysis showed that only a subset of these genes had been identified (marked by bullets). The heatmap shows relative expression levels, where white represents higher levels. Reproduced from Pavlidis et al. (2001) with permission.
Meta-analyses of brain expression data sets are also rare, thus far. Vazquez-Chona et al. (2005) performed a meta-analysis of published expression data sets to identify a common response to neuronal injury. This is a good example of the use of disparate published data sets to draw biological inferences, in this case that some genes are commonly upregulated in the acute response to injury in retina, brain and spinal cord. The only other case of any type of meta-analysis of brain expression data is that of (Mulligan et al., 2006). Mulligan combined data collected by three different groups to find a common expression signature correlated with alcohol preference in mice (Mulligan et al., 2006). We note that these data were not “re-analyzed” in the sense we have been using the term, as they had not been previously published separately. However, this study serves as another illustration of how data sets that differ in the details of their design and collection can be integrated to improve the quality of inference.
At this point it is reasonable to ask: What types of data sets are most re-usable? In our view, the data sets of the widest utility might be those which involve examining “baseline” expression across brain regions, cell types and/or developmental stages. The Sugino data set mentioned above (Sugino et al., 2006) is of this class, and there are a number of other “survey” data sets available, some of which examine many tissues outside the nervous system (Chin et al., 2007; Ge et al., 2005; Sandberg et al., 2000; Siddiqui et al., 2005; Stansberg et al., 2007; Su et al., 2004; Zapala et al., 2005; Zhang et al., 2004). Along the same lines, spatially-resolved expression atlases are important resources for gene expression studies in the brain and complement the high-coverage approach offered by microarrays. Examples include the comprehensive Allen Brain Atlas (ABA) of in situ hybridization patterns (Lein et al., 2007), the more focused Gene Expression Nervous System Atlas (GENSAT) which uses fluorescent protein reporters (Gong et al., 2003), and Brain Gene Expression Map (BGEM, which works closely with GENSAT) (Magdaleno et al., 2006) which employs radioactive in situ hybridization.
It is also safe to say that data sets that assay small brain regions or purified samples as opposed to analyzing “whole brain” are much more amenable to interpretation (“which cells was this signal coming from”), as well as affording higher sensitivity to detect expression of genes that might be diluted in a bulk sample. The interpretation issue, but not the dilution issue, might be addressed by comparing the patterns to in spatially resolved data (Lein et al., 2004; Ponomarev et al., 2006; Sunkin, 2006). Recent work from (Chin et al., 2007) examined 68 “voxels” from mouse brain using expression arrays and used the ABA for validation. Both the Chin study and the work of (Lein et al., 2004) provide evidence for a high degree of agreement between microarray and in situ data.
Tools for data re-analysis
We suspect that the reasons neuroscientists do not make frequent use of published expression studies are two-fold. First, because there are still relatively few studies available, it may be hard to identify data that are sufficiently relevant to a question at hand. Second, performing a re-analysis can be complex and difficult, a situation that can be ameliorated by the introduction of improved tools. The current state of re-analysis tools is reviewed in this section.
Both the NCBI and the EBI provide analysis tools (Barrett et al., 2007; Kapushesky et al., 2004), which greatly facilities re-use of data in GEO and ArrayExpress. Through GEO’s web interface, if a data set is sufficiently well-annotated (having reached “GDS” status), t-tests can be performed between groups of samples (e.g., between “hippocampus” and “cerebellum”. Clustering analyses can be performed on GEO data as well. The EBI offers Expression Profiler, an expanding toolkit that includes clustering, principal components analysis, between-group analyses and other options (Kapushesky et al., 2004). A major advantage of these tools is that they integrate directly with the databases, making the move from data set identification to analysis easy. While users are limited to using the algorithms they implement, a great deal can be accomplished by users with little bioinformatics or statistics experience. Naturally there are numerous other tools available for expression data analysis, all of which can be applied to data from public resources by downloading the data and converting it into a format that is suitable for import. Both GEO and ArrayExpress offer data download in a simplified “spreadsheet-like” tab-delimited format that makes this relatively straightforward.
For comparative or meta-analytical re-use of expression data to enter the every-day workflow of biologists, improved analytical tools and more precise annotations are needed. Tools are beginning to appear that aggregate published data under new covers, such as Oncomine (Rhodes et al., 2004) and the Gene Aging Nexus (Pan et al., 2007), both of which include data relevant to neuroscientists. Oncomine is, as the name suggests, entirely focused on human tumor data, and at this writing contains data from 264 analyzed studies (http://www.oncomine.org/). The Gene Aging Nexus (http://gan.usc.edu/) contains at least 42 aging-related data sets from multiple organisms. Oncomine is focused on differential expression analysis (Rhodes et al., 2004), while the Gene Aging Nexus offers the additional ability to analyze coexpression patterns or Gene Ontology categories (Pan et al., 2007). Stem cell researchers have developed a focused database system for meta-analysis (Assou et al., 2007).
For researchers interested in developing their own differential expression meta-analysis, a number of packages in Bioconductor (Gentleman et al., 2004) can be of assistance. GeneMeta implements algorithms described by (Choi et al., 2003). Other packages include RankProd (Breitling et al., 2004) and metaArray (Ghosh et all, unpublished; all packages available through the Bioconductor website, www.bioconductor.org).
A more specialized type of data re-use is offered by the GeneNetwork (http://www.genenetwork.org/). GeneNetwork is focused on the integration of genetic mapping information with expression profiles and behavioral traits (Li et al., 2005). GeneNetwork includes several brain gene expression data sets from well-studied recombinant inbred mouse strains, affording an unusual opportunity to treat expression levels as a quantitative trait that can also be related to behavior (Chesler et al., 2005). GeneNetwork offers a number of powerful query tools to access expression data and correlations with traits.
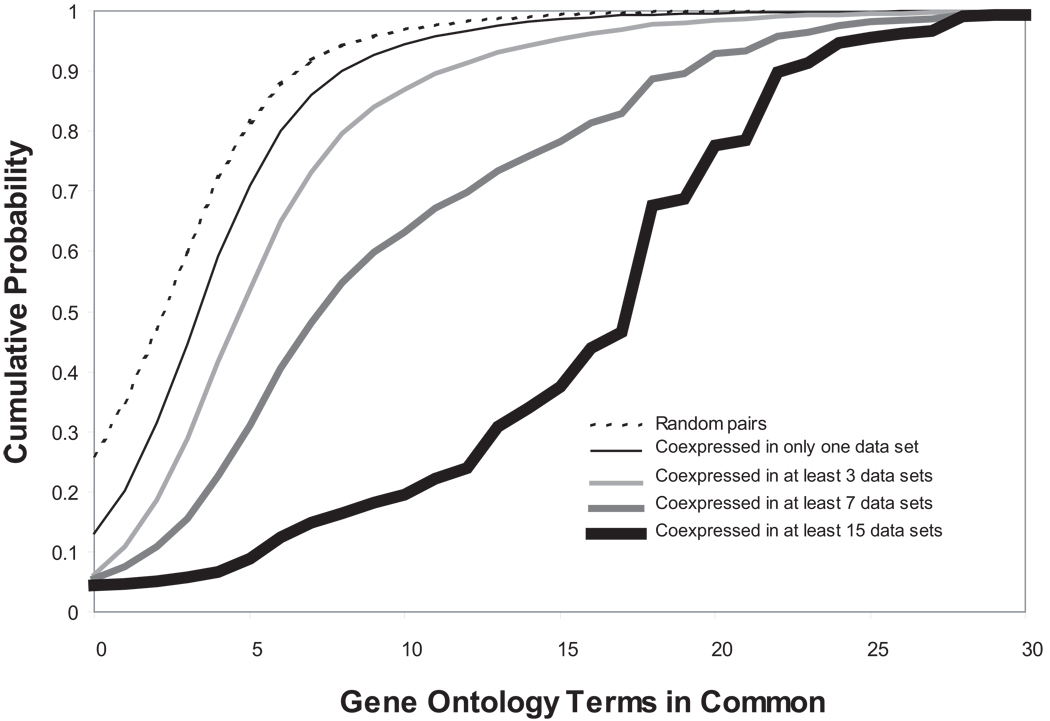
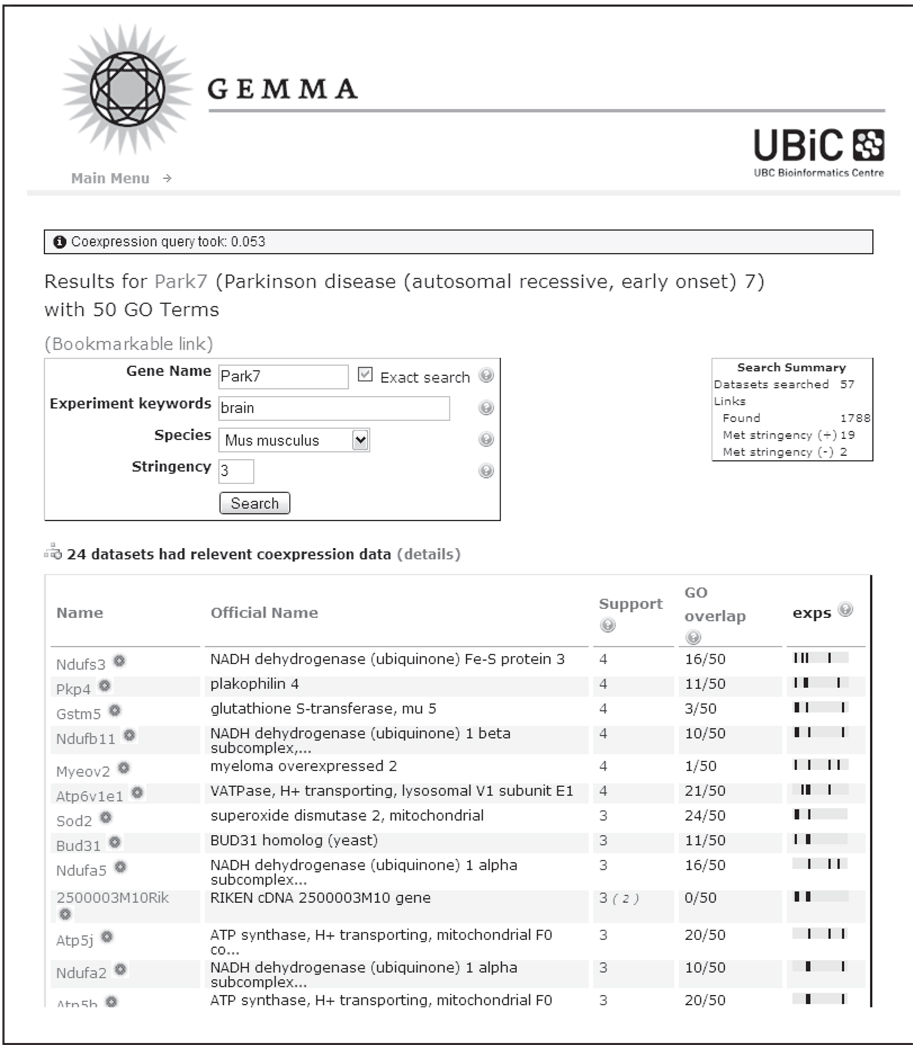
Our own entry into this field was TMM, a database of approximately 200 published mouse and human data sets (Lee et al., 2004), and focused on co-expression analysis. Recently we have replaced TMM with an updated database and software system, Gemma (http://www.bioinformatics.ubc.ca/Gemma/; details to be published elsewhere). We initially approached the problem of expression data re-analysis from a functional genomics standpoint, where the goal is to predict the function of genes. An approach that had been popularized from the early days of expression profiling was “guilt by association”, in which genes that have correlated expression patterns are inferred to have a closer functional relationship than genes which are uncorrelated (Eisen et al., 1998). This is a probabilistic statement, as transcripts which are not correlated can be functionally related, and transcripts that are highly correlated might reflect different levels of functional relatedness, ranging from protein co-localization to simply covariance in a mixed cell population. TMM and Gemma implement a type of co-expression meta-analysis that identifies expression patterns that repeatedly occur in multiple data sets. In a meta-analysis of 60 human data sets (mostly from tumors), we showed that combining information from multiple studies yielded much higher-quality functional predictions than single data sets ((Lee et al., 2004); Figure 5). Gemma includes a web-accessible tool that allows the exploration of coexpression patterns in an interactive fashion. A screenshot from the current version of Gemma is given in Figure 6.
Figure 5.
Recurring expression patterns yield higher-quality functional inferences. Each curve is a cumulative distribution of gene similarity as reflected in shared GO terms. Curves further to the right represent coexpression patterns that reoccur in more data sets, and exhibit higher functional similarity. Redrawn using data from (Lee et al., 2004).
Figure 6.
Screen shot of coexpression results from Gemma. Users enter a query gene of interest (PARK7 in this case) and are provided with a list of genes that are reproducibly coexpressed in multiple studies. The “support” is the number of data sets in which the pattern is found. “GO overlap” reflects the existing state of knowledge about the relatedness of the query gene to the result gene. The “Exps” column illustrates which data sets, of those searched, provide the support. A black line is shown for each data set where the pattern is found, giving a visual cue to which data sets are contributing most to the overall result. In this example, 57 data sets were searched, yielding 1788 patterns involving PARK7. Of these, nineteen positive and two negative correlation patterns were reproduced in at least 3 of the data sets.
At this writing, Gemma contains approximately 400 ‘series’ from GEO, and some additional data sets that were available only from other sources, typically as tab-delimited text from investigators’ personal web sites. An attempt was made to include as many data sets that are brain-related as possible, accounting for a large fraction of the data in the system. The balance includes data sets which were part of our older system (TMM) and a selection of other data sets from GEO. Additional data sets are being added on a frequent basis.
Challenges
The remainder of our discussion focuses on the difficulties facing researchers who want to re-use published expression data. Our focus is on issues surrounding the development of Gemma, but many of the problems apply to anyone who wants to use a published data set.
Data models and formats
Expression data has been the subject of extensive discussions about standards for data description and exchange formats. We review these in this section, highlighting issues relevant to the topic of data re-use.
The Microarray And Gene Experiment object model (MAGE-OM) represents an attempt to provide a detailed data model for gene expression microarray data (Spellman et al., 2002), and is an Object Management Group adopted specification (http://www.omg.org/cgi-bin/doc?formal/03-02–03). MAGE-OM does not support other types of expression measurement technologies such as Serial Analysis of Gene Expression (SAGE). MAGE-OM models array designs, array manufacture, experimental designs, as well as the expression data itself and other supporting data. The importance of MAGE-OM for data re-use lies, in large part, in its influence on database providers and exchange formats (described in the next paragraphs). MAGE-OM is fairly complex and is also designed for the archiving of data, not for direct use in re-analysis applications. Specifically, there is no simple concept of “the data for one probe across the conditions” (the most common use case), and such data can be extracted from the MAGE model only by fairly complex manipulations. Thus secondary users of data modeled in MAGE usually find it necessary to convert the data into another implicit or explicit model.
The Gene Expression Omnibus (GEO) was the first major expression data repository to go on-line, accepting its first submissions in 2001 (Edgar et al., 2002). One reason for the early appearance of GEO is apparently the simplified data model under which it operates, which eased its development. This is in contrast to ArrayExpress, which fully supports MAGE-OM, and which has trailed behind GEO in submissions. From our point of view, a major feature of the public databases is their support of data re-use. The use of data in ArrayExpress has been hampered by the challenges of handling MAGE-ML, a primary export format of the data. MAGE-ML is an XML representation of MAGE-OM, and parsing of MAGE-ML can be done with the help of software toolkits built for the purpose (MAGE-stk, http://mged.sourceforge.net/software/MAGEstk.php). However, MAGE-ML is still difficult to handle (Eisenstein, 2006), and the code we use to transform MAGE-ML into our native data model runs to thousands of lines. On the other hand, data in ArrayExpress is relatively well-annotated compared to data in GEO, and ArrayExpress recently started offering data in a simplified format, “MAGETAB”.
The Gene Expression Omnibus provided data in a simpler format from the start (SOFT; alternative formats including an XML format are now available) but without any software tool support. While writing a parser to handle SOFT files was not especially challenging, there were still a number of hurdles to allowing all GEO SOFT files to be successfully handled. To give an example, GEO ‘series’ (GSExxxx accessions) are eventually curated into ‘data sets’ (GDSxxxx) which have much more detailed annotations, but without all the raw data. However, it is not possible, based on the SOFT file for a GSE, to identify the matching GDS, and we resorted to screen-scraping the GEO web site for this information. The limitations of the GEO data model, while making data submission very simple, have presented a number of challenges. Perhaps most vexing is the failure of submitters to provide clear indications of which samples represent ‘technical replicates’ run on different platforms. Because GEO does not have the concept of an “RNA sample” (or similar concepts for which there are MAGE-OM classes), there is no way to determine that, for example, RNA from one tumor was run on both the Affymetrix HG-U133A and the HG-U133B platforms, except by the name of the sample and sometimes information on the experimental conditions which the samples shared, if available. This would not be so bad but for the tendency for submitters to give the same sample run on different platforms very different names. For example, instead of naming the replicates in a consistent manner (“52.32 HG-U133A” and “52.32 HG-U133B”), we have find constructs such as “lung-52.32a” paired with “52#32 – HG-U133B”. This leads to precarious attempts to match these samples up with software or time-consuming and sometimes still-uncertain curation efforts. We encourage potential data submitters to use consistent naming schemes for their samples.
An integral aspect of the offerings of these databases, independent of file formats, is the determination of what must be included in a submission. The aforementioned MIAME standard is generally accepted and is supported by the major databases, and by many journal publishers (Brazma et al., 2001; Spellman et al., 2002). To be MIAME compliant, a data set must fulfill several critical elements by making available: 1) the raw as well as processed expression data; 2) a detailed description of the experimental design; 3) a detailed description of the array designs used and 4) details of how the data were processed, such as normalization methods. Naturally the MAGE-ML and the GEO formats have “slots” for all of this information, but MIAME is independent of any specific format. While MIAME was in our view a major step forward in improving the usefulness of expression data, as discussed below it can be argued it does not go far enough in some areas (though others would argue the other way (Galbraith, 2006; Shields, 2006)). However, we recognize that the developers of MIAME tried to strike a balance between requiring detailed information and not being a burden on the submitter.
Data quality
A sticky issue that comes up when discussing the use of public data is quality. For our current purposes, we define quality as the extent to which the investigators have successfully limited the impact of technical sources of noise relative to biological signals. As described below, uniformly applying this definition is not trivial, and in practice the end (a validated result) can justify the means (even if low-quality data were used). At the same time, some other applicable aphorisms are “Garbage in, garbage out” and “Buyer beware”. Ignoring the issue of data quality risks turning an exercise in data re-use into a frustrating morass.
Most of the data available on the web has been in a sense vetted by reviewers of the relevant publications. However, peer review of a manuscript about microarray data does not generally entail close inspection of the raw data. Without further information we consider all available data to be of questionable quality. The easiest problem to spot is the presence of sample outliers, which show very different signal properties than the other samples (e.g., poor global correlations with other samples or much lower average signals). A variant of this problem is general high variance between samples, especially among biological or technical replicates. Some data sets contain unusually large numbers of missing values suggesting overall poor signals. The underlying causes of low data quality might not be recoverable from available information, but the usual suspects include degraded RNA or hybridization problems leading to high background.
Another source of noise is varying sample composition. It is common for studies of the nervous system to analyze RNA extracted from macroscopic chunks of tissue that are dissected from the brain. Unless very great care is taken, slight variation in the region that is taken can be a big source of noise. Taking smaller samples using microdissection, laser capture or cell sorting might ameliorate this problem by providing “purer” samples, but the cost of errors might actually be amplified: if one is trying to sample the amygdala, and accidentally sometimes get some neighboring areas, even in small amounts, the variability of the profiles obtained will be increased. The manipulations involved in isolating small samples are sometimes seen as a possible source of artifacts, but in general purer samples are valuable because they allow clearer interpretation of the results, and the ability to resolve genes expressed in small numbers of cells in enhanced.
The size of a data set can often limit its utility for re-use. Data sets that are highly re-usable, all else being equal, will be large (more than 10 and probably more than 20 samples). Small data sets yield results with either a very low sensitivity or unacceptable specificity. Larger data sets mean that signal can be more reliably distinguished from noise. This will always be true so long as other aspects of data quality are not relaxed in order to increase the sample size. In practice this means that results from very small data sets are not as trustworthy as those collected with larger sample sizes, and require independent validation before they can be used constructively. Other experimental design issues might also prove problematic for potential re-users, such as confounding variables. These can include batch effects that might not be well-advertised in the original publication.
We stress that the generator of data sets with quality problems might not have been bothered, because they obtained useful results anyway. This can easily be the case if, for example, there was sufficient signal to observe differential expression of some genes which were subsequently validated independently, despite an overall high level of noise. It is at this level that published studies are reviewed prior to publication, so underlying problems that might affect future re-purposing of the data might not be noticed.
Given the importance of data quality to re-use, who should be responsible for evaluating quality? Obviously, the investigators who generated the data stand to benefit by careful quality control. Just as obvious, the potential for re-use of data is enhanced if submitters are careful to include only high-quality data. However, we have to consider the case where the quality was good enough for the investigators to answer the question they were interested in asking, but not good enough for some other, unintended purpose that might be dreamed up by another investigator. It is not reasonable to hold submitters to an extreme high level of data quality standard, and it is similarly not reasonable to expect the curators of large databases to be able to clean all the data that is submitted. Therefore we are left with the conclusion that the data re-user must take quality into account, and if necessary remove data deemed to be of low quality. This has the potential to generate controversy or difficulties in interpretation, as the data are no longer tightly linked to the original study. Put another way, it is entirely possible that researchers who re-analyze a published data set with new quality control parameters will reach conflicting conclusions. When this happens, the submitter might feel that their data has been used against them, which discourages future data submissions. It is tempting to avoid such conflict by always using the data “as it was submitted”, but on the other hand it seems counterproductive to use low-quality data if it is avoidable.
Addressing quality would be easier if data submissions included quality data. In general quality control measures for each assay are not included in GEO or ArrayExpress submissions, though often they could be computed from the raw data that are provided. Unfortunately, MIAME appears to have too little to say about if, and how, quality control information is to be provided. MIAME requires “Quality control steps taken (e.g., replicates or dye swaps)” and “data selection procedures” (http://www.mged.org/Workgroups/MIAME/miame_checklist.html), but this falls short of getting data submitters to explicitly define the quality control measures they used to determine that an assay met inclusion criteria.
Reannotation of sequences on arrays
A major challenge to the interpretation of gene expression data is identifying what has been measured. This task is essentially impossible without knowing the sequence that was used on the array. Unfortunately, at least for the case of data in GEO, this information is often not available. This is in spite of the fact that the MIAME standards for microarray data submission require “unambiguous characteristics of the reporter molecule[s]” including sequences or precise accession numbers (http://www.mged.org/Workgroups/MIAME/miame_checklist.html). While there is almost always some type of sequence identifier given, this is sometimes clearly not the accession for the reporter (the sequence of the molecule that is actually on the array). For example, in GPL254, Genbank accessions are provided, but this is a platform of synthetic 70-base oligonucleotides. Instead the accessions appear to be an exemplar full-length transcript (e.g., RefSeq ID) for the gene the manufacturer claims to be assayed. The sequences for the oligonucleotides are not available from GEO. This is an area where the field can clearly improve. Repository maintainers could firm up their submission standards to require more exact sequence information submission. In cases where the actual sequence identifier is not provided, this should be clearly spelled out. In addition, purchasers of microarrays would benefit from using vendors which make sequences available, so we encourage experimenters to request the sequences up front.
An amusing (or frustrating) pastime in our lab is tracking down the sequences for array designs that are not immediately available from web-accessible resources. A typical exchange of emails involves contacting the investigator, who it seems sometimes has to spend considerable time tracking the information down, presumably digging through dusty hard drives to find the necessary information – or, unfortunately, not being able to find it. Alternatively, the sequences turn out to be “proprietary”, not because the investigator wants it that way, but because they got the probes from a company that considers the nucleotide sequences to be trade secrets. In some cases we have contacted firms with requests for sequences only to be told that “That division was sold”, “We got the probes from another company” or something similar. Despite doggedly following these trails, we are sometimes unable to find any living person who knows what was on the microarray, much less willing to make them public. Fortunately some searches for sequences have happy endings. For our purposes, we consider data that lacks reasonably high-quality sequence information to be unusable; we prefer not to take the word of the manufacturer for what was assayed, especially as it often turns out that sequences on arrays are not specific for transcripts of a single gene.
Probe specificity
As suggested above, with the sequences in hand we are not finished. By aligning the sequences to the genome or transcript sequences one can infer what gene (or genes) are assayed by each probe on a microarray. There are myriad approaches to doing this; in our work we have adopted the use of BLAT to align sequences to genome assemblies, and then relying on the annotation efforts of the UCSC or NCBI genome curators to tell us what gene lays in the aligned region (Barnes et al., 2005). In some cases, there is no simple answer, as many of the sequences on microarrays in our system have at least two high-quality alignments to the genome (human, mouse or rat, as appropriate). If only one of the alignments ‘hits’ a documented gene, then perhaps all is well. However, it is very common for probes to plausibly assay at least two different genes. In many cases this is hard to avoid, where duplicated genes are insufficiently diverged to design specific probes. But often the problem is a function of incomplete knowledge at the time of array design. The clones might have been selected at random or using fragmentary information, not a complete genome. The moral of the story is that the annotations provided by the manufacturer or submitter are very likely to be faulty if they do not rely on current, rigorous sequence alignment and annotation efforts.
Experiment annotation
High-quality experiment annotations are required to make re-use of data efficient. Two of the most important use-cases for annotations are locating data and labeling data. By locating data we mean being able to search for and successfully find, for example, all the experiments or hybridizations in a database that involved “mouse brain”. This is harder than it sounds because a data set might be annotated as involving “hippocampus”, but if the word “brain” is not used in the annotations, or a link to a relevant controlled vocabulary term is not provided (e.g., NeuroNames (Bowden and Dubach, 2003)), it is difficult for a computer program to infer that a “hippocampus” data set is also a “brain” data set. By labeling data we mean providing phenotypic or other data that is usable in computational analyses. For example, in order to identify genes that are differentially expressed in a data set, the samples belonging to different experimental groupings must be clearly identified.
The MIAME standard requires a good deal of information about the experimental design in a microarray experiment, such as (as appropriate) time, dose, or genetic variation information for each sample. However, other than recommending the use of the MGED Ontology (MO) terms there is not much said about how this information is to be structured. In GEO, annotations are primarily present in the form of free text submitted by the experimenter, associated with fields in the submission form (http://www.ncbi.nlm.nih.gov/projects/geo/info/soft2.html). This affords submitters a good deal of latitude in how they describe their data. This is an advantage for the submitter in that it makes their job easier (they can often use text from the “Materials and Methods” section of their paper), but a distinct disadvantage for those who are trying to locate data (e.g. by searching the GEO web site) or re-use it in bulk. Therefore it is almost inevitable that reusing expression data requires some reannotation effort. Even in ArrayExpress, which uses the MGED Ontology to provide “slots” for annotators to fill, the values in the “slots” are often free text, not terms in a controlled vocabulary. For example, one can find the annotation “Organism Part = ‘prefrontal cortex’” in on data set (E_AFMX-5) while in another the same concept might be expressed as “Organism Part = ‘brain, prefrontal cortex” (E-TAMB-136). There is clearly a limit to what data submitters can be expected to do with the tools that are available, and (understandably) to what the public database organizations (NCBI and EBI for GEO and ArrayExpress, respectively) are able to do with available curation resources. This suggests that better tools are needed for data submitters to assist them in providing useful annotations. A further challenge is the shifting landscape of terminologies: The MGED ontology is due to be made redundant by a revised ontology based on the Ontology for Biomedical Investigations (OBI; previously known as FuGO; http://mged.sourceforge.net/ontologies/MO_FAQ.htm).
Conclusions
While there are numerous challenges to re-using expression data, including availability, annotation, quality and comparability, in our view there is enormous potential to leverage existing data. Improvements in tools for the development of databases (submission and annotation) and for analysis will play a major role in making re-use of expression data as common an activity as re-using sequence data. Investigators who generate and then release data are the linchpin, and we hope that as the benefits become clearer, the motivation to release well-annotated data will increase. Organizations enforcing how data are released (e.g., journals) should work with standards organizations and data repositories (e.g., MGED, GEO and ArrayExpress) to help ensure that sufficiently detailed sequence and quality information are made available to help guide data re-users.
For neuroscientists, there are some unique opportunities and needs in integrating high-throughput expression profiling with other types of data. We predict that the next few years will see a rapid increase in the availability of tools integrating microarray data (which has a high degree of gene coverage but low spatial resolution) with spatially-resolved expression data, as well as information on neuronal function. Continuing efforts to make data available on the web and interoperable are essential (Koslow, 2005).
Acknowledgements
We are grateful to the Etienne Sibille and the anonymous reviewers for helpful suggestions, and to Tanya Barrett and the rest of the GEO staff for their assistance with the use of GEO. We are indebted to the many groups who generously provide expression data. Supported by NIH GM076990 and a Michael Smith Foundation for Health Research Career Award to P.P.
Footnotes
Information Sharing Statement
Supplemental information mentioned in this article is available at http://www.bioinformatics.ubc.ca/pavlidis/lab/reuse. Gemma is an open source project and is available at http://www.bioinformatics.ubc.ca/Gemma.
References
- Aarnio V, Paananen J, Wong G. Analysis of microarray studies performed in the neurosciences. J Mol Neurosci. 2005;27:261–268. doi: 10.1385/JMN:27:3:261. [DOI] [PubMed] [Google Scholar]
- Assou S, Le Carrour T, Tondeur S, Strom S, Gabelle A, Marty S, Nadal L, Pantesco V, Reme T, Hugnot JP, et al. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem cells (Dayton, Ohio) 2007;25:961–973. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes M, Freudenberg J, Thompson S, Aronow B, Pavlidis P. Experimental comparison and cross-validation of the Affymetrix and Illumina gene expression analysis platforms. Nucleic acids research. 2005;33:5914–5923. doi: 10.1093/nar/gki890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucleic acids research. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucl. Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota M, Dong HW, Swanson LW. Brain architecture management system. Neuroinformatics. 2005;3:15–48. doi: 10.1385/NI:3:1:015. [DOI] [PubMed] [Google Scholar]
- Bowden DM, Dubach MF. NeuroNames 2002. Neuroinformatics. 2003;1:43–59. doi: 10.1385/NI:1:1:043. [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature genetics. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS letters. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Brown PO, Eisen MB, Varmus HE. Why PLoS became a publisher. PLoS biology. 2003;1:E36. doi: 10.1371/journal.pbio.0000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nature genetics. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- Chin MH, Geng AB, Khan AH, Qian WJ, Petyuk VA, Boline J, Levy S, Toga AW, Smith RD, Leahy RM, Smith DJ. A genome-scale map of expression for a mouse brain section obtained using voxelation. Physiological genomics. 2007 doi: 10.1152/physiolgenomics.00287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Yu U, Kim S, Yoo OJ. Bioinformatics. Vol. 19. Oxford, England: 2003. Combining multiple microarray studies and modeling interstudy variation; pp. 84–90. [DOI] [PubMed] [Google Scholar]
- Cooper H, Hedges LV. Handbook of Research Synthesis. New York: Russell Sage Foundation; 1994. [Google Scholar]
- Crasto CJ, Marenco LN, Liu N, Morse TM, Cheung KH, Lai PC, Bahl G, Masiar P, Lam HY, Lim E, et al. SenseLab: new developments in disseminating neuroscience information. Briefings in bioinformatics. 2007;8:150–162. doi: 10.1093/bib/bbm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley P, Egan GF, Amari S, Beltrame F, Bennett R, Bjaalie JG, Dalkara T, De Schutter E, Gonzalez C, Grillner S, et al. Neuroscience data and tool sharing: a legal and policy framework for neuroinformatics. Neuroinformatics. 2003;1:149–165. doi: 10.1007/s12021-003-0002-1. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein M. Microarrays: quality control. Nature. 2006;442:1067–1070. doi: 10.1038/4421067a. [DOI] [PubMed] [Google Scholar]
- Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, et al. Science. Vol. 296. New York, N.Y.: 2002. Intra- and interspecific variation in primate gene expression patterns; pp. 340–343. [DOI] [PubMed] [Google Scholar]
- Erraji-Benchekroun L, Underwood MD, Arango V, Galfalvy H, Pavlidis P, Smyrniotopoulos P, Mann JJ, Sibille E. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biological psychiatry. 2005;57:549–558. doi: 10.1016/j.biopsych.2004.10.034. [DOI] [PubMed] [Google Scholar]
- Galbraith DW. The daunting process of MIAME. Nature. 2006;444:31. doi: 10.1038/444031b. [DOI] [PubMed] [Google Scholar]
- Ge X, Yamamoto S, Tsutsumi S, Midorikawa Y, Ihara S, Wang SM, Aburatani H. Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics. 2005;86:127–141. doi: 10.1016/j.ygeno.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorioso C, Sabatini M, Unger T, Hashimoto T, Monteggia LM, Lewis DA, Mirnics K. Specificity and timing of neocortical transcriptome changes in response to BDNF gene ablation during embryogenesis or adulthood. Mol Psychiatry. 2006;11:633–648. doi: 10.1038/sj.mp.4001835. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Science. Vol. 286. New York, N.Y.: 1999. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring; pp. 531–537. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gu J, Gu X. Induced gene expression in human brain after the split from chimpanzee. Trends Genet. 2003;19:63–65. doi: 10.1016/s0168-9525(02)00040-9. [DOI] [PubMed] [Google Scholar]
- Gu J, Gu X. Further statistical analysis for genome-wide expression evolution in primate brain/liver/fibroblast tissues. Human genomics. 2004;1:247–254. doi: 10.1186/1479-7364-1-4-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, Haroutunian V, Fienberg AA. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh WP, Chu TM, Wolfinger RD, Gibson G. Mixed-model reanalysis of primate data suggests tissue and species biases in oligonucleotide-based gene expression profiles. Genetics. 2003;165:747–757. doi: 10.1093/genetics/165.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JE, Schmidt FL. Methods of meta-analysis. London: Sage; 1990. [Google Scholar]
- Iwamoto K, Bundo M, Washizuka S, Kakiuchi C, Kato T. Expression of HSPF1 and LIM in the lymphoblastoid cells derived from patients with bipolar disorder and schizophrenia. Journal of human genetics. 2004a;49:227–231. doi: 10.1007/s10038-004-0136-5. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Kakiuchi C, Bundo M, Ikeda K, Kato T. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol. Psychiatr. 2004b;9:406–416. doi: 10.1038/sj.mp.4001437. [DOI] [PubMed] [Google Scholar]
- Jurata LW, Bukhman YV, Charles V, Capriglione F, Bullard J, Lemire AL, Mohammed A, Pham Q, Laeng P, Brockman JA, Altar CA. Comparison of microarray-based mRNA profiling technologies for identification of psychiatric disease and drug signatures. J. Neurosci. Methods. 2004;138:173–188. doi: 10.1016/j.jneumeth.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Kapushesky M, Kemmeren P, Culhane AC, Durinck S, Ihmels J, Korner C, Kull M, Torrente A, Sarkans U, Vilo J, Brazma A. Expression Profiler: next generation--an online platform for analysis of microarray data. Nucleic acids research. 2004;32:W465–W470. doi: 10.1093/nar/gkh470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslow SH. Should the neuroscience community make a paradigm shift to sharing primary data? Nature neuroscience. 2000;3:863–865. doi: 10.1038/78760. [DOI] [PubMed] [Google Scholar]
- Koslow SH. Discovery and integrative neuroscience. Clin EEG Neurosci. 2005;36:55–63. doi: 10.1177/155005940503600204. [DOI] [PubMed] [Google Scholar]
- Larsson O, Wennmalm K, Sandberg R. Comparative microarray analysis. Omics. 2006;10:381–397. doi: 10.1089/omi.2006.10.381. [DOI] [PubMed] [Google Scholar]
- Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome research. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lein ES, Zhao X, Gage FH. Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J Neurosci. 2004;24:3879–3889. doi: 10.1523/JNEUROSCI.4710-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lu L, Manly KF, Chesler EJ, Bao L, Wang J, Zhou M, Williams RW, Cui Y. Inferring gene transcriptional modulatory relations: a genetical genomics approach. Human molecular genetics. 2005;14:1119–1125. doi: 10.1093/hmg/ddi124. [DOI] [PubMed] [Google Scholar]
- Magdaleno S, Jensen P, Brumwell CL, Seal A, Lehman K, Asbury A, Cheung T, Cornelius T, Batten DM, Eden C, et al. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS biology. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, Kenyon C, Bargmann CI, Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nature genetics. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Pevsner J. Progress in the use of microarray technology to study the neurobiology of disease. Nature neuroscience. 2004;7:434–439. doi: 10.1038/nn1230. [DOI] [PubMed] [Google Scholar]
- Moreau Y, Aerts S, De Moor B, De Strooper B, Dabrowski M. Comparison and meta-analysis of microarray data: from the bench to the computer desk. Trends Genet. 2003;19:570–577. doi: 10.1016/j.tig.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Chiu CH, Pulapura S, Mehan MR, Nunez-Iglesias J, Zhang K, Kamath K, Waterman MS, Finch CE, Zhou XJ. Gene Aging Nexus: a web database and data mining platform for microarray data on aging. Nucleic acids research. 2007;35:D756–D759. doi: 10.1093/nar/gkl798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnykov N, Lilja P, Lukk M, et al. ArrayExpress--a public database of microarray experiments and gene expression profiles. Nucleic acids research. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Noble WS. Analysis of strain and regional variation in gene expression in mouse brain. Genome biology. 2001;2 doi: 10.1186/gb-2001-2-10-research0042. RESEARCH0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome biology. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya OO, Haroutunian V, Davis KL, Hernandez I, Sokolov BP. Novel putative nonprotein-coding RNA gene from 11q14 displays decreased expression in brains of patients with schizophrenia. J. Neurosci. Res. 2003;74:111–122. doi: 10.1002/jnr.10752. [DOI] [PubMed] [Google Scholar]
- Ponomarev I, Maiya R, Harnett MT, Schafer GL, Ryabinin AE, Blednov YA, Morikawa H, Boehm SL, Homanics GE, Berman A, et al. Transcriptional signatures of cellular plasticity in mice lacking the alpha 1 subunit of GABA(A) receptors. Journal of Neuroscience. 2006;26:5673–5683. doi: 10.1523/JNEUROSCI.0860-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer research. 2002;62:4427–4433. [PubMed] [Google Scholar]
- Rhodes DR, Chinnaiyan AM. Bioinformatics strategies for translating genome-wide expression analyses into clinically useful cancer markers. Annals of the New York Academy of Sciences. 2004;1020:32–40. doi: 10.1196/annals.1310.005. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Neoplasia. Vol. 6. New York, N.Y.: 2004. ONCOMINE: a cancer microarray database and integrated data-mining platform; pp. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Yasuda R, Pankratz DG, Carter TA, Del Rio JA, Wodicka L, Mayford M, Lockhart DJ, Barlow C. Regional and strain-specific gene expression mapping in the adult mouse brain. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11038–11043. doi: 10.1073/pnas.97.20.11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nature genetics. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields R. MIAME, we have a problem. Trends Genet. 2006;22:65–66. doi: 10.1016/j.tig.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Sibille E, Su J, Leman S, Le Guisquet AM, Ibarguen-Vargas Y, Joeyen-Waldorf J, Glorioso C, Tseng GC, Pezzone M, Hen R, Belzung C. Lack of serotonin(1B) receptor expression leads to age-related motor dysfunction, early onset of brain molecular aging and reduced longevity. Mol Psychiatry. 2007 doi: 10.1038/sj.mp.4001990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui AS, Khattra J, Delaney AD, Zhao Y, Astell C, Asano J, Babakaiff R, Barber S, Beland J, Bohacec S, et al. A mouse atlas of gene expression: large-scale digital gene-expression profiles from precisely defined developing C57BL/6J mouse tissues and cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18485–18490. doi: 10.1073/pnas.0509455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov BP, Jiang LX, Trivedi NS, Aston C. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. J. Neurosci. Res. 2003;72:756–767. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Miller M, Stewart J, Troup C, Sarkans U, Chervitz S, Bernhart D, Sherlock G, Ball C, Lepage M, et al. Design and implementation of microarray gene expression markup language (MAGE-ML) Genome biology. 2002;3 doi: 10.1186/gb-2002-3-9-research0046. RESEARCH0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansberg C, Vik-Mo AO, Holdhus R, Breilid H, Srebro B, Petersen K, Jorgensen HA, Jonassen I, Steen VM. Gene expression profiles in rat brain disclose CNS signature genes and regional patterns of functional specialisation. Bmc Genomics. 2007;8 doi: 10.1186/1471-2164-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. Science. Vol. 302. New York, N.Y.: 2003. A gene-coexpression network for global discovery of conserved genetic modules; pp. 249–255. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nature neuroscience. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Sunkin SM. Towards the integration of spatially and temporally resolved murine gene expression databases. Trends Genet. 2006;22:211–217. doi: 10.1016/j.tig.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Tkachev D, Mimmack ML, Ryan MM, Wayland M, Freeman T, Jones PB, Starkey M, Webster MJ, Yolken RH, Bahn S. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- Vazquez-Chona FR, Khan AN, Chan CK, Moore AN, Dash PK, Hernandez MR, Lu L, Chesler EJ, Manly KF, Williams RW, Geisert EE., Jr Genetic networks controlling retinal injury. Molecular vision. 2005;11:958–970. [PMC free article] [PubMed] [Google Scholar]
- Watson R. EC to promote open access publishing. BMJ. 2007;334:389. doi: 10.1136/bmj.39129.642130.DB. Clinical research ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW. Expression genetics and the phenotype revolution. Mamm Genome. 2006;17:496–502. doi: 10.1007/s00335-006-0006-x. [DOI] [PubMed] [Google Scholar]
- Zapala MA, Hovatta I, Ellison JA, Wodicka L, Del Rio JA, Tennant R, Tynan W, Broide RS, Helton R, Stoveken BS, et al. Adult mouse brain gene expression patterns bear an embryologic imprint. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10357–10362. doi: 10.1073/pnas.0503357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Morris QD, Chang R, Shai O, Bakowski MA, Mitsakakis N, Mohammad N, Robinson MD, Zirngibl R, Somogyi E, et al. The functional landscape of mouse gene expression. Journal of biology. 2004;3:21. doi: 10.1186/jbiol16. [DOI] [PMC free article] [PubMed] [Google Scholar]