Abstract
Background
Obesity is a strong risk factor for resistance to insulin-mediated glucose disposal, a precursor of type 2 diabetes and other disorders.
Objectives
To identify molecular pathways that may cause such obesity-associated insulin resistance in human subjects, we exploited the fact that not all obese individuals are prone to insulin resistance. Thus the degree of obesity as a variable was removed by studying morbidly obese human subjects of similar BMI values who are insulin-sensitive versus insulin-resistant.
Setting
University Medical Center, United States
Methods
Combining gene expression profiling with computational approaches, we determined the global gene expression signatures of omental and subcutaneous adipose tissue samples obtained from similarly obese patients undergoing gastric bypass surgery.
Results
Gene sets related to chemokine activity and chemokine receptor-binding were identified as most highly expressed in the omental tissue from insulin-resistant compared to insulin sensitive subjects, independent of BMI. These upregulated genes included chemokines CCL2, CCL3, CCL4, CCL18 and IL8/CXCL8, and were not differentially expressed in subcutaneous adipose tissues between the two groups of subjects. Strikingly, insulin resistance, but not BMI, was associated with increased macrophage infiltration in the omental adipose tissue, as was adipocyte size, in these morbidly obese subjects.
Conclusions
Our findings demonstrate that inflammation of omental adipose tissue is strongly associated with insulin resistance in human obesity even in subjects with similar BMI values. Increased omental fat mass may contribute to the amplified inflammatory response observed in this population.
Keywords: abdominal adipose tissue, insulin resistance, chemokines, inflammation, macrophages
Introduction
The prevalence and severity of obesity are increasing dramatically in the United States and throughout the world1. Obesity is closely associated with insulin resistance which is characterized by a decreased response to insulin signaling in several peripheral tissues including adipose, liver and muscle2. Interestingly, not all obese individuals are insulin-resistant3,4, highlighting the importance of identifying genetic and environmental factors that place obese individuals at the greatest risk for obesity-related complications. The role of adipose tissue in controlling whole-body metabolism is being increasingly recognized5. In addition to its role as an energy storage depot, the adipose tissue secretes numerous hormones and cytokines that function to regulate food intake and nutrient homeostasis6.
Obesity is associated with the activation of pro-inflammatory pathways and macrophage infiltration into adipose tissue, which ultimately impairs the energy storage capability and endocrine function of the adipose tissue7,8,9,10,11. Despite increasing awareness of the role inflamed adipose tissue plays in obesity related co-morbidities (i.e. insulin resistance, dyslipidemia, hypertension), there is limited understanding of the molecular signals that initiate the inflammatory process. To address this question, most gene expression profiling studies have examined differences in adipose tissue gene expression between obese and lean subjects and found that obesity is associated with an increase in the expression of proinflammatory genes12,13. These studies, however, cannot reveal genes responsible for the metabolically compromised status of some obese individuals compared to other obese individuals.
To identify genes potentially involved in mediating obesity-related insulin resistance, we determined the gene expression profiles of subcutaneous and visceral adipose tissue from obese insulin-resistant and obese insulin-sensitive adults that were matched for BMI. Assessing differences in adipose tissue gene expression between obese subjects who differ in insulin sensitivity, yet are matched for the extent of obesity has not been previously published. Our hypothesis was that candidate genes identified by differential expression in this screen would be directly related to insulin resistance and not simply a feature of the obese state. We found a set of parameters that were strikingly associated with omental adipose tissue of insulin resistant obese subjects compared to the BMI-matched insulin sensitive obese individuals: elevated expression of genes encoding chemokines, increased macrophage infiltration and increased adipocyte size. The data suggest that these changes may be primary rather than secondary to the occurrence of insulin resistance.
Materials and Methods
Patients
Adult patients undergoing a laparoscopic Roux-en-Y gastric bypass surgery between October 2005 and May 2009 were recruited from the University of Massachusetts Memorial Medical Center, a tertiary care academic hospital in a metropolitan setting in central New England. All patients were qualified for gastric bypass surgery under published NIH consensus guidelines for the treatment of morbidly obese patients14. Morbidly obese insulin-resistant patients were matched for body mass index (BMI) to morbidly obese insulin-sensitive patients. Demographic data including age, gender, height, weight, calculated BMI and fasting glucose and insulin levels were recorded at the time of gastric bypass surgery. A lipid profile (triglycerides, total cholesterol, low-density lipoprotein (LDL) cholesterol and high-density lipoprotein (HDL) cholesterol) was obtained prior to surgery. Patients with diabetes mellitus were excluded from this study because treatment of this metabolic disorder has been shown to alter the gene expression profile15. Adipose tissue samples were taken from lower abdominal wall (subcutaneous) and greater omentum (visceral). All subjects provided written informed consent before taking part in the study. The study was approved by the University of Massachusetts Medical School Institutional Review Board.
Insulin sensitivity
After an overnight fast, pre-operative serum levels of insulin and glucose were determined. A solid phase chemiluminescent immunoassay was used to determine serum insulin levels (IMMULITE 2000 Insulin), whereas levels of serum glucose were determined by an oxygen rate method employing a Beckman Oxygen electrode (SYNCHRON LX Systems). The homeostatic model of assessment (HOMA2-IR) was used to estimate insulin resistance (http://www.dtu.ox.ac.uk) by using fasting glucose (mg/dL) and fasting insulin levels (mIU/mL). A cut point of HOMA2-IR ≥ 2.3 was used to characterize subjects as being insulin resistant as validated by previous studies in this patient population16.
Microarray Study
Paired samples of omental and subcutaneous adipose were obtained from fasting patients during gastric bypass surgery. Whole adipose tissue was dissected to remove fibrotic tissue and obvious vasculature, and frozen in 0.5 cm3 sections in liquid nitrogen. Total RNA was isolated from frozen human tissues by homogenizing the tissue in TRIzol (Invitrogen, Carlsbad, CA). RNA quality was assessed using a denaturing agarose gel. Total RNA was purified using RNeasy Mini-Kit (Qiagen, Valencia, CA). Ten micrograms of purified total RNA was prepared according to Affymetrix protocols. Prepared cRNA was hybridized to Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays.
GeneChip Expression Array Analysis for individual genes was performed as previously published17 filtering for p-value <0.05 and fold change ≥ 2. This method was used to rank the genes expressed differentially between insulin sensitive and insulin resistant patients in both adipose depots. The gene lists that were obtained using the filtering parameters mentioned above were entered into the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics database (http://david.abcc.ncifcrf.gov/home.jsp)18, 19 to determine which molecular functions were most represented in the top-scoring genes using information obtained from the Gene Ontology Consortium (http://www.geneontology.org). The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE20950.
Real-time PCR
RNA isolated from the visceral adipose tissue from the original cohort of twenty patients was analyzed to confirm the microarray analysis findings. cDNA was synthesized using iScript cDNA Synthesis Kit (BioRad, Hercules, CA). Quantitative real-time PCR for specific genes was performed using SybrGreen normalized to beta 2 microglobulin. Primers were designed using PrimerBank which is a public resource for PCR primers using a primer design algorithm that has been tested by real-time PCR experiments for PCR specificity and efficiency (pga.mgh.harvard.edu/primerbank) 20. Melting curve data was collected to visualize the presence of primer dimers. We used the following primers for RT-PCR: CCL2-sense, 5′-CCC CAG TCA CCT GCT GTT AT-3′; CCL2-antisense, 5′-TGG AAT CCT GAA CCC ACT TC-3′; IL8-sense, 5′-ACT GAG AGT GAT TGA GAG TGG AC-3′; IL8-antisense, 5′-AAC CCT CTG CAC CCA GTT TTC-3′; CCL3-sense, 5′-AGT TCT CTG CAT CAC TTG CTG-3′; CCL3-antisense, 5′-CGG CTT CGC TTG GTT AGG AA-3′; CCL4-sense, 5′-CTG TGC TGA TCC CAG TGA ATC-3′; CCL4-antisense, 5′TCA GTT CAG TTC CAG GTC ATA CA-3′; CCL18-sense, 5′-GGG GGC TGG TTT CAG AAT A-3′, CCL18-antisense, 5′-CTC CTT GTC CTC GTC TGC AC-3′; Beta2M-sense, 5′-GGC TAT CCA GCG TAC TCC AAA-3′; Beta2M-antisense, 5′-CGG CAG GCA TAC TCA TCT TTT T-3′. Expression of specific mRNAs was quantified in duplicate samples on an iCycler IQ Real-Time PCR detection system (Bio-Rad Laboratories) using the ΔΔCT method.
Adipose tissue histology
Tissues were fixed in 10% buffered formalin and embedded in paraffin from which multiple 5-mm sections were obtained. Images were acquired using an Axioskop2 Plus (Zeiss, Göttingen, Germany) coupled to a Spot RTKe (Diagnostic Instruments, Inc) and were analyzed using SPOT 4.0.2 software. For each subject, adipocyte cell diameter was calculated from the perimeter measurement (Adobe Photoshop CS4 Extended) and averaged using 100 cells. Immunohistochemical detection of CD68 (Santa Cruz Biotechnologies, Heidelberg, Germany) diluted 1:50 was performed with the avidinbiotin peroxidase method. Adipocytes and CD68+ cells were counted at 10x magnification in ten different randomly chosen areas in each processed slide.
Statistical analysis
Baseline differences between insulin sensitive and insulin resistant subjects were evaluated using the Student's t test for continuous variables and Fisher's exact test for categorical variables. For genes analyzed by real time-PCR, significant differences between insulin sensitive and insulin resistant groups were determined by Student's t test. The results were considered significant at P ≤ 0.05. The correlations between subject clinical data (HOMA2-IR or BMI) and characteristics of adipose histology (macrophage infiltration or adipocyte diameter) were analyzed by Pearson's coefficient.
Results
Clinical and metabolic characteristics of the study population
Adipose tissue samples from 20 human subjects were used in this study. Fourteen of the twenty subjects were female and the mean age of the cohort was 42 years (Table 1). Using HOMA2-IR analysis, we divided subjects into two groups: insulin sensitive (n=10) and insulin resistant (n=10). Insulin resistant subjects showed significantly elevated HOMA2-IR as compared to insulin sensitive subjects (see Methods). In addition, both fasting insulin and fasting glucose levels tended to be higher in insulin resistant subjects. There was no statistically significant difference in the components of the lipid profile between the two groups, although the insulin resistant subjects tended to have higher triglyceride values.
Table 1.
Clinical and Biochemical Characteristics of the Study Participants*
| Insulin Sensitive HOMA < 2.4 (N= 10) |
Insulin Resistant HOMA ≥ 2.4 (N= 10) |
P Value† | |
|---|---|---|---|
| Age - yr | 0.2 | ||
| Mean | 39 ± 6 | 43 ± 9 | |
| Range | 31-54 | 33-57 | |
| Female sex - no. (%) | 8 (80) | 6 (60) | 0.4‡ |
| Weight (kg) | 0.2 | ||
| Mean | 137 ± 11 | 149 ± 18 | |
| Range | 119-177 | 120-188 | |
| BMI§ | 0.7 | ||
| Mean | 48 ± 3 | 49 ± 7 | |
| Range | 46-55 | 39-60 | |
| Total cholesterol (mg/dL)** | 0.9 | ||
| Mean | 176 ± 27 | 179 ± 50 | |
| Range | 115-211 | 111-276 | |
| LDL cholesterol (mg/dL)** | 0.6 | ||
| Mean | 109 ± 15 | 101 ± 45 | |
| Range | 76-127 | 45-190 | |
| HDL cholesterol (mg/dL)** | |||
| Mean | 44 ± 5 | 42 ± 9 | 0.5 |
| Range | 38-52 | 30-56 | |
| Triglycerides (mg/dL)** | 0.2 | ||
| Mean | 129 ± 33 | 166 ± 74 | |
| Range | 94-188 | 74-274 | |
| Lipid lowering therapy - no. (%) | 1 (10) | 3 (30) | 0.3‡ |
| Fasting glucose (mg/dL) | 0.02 | ||
| Mean | 88 ± 9 | 101 ± 12 | |
| Range | 75-100 | 82-127 | |
| Fasting insulin (mIU/mL) | <0.001 | ||
| Mean | 9 ± 4 | 24 ± 7 | |
| Range | 4-14 | 16-36 | |
| HOMA2 | <0.001 | ||
| Mean | 1.3 ± 0.5 | 3.6 ± 5.3 | |
| Range | 0.6-2.1 | 2.4-6.7 |
Plus-minus values are mean ± SD.
P values were estimated using Student's t-test unless otherwise specified.
P-value was estimated using Fisher's exact test.
The body-mass-index (BMI) is the weight in kilograms divided by the square of the height in meters.
Data was not available for three patients (1 IS, 2 IR)
Global gene expression profiling of adipose tissue identifies active chemokine signaling in obese insulin resistant patients
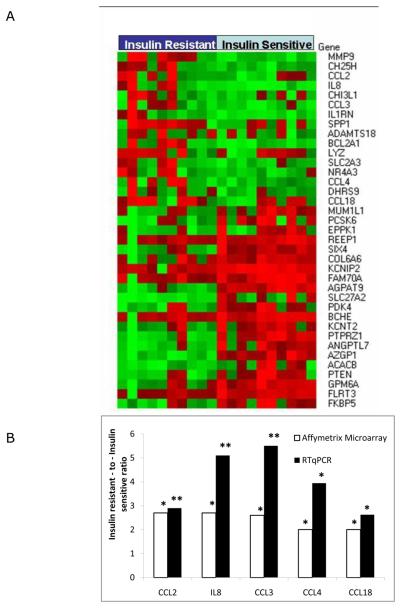
In order to identify genes related to insulin resistance rather than obesity per se, we first utilized cDNA microarrays to determine the gene expression profiles of 39 paired subcutaneous and omental samples from the 20 BMI-matched subjects (one insulin resistant subject did not have a subcutaneous sample available for microarray analysis). Using GeneChip Expression Array Analysis and filtering parameters p-value < 0.05 and fold change ≥ 2, we identified 37 genes differentially expressed in omental tissue of insulin resistant subjects as compared to insulin sensitive subjects, and three genes differentially expressed in subcutaneous tissue between the two subject groups (Table 2). In order to elucidate the molecular functions differentially regulated in the omental adipose tissue of our subjects, we entered the gene list into the DAVID bioinformatics database and identified 2 Gene Ontology Molecular Function gene sets as being differentially expressed between the insulin resistant and insulin sensitive subjects (Table 3). The gene sets representing chemokine activity and chemokine receptor binding were upregulated in omental tissue of insulin resistant subjects and reached statistical significance using the Bonferroni correction to address the problem of multiple comparisons. Both gene sets included the genes chemokine (C-C motif) ligand 18 (CCL18), chemokine (C-C motif) ligand 2 (CCL2), chemokine (C-C motif) ligand 3 (CCL3), chemokine (C-C motif) ligand 4 (CCL4) and interleukin 8 (IL8). These five genes were studied with quantitative real-time PCR analysis which revealed that the direction and magnitude of the differential expression in omental tissue was consistent with the data provided by microarray analysis, thereby confirming the computational approach (Fig 1B). These trends held up when the data were re-analyzed using just the female patients, confirming that the observed differences were not due to gender bias (Supplementary Figure 1).
Table 2.
Individual genes differentially expressed in adipose tissue from insulin resistant compared to insulin sensitive obese patients.
| Probe ID | Gene name | Symbol | FC | P value |
|---|---|---|---|---|
| OMENTAL DEPOT | ||||
| 203936_s_at | matrix metallopeptidase 9 | MMP9 | 2.9 | 0.001 |
| 206932_at | cholesterol 25-hydroxylase | CH25H | 2.8 | 0.014 |
| 216598_s_at | chemokine (C-C motif) ligand 2 | CCL2 | 2.7 | 0.013 |
| 202859_x_at | interleukin 8 | IL8 | 2.7 | 0.023 |
| 209395_at | chitinase 3-like 1 (cartilage glycoprotein-39) | CHI3L1 | 2.7 | 0.007 |
| 205114_s_at | chemokine (C-C motif) ligand 3 | CCL3 | 2.6 | 0.029 |
| 212657_s_at | interleukin 1 receptor antagonist | IL1RN | 2.4 | 0.005 |
| 209875_s_at | secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T- lymphocyte activation 1) |
SPP1 | 2.2 | 0.044 |
| 230040_at | ADAM metallopeptidase with thrombospondin type 1 motif, 18 | ADAMTS 18 |
2.2 | 0.044 |
| 205681 at | BCL2-related protein A1 | BCL2A1 | 2.1 | 0.004 |
| 213975 s at | lysozyme (renal amyloidosis) | LYZ | 2.1 | 0.016 |
| 202499_s_at | solute carrier family 2 (facilitated glucose transporter), member 3 | SLC2A3 | 2.1 | 0.021 |
| 209959_at | nuclear receptor subfamily 4, group A, member 3 | NR4A3 | 2.1 | 0.024 |
| 204103 at | chemokine (C-C motif) ligand 4 | CCL4 | 2.0 | 0.044 |
| 224009_x_at | dehydrogenase/reductase (SDR family) member 9 | DHRS9 | 2.0 | 0.005 |
| 32128 at | chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) | CCL18 | 2.0 | 0.043 |
| 229160 at | melanoma associated antigen (mutated) 1-like 1 | MUM1L1 | −2.0 | 0.019 |
| 242662 at | Proprotein convertase subtilisin/kexin type 6 | PCSK6 | −2.0 | 0.044 |
| 232164 s at | epiplakin 1 | EPPK1 | −2.0 | 0.024 |
| 204364 s at | receptor accessory protein 1 | REEP1 | −2.0 | 0.042 |
| 229796 at | SIX homeobox 4 | SIX4 | −2.0 | 0.002 |
| 230867 at | collagen type VI alpha 6 | COL6A6 | −2.0 | 0.015 |
| 1555230 a a | tKv channel interacting protein 2 | KCNIP2 | −2.0 | 0.015 |
| 219895 at | family with sequence similarity 70, member A | FAM70A | −2.0 | 0.034 |
| 224480_s_at | 1-acylglycerol-3-phosphate O-acyltransferase 9 | AGPAT9 | −2.1 | 0.000 |
| 205769_at | solute carrier family 27 (fatty acid transporter), member 2 | SLC27A2 | −2.1 | 0.013 |
| 205960 at | pyruvate dehydrogenase kinase, isozyme 4 | PDK4 | −2.2 | 0.025 |
| 205433 at | butyrylcholinesterase | BCHE | −2.2 | 0.046 |
| 234103 at | Potassium channel, subfamily T, member 2 | KCNT2 | −2.2 | 0.006 |
| 204469_at | protein tyrosine phosphatase, receptor-type, Z polypeptide 1 | PTPRZ1 | −2.3 | 0.001 |
| 206423_at | angiopoietin-like 7 | ANGPTL7 | −2.3 | 0.003 |
| 209309 at | alpha-2-glycoprotein 1, zinc-binding | AZGP1 | −2.3 | 0.000 |
| 214584 x at | acetyl-Coenzyme A carboxylase beta | ACACB | −2.4 | 0.001 |
| 233314_at | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) |
PTEN | −2.4 | 0.019 |
| 209470 s at | glycoprotein M6A | GPM6A | −2.4 | 0.002 |
| 222853_at | fibronectin leucine rich transmembrane protein 3 | FLRT3 | −2.5 | 0.019 |
| 204560 at | FK506 binding protein 5 | FKBP5 | −2.8 | 0.010 |
| SUBCUTANEOUS DEPOT | ||||
| 214587 at | collagen, type VIII, alpha 1 | COL8A1 | −2.0 | 0.031 |
| 230867 at | collagen type VI alpha 6 | COL6A6 | −2.1 | 0.024 |
| 1554062 at | Xg blood group | XG | −2.2 | 0.045 |
Analysis was performed from Affymetrix GeneChip data using Microarray Computational Enviroment 2.0 developed by the Diabetes and Endocrinology Research Center at the University of Massachusetts Medical School. Shown are all named genes significantly upregulated 2 fold or more. Fold change indicates the difference in expression in insulin resistant as compared to insulin sensitive obese subjects. The P values were not adjusted for multiple testing. P < 0.05 was considered statistically significant. Genes with an Absent call were excluded.
Table 3.
Gene Ontology Molecular Function Terms Enriched in Omental Adipose Tissue from Insulin Resistant Compared to Insulin Sensitive Obese Patients.*
| Term | Gene Count | % | P value | Bonferroni |
|---|---|---|---|---|
| chemokine activity | 5 | 13.5 | 5.3E-06 | 2.2E-04 |
| chemokine receptor binding | 5 | 13.5 | 5.8E-06 | 2.4E-04 |
Analysis was performed from Affymetrix GeneChip data using Database for Annotation Visualization and Integrated Discovery 6.7 available on the National Institute of Allergy and Infectious Diseases, NIH website. Shown are all molecular function gene terms with a significant enrichment. The P value was adjusted for multiple testing. Bonferroni P < 0.05 was considered statistically significant.
Figure 1.
Expression of inflammation-related genes in omental adipose tissue from insulin resistant and insulin sensitive obese human subjects using microarray and RTqPCR. A. Heatmap representing normalized expression of genes identified by Microarray Computational Environment (MACE) as being significantly increased in omental adipose tissue from insulin resistant subjects and insulin sensitive human subjects. The genes are listed on the right. Expression levels above the mean for the gene are shown in red and expression levels below the mean for the gene are shown in green. B. Fold change in mRNA level of genes in omental adipose tissue of obese, insulin-resistant subjects (n=10) relative to obese, insulin-sensitive subjects (n=10) based on microarray data (white bars) and quantitative real-time analysis (black bars) performed by RTqPCR. ** P < 0.01; * P < 0.05.
Adipose tissue macrophage infiltration and histology
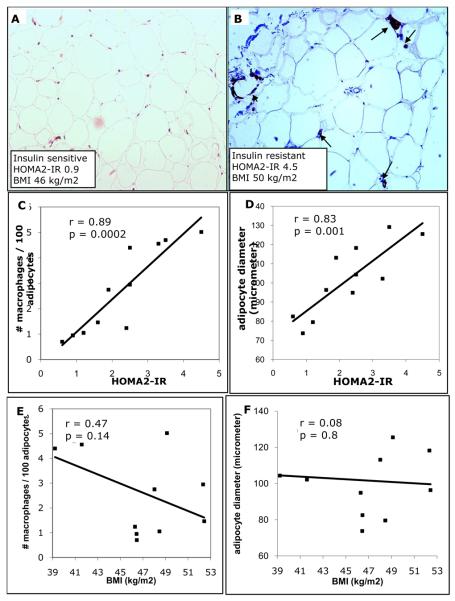
To determine whether the differences in chemokine expression with insulin resistance were related to changes in adipose tissue composition, we performed an immunohistochemical analysis of omental adipose tissue samples from eleven patients which comprised a subset of the original cohort of twenty patients. We observed strikingly larger adipocyte size and increased macrophage infiltration associated with insulin resistance (Fig 2A,B). There was a significant correlation between HOMA2-IR and macrophage infiltration (Fig 2C), yet, correlation analysis of CD68 staining with BMI in the morbidly obese range showed no association (Fig 2E). Adipocyte diameter was inversely correlated with HOMA2-IR, but not with BMI in our obese subjects (Fig 2D,F). Thus, the striking morphological appearance and cellular composition of the adipose tissue relates significantly to insulin resistance but not the obese state per se in this cohort.
Figure 2.
Relationship between macrophage infiltration, adipocyte diameter and HOMA2-IR in omental adipose tissue. Immunohistochemical detection of CD68+ macrophages performed on omental adipose tissue samples obtained from obese human subjects undergoing gastric bypass surgery. A-B. CD68 staining of omental adipose tissue from a representative insulin sensitive (A) and insulin resistant (B) subject. Unlike in adipose tissue from insulin sensitive subject (A) macrophages are observed throughout the tissue (arrows) and arranged in crown-like structures (arrowhead) in the insulin resistant subject (B). Sections taken at 20x magnification. CF, Quantitative analysis of adipocyte diameter and macrophage infiltration in adipose tissue of insulin resistant and insulin sensitive obese human subjects. Adipocyte diameter was calculated from the perimeter measurement of 100 cells. Insulin sensitivity, as determined by HOMA2-IR, correlates with CD68+ macrophage infiltration (C) and adipocyte diameter (D). Body mass index (BMI) shows no correlation with macrophage infiltration (E) or adipocyte diameter (F). Data is an average of 10 histological fields using 10X objective (C or E).
Discussion
In this study we aimed to identify molecular pathways in adipose tissue related directly to insulin resistance in obese individuals. By combining expression profiling with computational approaches, we found that elevated chemokine and chemokine receptor activity distinguishes the omental adipose tissue of insulin resistant obese patients from omental adipose of insulin sensitive obese individuals. In addition, our histological analysis determines that increased adipocyte size and increased macrophage infiltration of the adipose tissue is associated with insulin resistance and not simply BMI in morbidly obese human subjects.
Both human and animal studies demonstrate that hypertrophied adipocytes secrete chemokine (C-C motif) ligand-2 (CCL2), which is a chemoattractant that increases macrophage infiltration7,11,21. As such, CCL2 is thought to be the primary chemokine secreted from adipose tissue in the obese state, and responsible for adipose inflammation leading to subsequent metabolic alterations12. However, CCL2-deficient mice do not have diminished macrophage infiltration or improved glucose metabolism compared to control mice22, suggesting that other chemokines may play critical a role in the adipose inflammation associated with obesity. The increased expression of five chemokines in the visceral adipose tissue of our insulin resistant cohort supports this theory, which has been previously demonstrated in studies of obese subjects with lean controls23. Of note, CCL2 did not have the highest fold change of the chemokines we identified as being upregulated in insulin resistance. Interleukin 8/CXCL8, another chemokine identified in our insulin resistant sample, is differentially secreted by visceral adipose tissue and implicated in coronary heart disease24. Recently, CD11c+CD206+ adipose tissue macrophages obtained from human donor subjects were shown to display high secretion of IL825. Taken together with our findings, we suggest IL-8 may be a determinant of obesity-related sequelae.
Enlarged adipocyte size is associated with insulin resistance26 and increased adipokine production and secretion27,28. In our study population, insulin resistance was associated with an increase in adipocyte size. Chemokines, secreted by hypertrophied adipocytes, attract macrophages that can be seen dispersed throughout the tissue, or clustered around adipocytes in “crown like structures” and secrete cytokines that contribute to the low-grade inflammation seen in obesity. Omental fat appears to have more macrophage infiltration than subcutaneous fat, a finding that is exaggerated by central obesity29. Our data agree with this relationship between insulin resistance, adipocyte size and macrophage infiltration and support the hypothesis that this relationship is not a function of overall weight, but perhaps related to one or more other unknown factors including adipose distribution or increased omental fat mass.
Our data are consistent with previous analyses using large-scale gene expression studies performed in the subcutaneous and visceral adipose tissues of lean and obese subjects. Arner, et al showed that obesity was associated with pathways involved in immune and defense responses12. Dolinkova, et al reported an increase in expression of proinflammatory and adipogenic genes in obese compared to lean women13. However, in both studies, limited information was provided regarding insulin sensitivity of the participants, so the obese group may be heterogeneous with respect to obesity related co-morbidities. Maclaren, et al showed that changes in expression of genes encoding proteins in the insulin signaling pathway were greater in omental than in subcutaneous tissue, and were related to insulin resistance rather than obesity30.
The strengths of our study consist of the inclusion of BMI-matched obese subjects with a range of insulin sensitivity and the use of microarray data, which allows for the unbiased analysis of the expression of multiple genes in different pathways. The use of BMI-matched subjects reduces the influence of obesity itself as a variable in correlating insulin resistance with altered gene expression. One may argue that the measurement of BMI may not accurately reflect the degree of body fat content and may be strengthened by more objective data on body fat composition or waist circumference. Recent evidence31 suggests that waist circumference is not a useful measurement in severe obesity (BMI ≥ 40), so an abdominal MRI would be the best way to quantify visceral adipose tissue in this population. Acquisition of this data may have shown that our results are secondary to increased omental fat mass in the insulin resistant cohort. Due to our limited group of surgical patients we included both female and male subjects in both the insulin resistant and insulin sensitive cohorts. Yet, the enrichment of genes encoding chemokines in the omental adipose tissue of insulin resistant patients was observed even when we limited our analysis to the female patients (Figure S1), making it unlikely that our observed differences were due to gender bias. Our data are consistent with a growing body of evidence suggesting that adipose inflammation plays a role in mediating the insulin resistance itself, as it is manifest in obesity. Intact adipose tissue is comprised of a pool of heterogeneous cells (i.e. adipocytes, preadipocytes, macrophages, and endothelial cells) and future studies using isolated cell fractions will be useful in determining which cell type is the primary source of the inflammatory signals.
Conclusions
We show that omental adipose chemokine expression and macrophage infiltration is associated with insulin resistance, independent of obesity in a morbidly obese population. This study raises the important question: why do some obese patients display such inflammation in their omental adipose tissue, while other equally obese patients do not? Pharmaceutical agents aimed at interrupting the chemokine pathway may be able to diminish the metabolic sequelae associated with obesity.
Supplementary Material
Expression of inflammation-related genes in omental adipose tissue from insulin resistant and insulin sensitive female obese human subjects using microarray and RTqPCR. Fold change in mRNA level of genes in omental adipose tissue of female obese, insulin-resistant subjects (n=6) relative to female obese, insulin-sensitive subjects (n=8) based on microarray data (white bars) and quantitative real-time analysis (black bars) performed by RTqPCR. * P < 0.05.
Acknowledgments
This work was supported in part by the National Institute of Diabetes and Digestive and Kidney Diseases Grant DK30898 (MPC), Worcester Foundation for Biomedical Research Grant Award (RAP), and the UMass Center for Clinical and Translational Science (RAP, OTH). Core resources (Bioinformatics, Genomics and Morphology) and a Pilot Grant supported by the Diabetes Endocrinology Research Center grant DK32520 were also used.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: All authors have read and approved the manuscript and none of them have any potential conflicts of interest to report
References
- 1.Flegal KM, Carroll MD, Ogden CL, Johnson CL. Prevalence and trends in obesity among US adults, 1999-2000. Jama. 2002;288:1723–1727. doi: 10.1001/jama.288.14.1723. [DOI] [PubMed] [Google Scholar]
- 2.Liang CP, Han S, Senokuchi T, Tall AR. The macrophage at the crossroads of insulin resistance and atherosclerosis. Circ Res. 2007;100:1546–1555. doi: 10.1161/CIRCRESAHA.107.152165. [DOI] [PubMed] [Google Scholar]
- 3.Reaven G. All obese individuals are not created equal: insulin resistance is the major determinant of cardiovascular disease in overweight/obese individuals. Diab Vasc Dis Res. 2005;2:105–112. doi: 10.3132/dvdr.2005.017. [DOI] [PubMed] [Google Scholar]
- 4.Sims EA. Are there persons who are obese, but metabolically healthy? Metabolism. 2001;50:1499–1504. doi: 10.1053/meta.2001.27213. [DOI] [PubMed] [Google Scholar]
- 5.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sartipy P, Loskutoff DJ. Monocyte chemoattractant protein 1 in obesity and insulin resistance. Proc Natl Acad Sci U S A. 2003;100:7265–7270. doi: 10.1073/pnas.1133870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shoelson SE, Herrero L, Naaz A. Obesity, inflammation, and insulin resistance. Gastroenterology. 2007;132:2169–2180. doi: 10.1053/j.gastro.2007.03.059. [DOI] [PubMed] [Google Scholar]
- 9.Weisberg SP, McCann D, Desai M, et al. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wellen KE, Hotamisligil GS. Obesity-induced inflammatory changes in adipose tissue. J Clin Invest. 2003;112:1785–1788. doi: 10.1172/JCI20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu H, Barnes GT, Yang Q, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahlman I, Kaaman M, Olsson T, et al. A unique role of monocyte chemoattractant protein 1 among chemokines in adipose tissue of obese subjects. J Clin Endocrinol Metab. 2005;90:5834–5840. doi: 10.1210/jc.2005-0369. [DOI] [PubMed] [Google Scholar]
- 13.Dolinkova M, Dostalova I, Lacinova Z, et al. The endocrine profile of subcutaneous and visceral adipose tissue of obese patients. Mol Cell Endocrinol. 2008;291:63–70. doi: 10.1016/j.mce.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 14.NIH conference Gastrointestinal surgery for severe obesity. Consensus Development Conference Panel. Ann Intern Med. 1991;115:956–961. [PubMed] [Google Scholar]
- 15.Di Gregorio GB, Yao-Borengasser A, Rasouli N, et al. Expression of CD68 and macrophage chemoattractant protein-1 genes in human adipose and muscle tissues: association with cytokine expression, insulin resistance, and reduction by pioglitazone. Diabetes. 2005;54:2305–2313. doi: 10.2337/diabetes.54.8.2305. [DOI] [PubMed] [Google Scholar]
- 16.Perugini RA, Quarfordt SH, Baker S, Czerniach DR, Litwin DE, Kelly JJ. Metabolic characterization of nondiabetic severely obese patients undergoing Roux-en-Y gastric bypass: preoperative classification predicts the effects of gastric bypass on insulin-glucose homeostasis. J Gastrointest Surg. 2007;11:1083–1090. doi: 10.1007/s11605-007-0158-3. [DOI] [PubMed] [Google Scholar]
- 17.Tang X, Guilherme A, Chakladar A, et al. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPARgamma, adipogenesis, and insulin-responsive hexose transport. Proc Natl Acad Sci U S A. 2006;103:2087–2092. doi: 10.1073/pnas.0507660103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 20.Xiaowei W, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Research. 2003;31(24):e154. doi: 10.1093/nar/gng154. 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curat CA, Miranville A, Sengenes C, et al. From blood monocytes to adipose tissue-resident macrophages: induction of diapedesis by human mature adipocytes. Diabetes. 2004;53:1285–1292. doi: 10.2337/diabetes.53.5.1285. [DOI] [PubMed] [Google Scholar]
- 22.Kirk EA, Sagawa ZK, McDonald TO, O'Brien KD, Heinecke JW. Monocyte chemoattractant protein deficiency fails to restrain macrophage infiltration into adipose tissue [corrected] Diabetes. 2008;57:1254–1261. doi: 10.2337/db07-1061. [DOI] [PubMed] [Google Scholar]
- 23.Huber J, Kiefer FW, Zeyda M, et al. CC chemokine and CC chemokine receptor profiles in visceral and subcutaneous adipose tissue are altered in human obesity. J Clin Endocrinol Metab. 2008;93:3215–3221. doi: 10.1210/jc.2007-2630. [DOI] [PubMed] [Google Scholar]
- 24.Bruun JM, Lihn AS, Madan AK, et al. Higher production of IL-8 in visceral vs. subcutaneous adipose tissue. Implication of nonadipose cells in adipose tissue. Am J Physiol Endocrinol Metab. 2004;286:E8–13. doi: 10.1152/ajpendo.00269.2003. [DOI] [PubMed] [Google Scholar]
- 25.Wentworth JM, Naselli G, Brown WA, Doyle L, Phipson B, Smyth GK, Wabitsch M, O'Brien PE, Harrison LE. Pro-inflammatory CD11c+CD206+ adipose tissue macrophages are associated with insulin resistance in human obesity. Diabetes. 2010 doi: 10.2337/db09-0287. Epub March 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weyer C, Foley JE, Bogardus C, Tataranni PA, Pratley RE. Enlarged subcutaneous abdominal adipocyte size, but not obesity itself, predicts type II diabetes independent of insulin resistance. Diabetologia. 2000;43:1498–1506. doi: 10.1007/s001250051560. [DOI] [PubMed] [Google Scholar]
- 27.Skurk T, Alberti-Huber C, Herder C, Hauner H. Relationship between adipocyte size and adipokine expression and secretion. J Clin Endocrinol Metab. 2007;92:1023–1033. doi: 10.1210/jc.2006-1055. [DOI] [PubMed] [Google Scholar]
- 28.Winkler G, Kiss S, Keszthelyi L, et al. Expression of tumor necrosis factor (TNF)-alpha protein in the subcutaneous and visceral adipose tissue in correlation with adipocyte cell volume, serum TNF-alpha, soluble serum TNF-receptor-2 concentrations and C-peptide level. Eur J Endocrinol. 2003;149:129–135. doi: 10.1530/eje.0.1490129. [DOI] [PubMed] [Google Scholar]
- 29.Harman-Boehm I, Bluher M, Redel H, et al. Macrophage infiltration into omental versus subcutaneous fat across different populations: effect of regional adiposity and the comorbidities of obesity. J Clin Endocrinol Metab. 2007;92:2240–2247. doi: 10.1210/jc.2006-1811. [DOI] [PubMed] [Google Scholar]
- 30.MacLaren R, Cui W, Simard S, Cianflone K. Influence of obesity and insulin sensitivity on insulin signaling genes in human omental and subcutaneous adipose tissue. J Lipid Res. 2008;49:308–323. doi: 10.1194/jlr.M700199-JLR200. [DOI] [PubMed] [Google Scholar]
- 31.Drapeau V, Lemieux I, Richard D, et al. Waist circumference is useless to assess the prevalence of metabolic abnormalities in severely obese women. Obesity Surgery. 2007;17:905–909. doi: 10.1007/s11695-007-9168-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of inflammation-related genes in omental adipose tissue from insulin resistant and insulin sensitive female obese human subjects using microarray and RTqPCR. Fold change in mRNA level of genes in omental adipose tissue of female obese, insulin-resistant subjects (n=6) relative to female obese, insulin-sensitive subjects (n=8) based on microarray data (white bars) and quantitative real-time analysis (black bars) performed by RTqPCR. * P < 0.05.