Abstract
We have studied the shape of myosin VI, the actin minus-end directed motor, by negative stain and metal shadow electron microscopy. Single particle processing was used to make two-dimensional averages of the stain images, which greatly increases the clarity and allows detailed comparisons with crystal structures. A total of 169,964 particle images were obtained from two different constructs in six different states (four nucleotide states and with and without Ca2+). The shape of truncated apo myosin VI was very similar to the apo crystal structure, with the lever arm bent strongly backward and around the motor domain. In the full-length molecule, the C-terminal part of the tail has an additional bend taking it back across the motor domain, which may reflect a regulated state. Addition of ATP, ADP, or ATP-γS resulted in a large change, straightening the molecule from the bent shape and swinging the lever by ∼140°. Although these nucleotides would not be expected to produce the pre-powerstroke state, myosin VI in their presence was most similar to the truncated crystal structure with bound ADP-VO4, which is thought to show the pre-powerstroke shape. The nucleotide data were therefore substantially different from expectation based on crystal structures. The full-length molecule was almost completely monomeric; only ∼1% were dimers, joined through the ends of the tail. Addition of calcium ions appeared to result in release of the second calmodulin light chain. In negatively stained molecules there was little indication of extended α-helical structure in the tail, but molecules viewed by metal shadowing had a tail ∼3× longer, 29 vs. 9 nm, part of which is likely to be a single α-helix.
Introduction
The myosin VI molecular motor is ubiquitous in eukaryotes and is involved in a wide variety of important cellular processes, including oogenesis, spermatogenesis, hearing, and secretion. Myosin VI is unique in being the only myosin of the 40 or so now known in humans that walks towards the pointed end of actin filaments, in the opposite direction to all the other myosins so far studied. Myosin VI consists of a motor domain, a lever arm, and a helical tail region ending in a C-terminal cargo-binding domain. The cargo domain binds a wide range of adaptor proteins that mediate the diverse functions of myosin VI. Several crystal structures of myosin VI have been solved in different long-lived nucleotide states, using constructs truncated after the lever arm (subfragment 1, or S1) or with only the motor domain present (1–3). These structures, together with single molecule data (4,5), show that reverse movement is achieved by a 53-residue unique sequence called Insert 2 (aa 761–813) present after the converter region of the motor domain; this redirects the lever arm through ∼120°, although the details of the reverse mechanism are not fully understood. Also unique to myosin VI is a sequence, Insert 1 (aa 276–297), near the nucleotide binding site that reduces the rate of ATP binding >10-fold compared to other myosins (6).
Myosin VI is also unusual in having an exceptionally large powerstroke considering the length of its lever arm (7,8). The sequence of the lever shows a single IQ motif, which suggested only one calmodulin would be bound. Although a second calmodulin was subsequently found associated with Insert 2 (9), the working stroke is much larger than expected for a lever with only two calmodulins: the powerstroke is ∼18 nm, whereas myosin II, which also has two light chains, has only a ∼5 nm step (10).
Initial analysis of the sequence of the myosin VI tail showed regions of heptad repeats of hydrophobic residues, which suggested that the molecule would dimerize by forming a coiled-coil α-helix (11). As a consequence of this presumed dimerization, many articles have appeared on the properties of myosin VI artificially dimerized by inclusion of leucine zipper sequence. Myosin VI has subsequently been demonstrated to dimerize without inclusion of a leucine zipper, either by being held in close proximity or by C-terminal binding partners that are themselves dimeric (12). However, both the expressed and native molecules have been shown to be monomeric (7), and whether myosin VI is dimeric in vivo is controversial (7,13).
More recently, at least part of the myosin VI tail has been suggested to be a single α-helical (SAH) domain, rather than forming a coiled-coil. A SAH domain in myosin VI was suggested due to the presence of regular repeats of charged E, R, and K residues in the tail (7). Subsequently, similar sequence in myosin X was synthesized and shown to form a single helix, which led to the proposal that a SAH domain could act to increase the effective length of the myosin VI lever arm and thereby explain the unexpectedly large powerstroke (14). However, the tail immediately after the IQ motif but before the predicted SAH domain was then shown to be a three-helix bundle, which seems to unfold only when myosin VI is dimerized (15). Thus, the structure and functions of the different regions of the tail and cargo-binding domain remain to be established.
A different explanation to an extension of the lever arm lengthening the powerstroke is that the angular throw of the myosin VI lever is greater than the 70–90° seen in myosins II and V. This idea resulted from the myosin VI crystal structure with ADP.Vi bound (representing the prepowerstroke ADP.Pi state) in which the lever arm was predicted to emerge from the motor domain at 140° or more from the angle seen in the apo state (2). Recently, single molecule fluorescence measurements indicated a swing of ∼180° in artificially dimerized myosin VI (16,17). There have also been suggestions that the converter can decouple from the rest of the motor domain under some circumstances, which would allow the lever to swing freely (1,16).
Many questions remain concerning how myosin VI generates force, how its shape changes in different nucleotide states, and what the structures of its tail- and cargo-binding domains are and their propensity to dimerize. To better understand the properties and functions of myosin VI, more information is needed about the shape and flexibility of the entire molecule, especially in response to different conditions. In this article, we report electron microscopy of both truncated and full-length myosin VI under a variety of conditions, including different nucleotide states. We mainly used the negative stain method as this provides ∼2 nm resolution (see, for example, (18)) which, after two-dimensional averaging by single particle analysis, allows detailed shape comparisons with crystal structures (19–22). The strongly bent shape of truncated apo myosin VI agrees well with the corresponding crystal structure. A large change to a straight shape involving a lever swing of ∼140° was seen in the presence of ATP, ATP-γS, and ADP. These shapes are different from expectation based on crystal structures in long-lived analog nucleotide states. A few dimers were observed, but these were only ∼1% of the total. Molecules exposed to calcium appeared to have lost the IQ calmodulin. We also used the rotary metal shadowing method, which resulted in a different appearance in the tail/cargo binding domain. A preliminary report of some of this work has already appeared (23).
Materials and Methods
Preparation of myosin VI and its S-1subfragment
Full-length myosin VI (aa 1–1276) from chicken intestinal brush border cells containing the large tail insert (24) was cloned into the Bac-to-Bac (Invitrogen, UK) vector FasBacHT. Protein with an N-terminal hexa-His tag was coexpressed with calmodulin in insect Sf9 cells and purified as described previously (7). The truncated myosin VI S-1 subfragment (aa 1–839) containing the motor and IQ domain with either a N-terminal or C-terminal hexa-His tag was cloned into the same vector, coexpressed with calmodulin and purified by the same procedures used for the full-length molecule. Proteins were stored in 30 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 20 mM MOPS (pH 7.5), and 1 mM DTT. Long-term storage used dropwise freezing into liquid nitrogen. The ATPase activity of the frozen and thawed myosin VI was routinely tested by the Kendrick-Jones laboratory (J.K.-J.) in Cambridge and by Claudia Veigel and colleagues in London using an optical trap and in vitro motility assays (in vitro motility assays are especially sensitive to inhibitions by small numbers of inactive myosin molecules). Moreover, no differences were observed in the appearances of the molecules with or without freezing; thus, image averages of the apo S1 molecule remained very similar to projections of the corresponding crystal structure (see Results). The hexa-His tags were sometimes removed using rTEV protease (Invitrogen, Paisley, UK) specific for a cleavage site located between the His tag and the myosin VI reading frame. No differences were detected in activity, trap, or motility assays with or without the His tag.
Negative staining
Negative staining was performed as described (25). A droplet of myosin solution diluted just before use to ∼10 μg/mL in the same buffer as above, but without DTT, was put on an EM grid covered with a thin (5–10 nm) carbon foil. In different experiments, the buffer also contained ATP, ATP-γS, or ADP (0.5 mM) or 0.1 mM CaCl2 (with 0.5 mM ATP). The grid was first rinsed with a few drops of this buffer (with the appropriate nucleotide), then with 1% uranyl acetate (pH 4.5), and then blotted from its edge and dried. A key element of the staining method is that before use, the carbon-coated grids are treated with ultraviolet (UV) light (mostly ∼250 nm) from a mercury vapor lamp for ∼40 min. This makes the carbon hydrophilic, which results in a thin layer of stain after drying. The action of the UV on the carbon is not understood but it is noteworthy that irradiation is accompanied by a strong smell of ozone and is therefore carried out in an enclosure in a fume hood. We also observed that when our original lamp (Type R51; UV Products, Cambridge, UK) failed, a replacement (Type R52) that did not give off ozone did not result in successful negative staining of myosin. Consultation with UV Products revealed that the new Type R52 emission tube had been engineered to eliminate ozone. When a replacement tube that was without this modification and did give off ozone was used, successful staining was restored. It may also be mentioned that UV treatment is extensively used in industrial processes to prepare surfaces other than carbon.
Rotary shadowing
Rotary shadowing was carried out as described (26), which involves evaporation of platinum on to proteins dried in vacuo from glycerol-containing buffers on freshly cleaved mica. One requirement is to minimize the amount of nonvolatile salts by reducing the solution to a thin layer before drying, which results in uniform image background. This is usually done by spraying the protein onto mica, but in our case, we layered the solution on to the mica, which was then thinned by spinning the mica on a horizontal disc, throwing off excess liquid.
Electron microscopy and single particle image processing
Images defocused by 100–200 nm were recorded at a dose of ∼100 e/Å2 on SO-163 film (Kodak, Rochester, NY) in a model No. 1200ex microscope with a LaB6 electron source (JEOL USA, Peabody, MA). Micrographs were digitized at a step size corresponding to 5 Å with an Imacon 848 scanner (Hasselblad, Gothenburg, Sweden) and processed using the SPIDER/WEB software suite (27). Image stacks of 5000–25,000 particles were windowed out in WEB and brought into alignment by multiple rounds of translation and rotation. The stacks were then classified into homogeneous groups with the k-means clustering algorithm and averaged. Many trials were done generating different numbers of classes, with the object of maximizing the number of homogeneous images in a class, which improves clarity and detail. This was balanced against heterogeneity which results in blurring. Heterogeneity therefore requires more classes in order to achieve homogeneity within classes. Complete classifications are shown in the Supporting Material. Further details are given in Burgess et al. (28).
Results
Electron microscopy of individual myosin molecules has almost exclusively employed shadowing with platinum to visualize them. Here, instead we used mainly negative staining because it achieves ∼2 nm resolution, which is approximately twofold higher than shadowing. A total of 169,964 stain particle images were obtained under several different conditions. These were classified into homogeneous groups and averaged. Two-dimensional averages containing 50–100 images generally showed greatly improved detail and signal-to-noise (averages of >100 particles showed little further improvement). The following sections and figures show representative class averages, but complete classifications are in the Supporting Material, which allows the variability between classes to be assessed.
Shape of apo myosin VI
Image averages of negatively stained myosin VI molecules truncated after the second calmodulin light chain (subfragment 1, S1) are shown in the absence of nucleotide in Fig. 1. Alongside for comparison is the apo myosin VI S1 crystal structure (1). The micrographs show mainly one view, indicating a preferred orientation on the grids. We call this view the face-profile because the motor domain has two indentations on one side and is smoothly curved on the other. The indentation near the tip of the motor is the large cleft between the upper and lower 50 kDa domains. The second indentation is the gap between the lower 50 kDa domain and the SH3 subdomain (see also Fig. 2, b and d, for particularly clear examples of these details). Insert 2 is visible protruding from the motor domain and extending from this the lever arm curves around the side of the motor domain opposite the SH3 region. In ∼50% of cases, both calmodulins are resolved in the lever, with their lobes characteristically straddling the lever diagonally, such as in the non-IQ calmodulin in the third average panel. That the calmodulins are not always clear may reflect flexibility. There is thus good agreement between the image averages and the crystal structure, demonstrating that negative staining can faithfully reveal considerable detail in myosin VI molecules.
Figure 1.
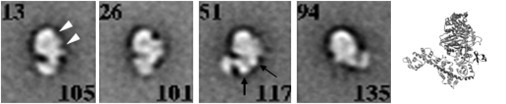
Image averages of truncated myosin VI (S-1) without nucleotide. The characteristic face-profile of the motor domain is seen in each panel (facing right except in panel 4). On one side, the motor shows two indentations that are the cleft between the upper and lower 50 kDa subdomains and the gap between the lower 50 kDa and SH3 subdomains (open arrowheads); on the other side, the motor curves smoothly. Insert 2 is visible (thin arrow), extending from which is the lever arm with two calmodulins, with their diagonally positioned lobes resolved in some cases (thick arrow). The numbers at the bottom right in each panel are the number of images in the average and at the top left are the class identification numbers. To the right is the crystal structure of a similar apo myosin VI S1 molecule (15) showing a very similar shape (see particularly stain panel 3). Scale bar, 5 nm.
Figure 2.
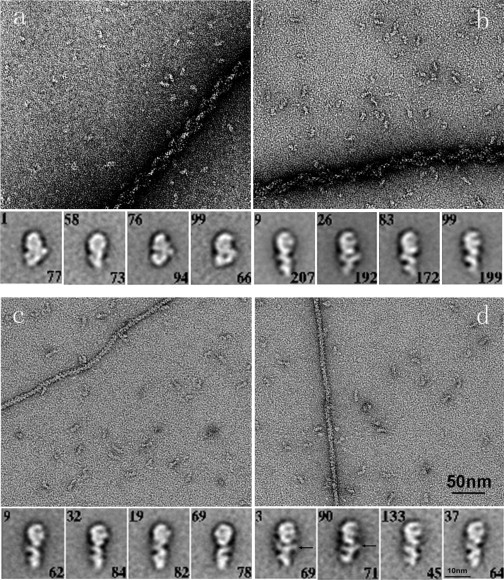
Full-length myosin VI molecules. The four panels a–d show molecules under different conditions: (a) apo, (b) ADP, (c) ATP and (d) ATP-γS (all nucleotides 0.5 mM). The upper part of each panel shows the field view with individual myosin VI molecules and residual actin filaments that are used in the myosin purification. Note the straightened shapes of the myosin VI in the presence of nucleotides; also, the heavy decoration of the actin filaments by the myosin VI without nucleotide or in ADP, and the bareness of the actin filaments in ATP and ATP-γS. Arrows in panel d indicate possible detachment of calmodulin lobes.
We also prepared image averages of full-length myosin VI, which has not been crystallized. Fig. 2 a shows field and windowed image averages of full-length myosin VI without nucleotide. As in our previous work (7), averaging showed the tail of the molecule bending sharply after the IQ-calmodulin region; as a result, the tail/cargo binding domain appears to fold back across the N-terminal part of the motor domain, perhaps binding to it. Such folding may be part of an inactivation/regulatory mechanism, such as occurs in myosins II and V.
Shape of myosin VI in the presence of nucleotides
Field images, together with image averages, of full-length myosin VI in the presence of ADP, ATP, and ATP-γS are shown in Fig. 2, b–d. Also visible in the field images are actin filaments, which remain present at low levels as a result of the myosin VI purification protocol. As would be expected for functional myosin molecules, the actin filaments are heavily decorated by the myosin in the apo and ADP conditions, whereas they are bare in the presence of ATP and ATP-γS. These data are therefore consistent with the myosin VI being functionally competent.
Strikingly, the conformation of the myosin VI in the image averages was similar in all three added nucleotides and very different from the strongly bent apo state. In the presence of all three nucleotides, the molecules were essentially straight with the lever and tail in line with the long axis of the motor domain. The lever swing producing this change was ∼140°. Although there were a few bent classes these were a very small proportion of the total (<5%, see full classifications in the Supporting Material). The same face-view of the motor domain seen without nucleotide predominates, confirming a preferred orientation on the substrate, although sometimes facing in the opposite direction. In the straight tails, three distinct regions can usually be seen, two of which are alternately angled with lobes on opposite sides of the lever indicating they are calmodulin. Interestingly, in some cases, the lobe of the calmodulin associated with Insert 2 and nearest the motor SH3 subdomain sometimes appears to extend as though it has detached from the backbone α-helix (arrowed). Similar calmodulin lobe detachment has previously been reported (29).
The third C-terminal region of the tail is relatively featureless. There is no evidence of a thin, extended helical region (SAH domain), as predicted (7,14). The C-terminal cargo binding domain is also not resolved, possibly because it is extremely mobile; it has been suggested to act as the end of a flexible lasso to capture its adaptor binding partners (30). Support for this suggestion is the observation that this domain binds to PIP2-containing liposomes and binding induces a large structural change (31% increase in helicity) (31).
Rotary shadowed myosin VI molecules
Full-length myosin VI with and without ATP present was also examined by rotary metal shadowing because, although this method achieves lower resolution, there was evidence that the length of the tail observed by this technique (32) was longer than seen by negative staining. Fig. 3 shows a montage of full-length molecules dried in the presence of ATP to give the straightened conformation described above, to make length measurements simpler (apo shadowed molecules were more compact, as expected (not shown)). Much less detail is visible than in the previous figures and this was not improved by image averaging (not shown). The large, globular motor domain is still clearly identifiable; however, the overall length excluding the motor domain, 29 ± 3 nm, is >3 times that measured in the stain image averages (9 ± 2 nm), but no details of the individual tails can be discerned.
Figure 3.
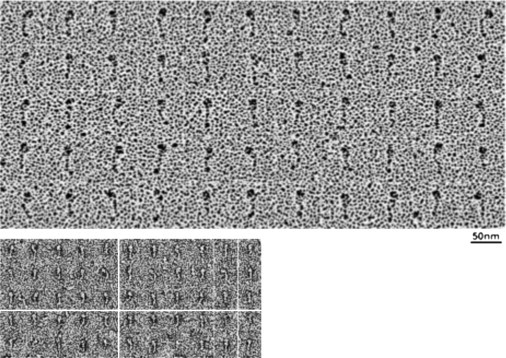
Rotary shadowed myosin VI. Shadowed full-length molecules (in the presence of 0.5 mM ATP) are in the upper panel. Note the bulbous, globular motor domain toward the top of each molecule, but little other detail is visible. Below to the same scale are negatively stained molecules. The distance between the motor domain and the C-terminal end of the molecule, 29 nm, was ∼3× that in stained data, 9 nm, suggesting a difference in preservation between the two methods.
Myosin VI dimers
We previously reported that, contrary to prediction, expressed and purified myosin VI molecules were almost completely monomeric (7). Although small amounts of myosin VI molecules have subsequently been shown to dimerize without inclusion of a leucine zipper (33), there is little evidence of dimers in situ. We do, however, find a very small population (∼1%) of dimers in the intact molecule preparations and examples are shown in Fig. 4. The dimers appear to form through interactions at the ends of their tails, rather than through a 2-nm-wide coiled-coil tail, as in myosins II and V. There was no evidence of extended helix or coiled-coil structure in the tails in these stained images, as has been predicted. Based on negatively stained images of myosins II (19) and V (22), a coiled-coil, if present, should have been visible.
Figure 4.
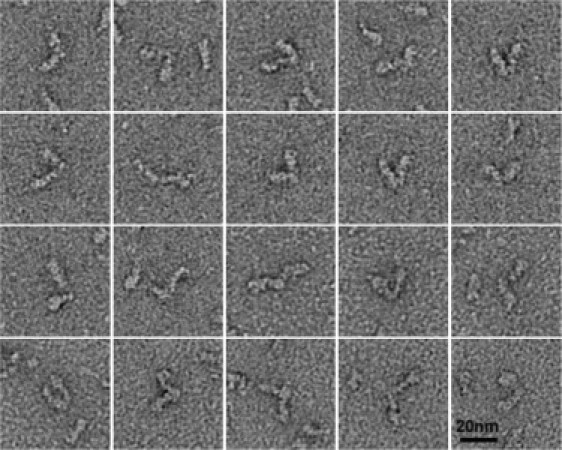
Myosin VI dimers. The majority of myosin VI molecules observed were monomeric but 1% were dimers joined through interactions at the ends of their tails. Note the absence of extended helical or coiled-coil regions in the tails. The grids for these images were made with 0.5 mM ATP present.
Effect of calcium
All the images shown so far were of myosin VI in low calcium with EGTA present. Fig. 5 shows image averages of whole myosin VI with 0.1 mM Ca2+ present. Consistent with expectation, the tail is substantially shorter and less complex, indicating the second (IQ) calmodulin has dissociated. The first calmodulin, which binds to Insert 2, appears still to be present. However, only one region is visible extending from the motor domain; this suggests the remainder of the tail and cargo binding domain coalesced with the calmodulin bound to Insert 2 (arrowed). Note also that the motor domain appears narrower in these averages compared to Figs. 1 and 2, which is consistent with a motor domain orientation rotated by ∼20° on the carbon substrate compared to the earlier figures.
Figure 5.
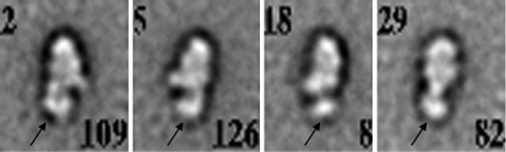
Myosin VI in calcium. Full-length myosin VI was treated with 0.1 mM CaCl2 (in the presence of 0.5 mM ATP). Note the apparent absence of the second (IQ) calmodulin (compare with Fig. 2B). Arrows mark the remaining (Insert 2) calmodulin. Scale bar, 5 nm.
Discussion
Myosin VI is arguably the most interesting and puzzling of the ∼40 myosins now identified in humans. This is due to its unique ability to walk backward and to its unexpectedly large powerstroke. These properties arise from the complex structure of the ∼800 residue motor domain and neither of these mechanisms is fully understood. Moreover, the structures and functions of the different regions of the tail and the extent to which the molecule dimerizes in vivo also remain unclear.
Unconventional myosin VI powerstroke
The powerstrokes of myosins II and V are both similar and relatively well characterized, primarily as a result of head (motor domain ± lever) crystal structures in a variety of long-lived nucleotide analog states. These structures have also been docked into lower resolution electron microscopy reconstructions with actin filaments in tightly bound states (e.g., (34)). Both myosin II and V powerstrokes involve a swing of the lever against the motor domain of ∼80°, from a sharply bent shape with ADP and Pi bound to a much straighter shape with either ADP or no ligand bound. In many myosin subclasses, this transition takes place in two stages as Pi and ADP are successively released. The shapes and relative angles of the motor domains and lever arms of myosins II and V in the apo and ADP.Pi states visualized by the same negative staining and image-averaging protocols used here agree well with corresponding crystal structures (19–22).
The myosin VI powerstroke appears to be very different; in the positions of its lever, the extent of its swing and probably also its effective length. Movement of the converter similar to that seen in myosins II and V would take the lever from the already highly bent shape of apo-myosin VI to one even more bent (2), so that the lever would cross the motor domain. Instead, with ADP.Vi in the active site, which is thought to represent the ADP.Pi state, the converter rearranges itself to a structure not seen in other myosins; the heavy chain α-helix to which the calmodulins would attach then emerges from the motor domain roughly parallel to its long axis (2). With the lever attached, this structure would produce an approximately straight shape similar to that seen here with ATP, ADP, and ATP-γS. The swing of the lever under these conditions from the apo state is roughly 140°, i.e., approximately twice that seen in the myosins II and V powerstrokes. This is somewhat smaller than the ∼180° swing reported from recent single molecule fluorescence measurements (16,17). However, the fluorescence determination used molecules that were artificially dimerized. Tethering between the heads may therefore have affected the lever swing. For instance, the levers in the dimer may be forced to make increased swings in order for the lead head to reach out to find an actin subunit at the appropriate orientation to bind to. Increased swinging could also be facilitated if the converter is decoupled from the rest of the motor, as was claimed (16).
Despite several attempts, we have not obtained images in the ADP.Pi analog states, ADP.AlF4 or ADP.Vi, because myosin VI aggregated in the presence of these ligands. However, the bent apo conformation is dramatically different from the straight conformations we observed in the presence of nucleotide, and this swing is therefore likely to be the powerstroke. The fact that the ∼140° swing from the apo state is similar to that seen by crystallography is also consistent with the rearrangement of the converter to that seen in the crystal structure with ADP.Vi bound. This novel structure of the converter is not seen in myosins II and V.
The straight myosin VI shape we see in the presence of nucleotides can therefore be argued to show the start of the powerstroke. In this respect, our results superficially correspond to those from myosins II and V in the presence of ATP, where the pre-powerstroke ADP.Pi state is the predominant intermediate. Myosins II and V in the presence of ATP are mostly in the ADP.Pi state, because the equilibrium constants of their hydrolysis steps, KH, are > 1, and because there is slow phosphate dissociation, k-P, making M-ADP-Pi the predominant steady-state intermediate during hydrolysis (Eq. 1 in Table 1) (33). In myosin VI, KH is 0.3–0.5 and 70–85% of the molecules would be expected to be in the M-ATP state during steady-state hydrolysis and 15–30% M-ADP-Pi (35). Less than 5% of the myosin VI images have a bent conformation (see the Supporting Material) and the most likely interpretation of the data is that the ADP-Pi also has a straight conformation, in agreement with the crystal structure in ADP.Vi.
Table 1.
Conformations of myosins II, V, and VI at stages in their reaction schemes are compared
 |
The cartoon in Table 1 compares the conformations of myosin VI observed here and by crystallography with consensus structures observed for myosins II and V. In the presence of ATP, ATPγS, and ADP we observed similar overall shapes, with the lever arm extending straight from the motor domain. In myosin II in the presence of ATPγS, hydrolysis is the rate-limiting step (36) and M-ATPγS is the predominant intermediate. The same is expected with ATPγS and myosin VI and therefore, in the presence of either ATP or ATPγS, myosin VI would be expected to have primarily triphosphate in the active site and produce similar conformations to those shown in Table 1. The ATP and ADP states of myosins II and V have a similar straight shape to the apo states of these molecules; however, the crystal structure of myosin VI with ADP.BeF bound is similar to the strongly bent apo state. Therefore, in the presence of ATP or ATP-γS, myosin VI might also be predicted to have a bent shape like the apo state, rather than the straight one observed. The shape of myosin VI in ATP is therefore quite different from expectation based on crystal structures, assuming that each active site ligand produces a single conformation. Note also that Eq. 1 of Table 1 should not be interpreted as evidence the powerstroke is associated with ADP dissociation from myosin VI, as binding to actin is also likely to have a significant effect upon the equilibrium between the different conformers.
The straight shape of myosin VI in the presence of ADP is also unexpected, because myosins I, II, and V with ADP bound have shapes not greatly different from their apo states. There is no myosin VI crystal structure with ADP bound, but in cryo-EM three-dimensional reconstructions of actin filaments saturated with myosin VI heads in the presence of ADP, the lever arm was only 15–20° different to the angle without nucleotide (11), rather than the ∼140° difference seen here. Thus, in ADP, myosin VI might be expected to be strongly bent and not greatly different in shape from the apo state.
We considered the possibility that the ADP used in these experiments might not have been pure. ADP typically contains ∼1% ATP and AMP, but this should not have been a problem as the myosin will have hydrolyzed the small amount of ATP by the time the grids were made. The affinity for AMP is several orders-of-magnitude weaker than ADP. Thus, the molecules will have had ADP bound. We also checked the ATP-γ-S by HPLC and found it contained in addition 2% ATP, 5% ADP, and 1% AMP, so ATP-γS should have been bound in this case. Our observations in Fig. 2 that myosin VI fully decorated actin in the presence of ADP and was completely dissociated from actin by ATP and ATP-γS are further evidence that the intended ligands were bound to the active site.
Overall, the myosin VI microscopy is consistent with the crystal structures in showing a much larger lever arm swing than is observed in myosins II and V. However, the straight shapes observed in ATP, ATP-S, and ADP, similar to what is thought to be the prepowerstroke conformation, were unexpected. Factors that could have resulted in this lack of agreement between the crystallography and microscopy are:
-
1.
It is certainly conceivable that the class averages of the molecules in the presence of nucleotides are not representative. However, we think that this is unlikely, because the entire dataset contained nearly 200,000 particle images. Individual experiment datasets generally contained a minimum of 5000 particle images and in some cases >20,000, classified into 100 or more averages. As can be seen in full classifications in the Supporting Material, the great majority of classes with bound nucleotide were straight. We also cannot rule out that the conformation is perturbed by immobilization on the carbon substrate or exposure to uranyl acetate, but in other respects, the correspondence between images and the apo and ADP-BeF crystal structures is good.
-
2.
There is considerable evidence for multiple conformations of myosin with analogs of ATP and ADP-Pi. In myosin II with ADP and the phosphate analog BeF bound to the active site, the equilibrium constant is near one for the conformation transition between the bent and straight converter helix measured spectroscopically (37) and by the the order-to-disorder transition of heads observed by x-ray diffraction in muscle fibers (38). This explains the different head crystal structures (39,40). ADP-AlF4 and ADP-Vi increase the equilibrium constant in favor of the bent (postpowerstroke) helix by a factor of 2–4, but there are still significant concentrations of the straight shape in both cases (37). The fact that both conformations of myosin II heads have been found in crystal structures with ADP-BeF bound shows that there is not a one-to-one correlation between biochemical and structural states with nucleotide analogs in the active site. The native substrate, ATP, produces a stronger shift toward the ordered (bent) conformation than observed with other analogs in both muscle (41) and nonmuscle myosin II (42). Two conformations of myosin II are observed in the presence of ATP; however, in this case, there are also two states, M-ATP and M-ADP-Pi. Thus for myosin II, there is a close correlation between the proportions of the two biochemical states and the proportions of the two conformations (41), consistent with there being a predominant conformation associated with each biochemical state, but the possibility of multiple conformations in particular states cannot be ruled out for myosin VI.
-
3.
Single molecule EM may reveal conformations not seen by crystallography and is not subject to crystallization conditions or lattice forces. Thus, both EM and crystallography have potential for artifacts. Finally, it seems clear that the force producing mechanism of myosin VI is very different from the more extensively characterized myosins II and V, and so may behave in unexpected ways. At present then, the discrepancy between the microscopy and crystallography data remains unresolved.
Tail bending
The strongly bent shape in apo full-length myosin VI image averages has a further bend of ∼90° after the second (IQ) calmodulin. This region, which is not present in S1, appears to direct the tail across the N-terminal region of the motor domain. However, there is no large bend after the IQ calmodulin in the straight molecules seen in ATP, ATP-γS, and ADP. This appears to indicate a nucleotide-driven shape change in the tail. A possible explanation for this is that in the apo state there is an interaction between the C-terminal part of the tail and the motor domain that is abolished by nucleotide in the active site. Such an interaction could be part of a regulatory mechanism that inactivates myosin VI, folding the molecule in a way reminiscent of the inactive folded states of nonmuscle myosins II and V (43–45). Consistent with this, preliminary pull-down data indicates that the expressed tail binds to S1 (motor domain) in myosin VI (not shown). Such a regulatory mechanism could be important in controlling myosin VI in situ.
Tail structure
The distance between the motor domain and the end of the molecule in shadowed images was 29 nm, which is ∼3× longer than that seen in the stain image averages. The number of residues between the IQ motif and the start of the globular C-terminal cargo binding domain is ∼220. However, ∼80 of these fold in a three-helix bundle whose length is ∼4 nm. The three-helix bundle appears not to be very stable and may unfold if myosin VI is induced to dimerize (15). If the remaining 140 residues were entirely α-helical, they would span 21 nm. Because each IQ-calmodulin lengthens the lever by 4 nm, the overall length of the lever, three-helix bundle, and presumed helical tail (including SAH domain) could be expected to be ∼33 nm. This is slightly longer than that measured by shadowing and neglects the size of the cargo-binding domain, but it is much closer than the estimate from the stain image averages.
The negative stain technique has been successfully used across biology. Very few artifacts produced by the method have been reported, despite the high concentration of uranium ions and low pH it involves. We cannot rule out that the shadowed tails are lengthened by the combing force of the receding meniscus during drying (46), perhaps unfolding the three-helix bundle; however, this would probably also result in a similar alignment between molecules locally, which was not observed. Moreover, unfolding the three-helix bundle would only lengthen this region by 8 nm, which is much smaller then the amount observed. Our data therefore suggest that the myosin VI tail collapses in stain, resulting in an artificially shortened structure.
Conclusions
The resolution attained by negative staining and image averaging is approximately twofold better than the shadowing method that has been used for most microscopy of myosin molecules, including myosin VI. Although this is still ∼10× lower than achieved by crystallography, it is sufficient to allow detailed comparisons to be made with crystal structure shapes. The microscopy has the advantages that native ligands such as ATP can be added, whole molecules can easily be observed, and a range of conformations can be observed.
Our data support the idea that force production in myosin VI is significantly different from other myosin classes, both in the angular throw and direction of its lever and in the rearrangement of its converter subdomain. There is good agreement between the image averages and the crystal structure of the apo S1 molecule, both of which show a similar strongly bent lever arm emerging from the converter. Microscopy in the presence of ATP, ADP, and ATP-γS all show a straightened molecule similar to the crystal structure with ADP.VO4 bound. Differences from the expected conformations with different nucleotide and nucleotide analogs bound may be explained by multiple nucleotide conformations that are present with nucleotide analogs bound to the active site.
Acknowledgments
Supported by National Institutes of Health grant No. EB00209 and the Biotechnology and Biological Sciences Research Council (UK).
Supporting Material
References
- 1.Ménétrey J., Bahloul A., Houdusse A. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435:779–785. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ménétrey J., Llinas P., Houdusse A. The structural basis for the large powerstroke of myosin VI. Cell. 2007;131:300–308. doi: 10.1016/j.cell.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 3.Ménétrey J., Llinas P., Houdusse A. The post-rigor structure of myosin VI and implications for the recovery stroke. EMBO J. 2008;27:244–252. doi: 10.1038/sj.emboj.7601937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yildiz A., Park H., Sweeney H.L. Myosin VI steps via a hand-over-hand mechanism with its lever arm undergoing fluctuations when attached to actin. J. Biol. Chem. 2004;279:37223–37226. doi: 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
- 5.Bryant Z., Altman D., Spudich J.A. The power stroke of myosin VI and the basis of reverse directionality. Proc. Natl. Acad. Sci. USA. 2007;104:772–777. doi: 10.1073/pnas.0610144104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De La Cruz E.M., Ostap E.M., Sweeney H.L. Kinetic mechanism and regulation of myosin VI. J. Biol. Chem. 2001;276:32373–32381. doi: 10.1074/jbc.M104136200. [DOI] [PubMed] [Google Scholar]
- 7.Lister I., Schmitz S., Kendrick-Jones J. A monomeric myosin VI with a large working stroke. EMBO J. 2004;23:1729–1738. doi: 10.1038/sj.emboj.7600180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altman D., Sweeney H.L., Spudich J.A. The mechanism of myosin VI translocation and its load-induced anchoring. Cell. 2004;116:737–749. doi: 10.1016/s0092-8674(04)00211-9. [DOI] [PubMed] [Google Scholar]
- 9.Chevreux G., Potier N., Sweeney H.L. Electrospray ionization mass spectrometry studies of noncovalent myosin VI complexes reveal a new specific calmodulin binding site. J. Am. Soc. Mass Spectrom. 2005;16:1367–1376. doi: 10.1016/j.jasms.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 10.Molloy J.E., Burns J.E., White D.C. Movement and force produced by a single myosin head. Nature. 1995;378:209–212. doi: 10.1038/378209a0. [DOI] [PubMed] [Google Scholar]
- 11.Wells A.L., Lin A.W., Sweeney H.L. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 12.Phichith D., Travaglia M., Sweeney H.L. Cargo binding induces dimerization of myosin VI. Proc. Natl. Acad. Sci. USA. 2009;106:17320–17324. doi: 10.1073/pnas.0909748106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altman D., Goswami D., Mayor S. Precise positioning of myosin VI on endocytic vesicles in vivo. PLoS Biol. 2007;5:e210. doi: 10.1371/journal.pbio.0050210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knight P.J., Thirumurugan K., Peckham M. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J. Biol. Chem. 2005;280:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 15.Mukherjea M., Llinas P., Sweeney H.L. Myosin VI dimerization triggers an unfolding of a three-helix bundle in order to extend its reach. Mol. Cell. 2009;35:305–315. doi: 10.1016/j.molcel.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reifenberger J.G., Toprak E., Selvin P.R. Myosin VI undergoes a 180 degrees power stroke implying an uncoupling of the front lever arm. Proc. Natl. Acad. Sci. USA. 2009;106:18255–18260. doi: 10.1073/pnas.0900005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y.J., Schroeder H.W., 3rd, Goldman Y.E. Myosin VI walks “wiggly” on actin with large and variable tilting. Mol. Cell. 2007;28:954–964. doi: 10.1016/j.molcel.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohi M., Li Y., Walz T. Negative staining and image classification—powerful tools in modern electron microscopy. Biol. Proc. Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgess S.A., Walker M.L., Trinick J. Flexibility within myosin heads revealed by negative stain and single-particle analysis. J. Cell Biol. 1997;139:675–681. doi: 10.1083/jcb.139.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgess S., Walker M., Trinick J. The prepower stroke conformation of myosin V. J. Cell Biol. 2002;159:983–991. doi: 10.1083/jcb.200208172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oke O.A., Burgess S.A., Trinick J. Influence of lever structure on myosin 5a walking. Proc. Natl. Acad. Sci. USA. 2010;107:2509–2514. doi: 10.1073/pnas.0906907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker M.L., Burgess S.A., Knight P.J. Two-headed binding of a processive myosin to F-actin. Nature. 2000;405:804–807. doi: 10.1038/35015592. [DOI] [PubMed] [Google Scholar]
- 23.Song C.F., Kendrick-Jones J., Trinick J. Electron microscopy of negatively stained myosin VI molecules. Biophys. J. 2007:494A. [Google Scholar]
- 24.Buss F., Kendrick-Jones J., Paul Luzio J. The localization of myosin VI at the Golgi complex and leading edge of fibroblasts and its phosphorylation and recruitment into membrane ruffles of A431 cells after growth factor stimulation. J. Cell Biol. 1998;143:1535–1545. doi: 10.1083/jcb.143.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker M., Knight P., Trinick J. Negative staining of myosin molecules. J. Mol. Biol. 1985;184:535–542. doi: 10.1016/0022-2836(85)90300-6. [DOI] [PubMed] [Google Scholar]
- 26.Trinick J., Knight P., Whiting A. Purification and properties of native titin. J. Mol. Biol. 1984;180:331–356. doi: 10.1016/s0022-2836(84)80007-8. [DOI] [PubMed] [Google Scholar]
- 27.Frank J., Radermacher M., Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 28.Burgess S.A., Walker M.L., Knight P.J. Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J. Struct. Biol. 2004;147:247–258. doi: 10.1016/j.jsb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Terrak M., Rebowski G., Dominguez R. Structure of the light chain-binding domain of myosin V. Proc. Natl. Acad. Sci. USA. 2005;102:12718–12723. doi: 10.1073/pnas.0503899102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buss F., Kendrick-Jones J. How are the cellular functions of myosin VI regulated within the cell? Biochem. Biophys. Res. Commun. 2008;369:165–175. doi: 10.1016/j.bbrc.2007.11.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spudich G., Chibalina M.V., Kendrick-Jones J. Myosin VI targeting to clathrin-coated structures and dimerization is mediated by binding to Disabled-2 and PtdIns(4,5)P2. Nat. Cell Biol. 2007;9:176–183. doi: 10.1038/ncb1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwaki M., Tanaka H., Yanagida T. Cargo-binding makes a wild-type single-headed myosin-VI move processively. Biophys. J. 2006;90:3643–3652. doi: 10.1529/biophysj.105.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park H., Ramamurthy B., Sweeney H.L. Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol. Cell. 2006;21:331–336. doi: 10.1016/j.molcel.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 34.Holmes K.C., Angert I., Schröder R.R. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–427. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- 35.Bagshaw C.R., Trentham D.R. The reversibility of adenosine triphosphate cleavage by myosin. Biochem. J. 1973;133:323–328. doi: 10.1042/bj1330323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bagshaw C.R., Eccleston J.F., Goody R.S. Transient kinetic studies of Mg++-dependent ATPase of myosin and its proteolytic subfragments. Cold Spring Harb. Symp. Quant. Biol. 1973;37:127–135. [Google Scholar]
- 37.Agafonov R.V., Negrashov I.V., Nesmelov Y.E. Structural dynamics of the myosin relay helix by time-resolved EPR and FRET. Proc. Natl. Acad. Sci. USA. 2009;106:21625–21630. doi: 10.1073/pnas.0909757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu S., Offer G., Yu L.C. Temperature and ligand dependence of conformation and helical order in myosin filaments. Biochemistry. 2003;42:390–401. doi: 10.1021/bi026085t. [DOI] [PubMed] [Google Scholar]
- 39.Holmes K.C. Muscle Contraction. Novartis Found. Symp. 1998;213:89–92. [PubMed] [Google Scholar]
- 40.Fisher A.J., Smith C.A., Rayment I. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4- Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 41.Xu S., Gu J., Yu L.C. The M.ADP.Pi state is required for helical order in the thick filaments of skeletal muscle. Biophys. J. 1999;77:2665–2676. doi: 10.1016/s0006-3495(99)77101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Málnási-Csizmadia A., Pearson D.S., Bagshaw C.R. Kinetic resolution of a conformational transition and the ATP hydrolysis step using relaxation methods with a Dictyostelium myosin II mutant containing a single tryptophan residue. Biochemistry. 2001;40:12727–12737. doi: 10.1021/bi010963q. [DOI] [PubMed] [Google Scholar]
- 43.Craig R., Smith R., Kendrick-Jones J. Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature. 1983;302:436–439. doi: 10.1038/302436a0. [DOI] [PubMed] [Google Scholar]
- 44.Liu J., Taylor D.W., Taylor K.A. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature. 2006;442:208–211. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- 45.Thirumurugan K., Sakamoto T., Knight P.J. The cargo-binding domain regulates structure and activity of myosin 5. Nature. 2006;442:212–215. doi: 10.1038/nature04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tskhovrebova L., Trinick J. Flexibility and extensibility in the titin molecule: analysis of electron microscope data. J. Mol. Biol. 2001;310:755–771. doi: 10.1006/jmbi.2001.4700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


