Abstract
The Zebrafish Information Network, ZFIN, is a WWW community resource of zebrafish genetic, genomic and developmental research information (http://zfin.org). ZFIN provides an anatomical atlas and dictionary, developmental staging criteria, research methods, pathology information and a link to the ZFIN relational database (http://zfin.org/ZFIN/). The database, built on a relational, object-oriented model, provides integrated information about mutants, genes, genetic markers, mapping panels, publications and contact information for the zebrafish research community. The database is populated with curated published data, user submitted data and large dataset uploads. A broad range of data types including text, images, graphical representations and genetic maps supports the data. ZFIN incorporates links to other genomic resources that provide sequence and ortholog data. Zebrafish nomenclature guidelines and an automated registration mechanism for new names are provided. Extensive usability testing has resulted in an easy to learn and use forms interface with complex searching capabilities.
INTRODUCTION
The zebrafish has recently emerged as an important model organism for studies of vertebrate genetics, genomics and development. Many features of the zebrafish make it desirable for genetic and developmental studies. The embryo is transparent and contains many identifiable cells and detailed anatomical, physiological and developmental properties are known for some of these cells. Individual cells can be transplanted and followed over time. Gene expression can be monitored and altered in individual cells. In addition, efficient methods for generating, isolating and characterizing zebrafish mutations are available (1–3). It is possible to generate haploid progeny that are viable for up to 5 days. Homozygous diploid progeny that carry only maternal or paternal genes can also be generated. Mutations have been isolated affecting a wide range of genes including genes that regulate developmental patterning, organogenesis, physiology and behavior. Given that many fundamental molecular mechanisms appear to be conserved among vertebrates groups (4–7), zebrafish mutations will likely provide insights into gene functions in other vertebrates including humans.
The number of laboratories participating in zebrafish research and the data generated by these laboratories are growing at a phenomenal rate. ZFIN has been mandated to serve as the central location for the integration of this rapidly expanding set of data. The goals of ZFIN are to (i) establish a database system for genetic, genomic and developmental zebrafish data, (ii) provide meaningful ways of relating genes to mutations to facilitate the understanding of gene regulation and function, (iii) integrate with other species databases to allow comparisons of syntenic relationships, mutant phenotypes and gene expression, and (iv) integrate information from diverse laboratories in a timely fashion and make it easily accessible to everyone in a manner that promotes scientific discovery.
Both biologists and computer scientists are involved in the development and curation of ZFIN. The usefulness and usability of a database depend upon careful assessment of the requirements, detailed testing of prototypes by real users, subsequent analysis of users’ interactive behavior while using the database and data integrity maintained through proper database implementation and professional data curation. ZFIN has been developed to meet these goals by using user-centered (8) and participatory design (9) techniques.
SCOPE
To facilitate information exchange within the zebrafish community, ZFIN maintains both data about the zebrafish research community and experimental data generated by the research community. Data are acquired by manual curation of literature-derived data, by uploads of large datasets and by user submissions. The data contained within the database are associated with their primary sources. The ZFIN home page (Fig. 1) provides multiple points of entry to the database. Users select their area of interest. Query forms return summary results that contain links to more detailed information.
Figure 1.

ZFIN home page (http://zfin.org).
The Zebrafish research community
Research community data include researchers, laboratories, companies and publications. ZFIN provides easy access to contact information consisting of address, telephone, fax, email, URL, biographical data, research interests and publications for scientists active in zebrafish research. Similarly, zebrafish research laboratory information may be accessed based on laboratory members, contact information, research interests and publications.
Company data consist of contact information, products and services. Companies are listed exclusively to provide information to the research community, not as endorsements. Companies that provide reagents or materials used in protocols described in ZFIN are included.
A current bibliography of zebrafish-related publications is maintained by ZFIN. The database staff routinely search public sources such as MEDLINE and upload information into the ZFIN database. Publications are associated with author, laboratory, fish and genetic records within the database. Users may retrieve publication abstracts by specifying author, title, abstract contents, source, MEDLINE number, keywords, year and publication type. As of August 2000, ZFIN contains ∼2700 publications.
Zebrafish mutations
Mutational studies combined with genetic and phenotypic analyses can identify genes involved in developmental, physiological and behavioral processes and help elucidate gene functions and the pathways in which they function. To this end, ZFIN maintains extensive curated mutant and wild-type fish data as well as curated genomic data.
The ZFIN fish page allows the researcher to retrieve a large amount of data about a fish line with minimum navigation. Each mutant or wild-type line is described by its name, abbreviated name, discoverer, current sources, availability, parental lineage, segregation and phenotype (Fig. 2). Allelic information such as the mutagenesis protocol, mutation type, linkage group and known linkages are also included. In cases where the mutant gene has been molecularly cloned, links to the gene record are included. Links are also provided to abstracts of primary and related publications. Images illustrating the phenotypes of many mutations are available, annotated with the developmental stage of the fish, the type and orientation of the image, the magnification and the anatomical structures shown. A user may search ZFIN for mutant and wild-type lines by specifying allele name, locus name, three-letter abbreviation, linkage group, mutation type, mutagen, phenotype, developmental stage and affected structures or defects from an anatomical parts list. These attributes may be used to form complex searches, such as finding a translocation mutant with a gastrulation defect and a short axis.
Figure 2.

ZFIN mutant page. Links to images, genetics and phenotypic information at the top, links to publications at the bottom. Navigation bar along the left side.
Genes
Curated research data about zebrafish genes are displayed on the ZFIN gene page (Fig. 3). This page summarizes information about a gene including its name, symbol, aliases, previous names, linkage group and description. Mapping information, independent linkages, PCR primer sets and primary and related publications are also shown. Links are provided to external databases providing sequence and homology information.
Figure 3.
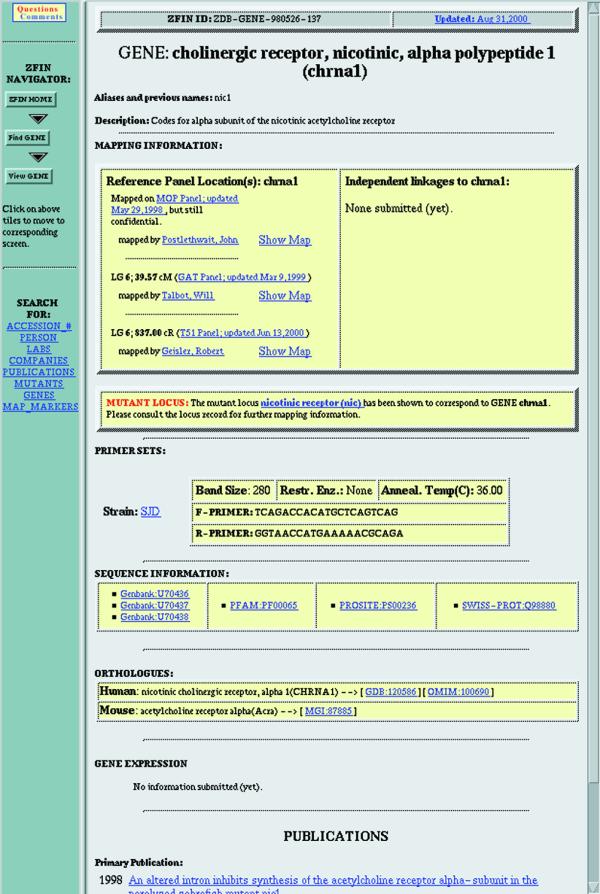
ZFIN gene page. Mapping information at the top, links to sequences and orthologs in outside databases in the middle and links to publications at the bottom. Navigation bar along the left side.
Genetic markers and mapping panels
Genetic maps are an invaluable resource to facilitate the identification of genes affected by mutations. The NIH zebrafish genome initiative currently funds establishment of a high density map of zebrafish genes and anonymous markers. Six mapping panels are currently represented in ZFIN. They include the Boston MGH Cross (MGH), Gates et al. (GAT), Heat Shock (HS) and Mother of Pearl (MOP) meiotic panels and two radiation hybrid panels, Goodfellow T51 (T51) and Loeb/NIH/5000/4000 (LN54). ZFIN uploads panel updates from the laboratories mapping these panels. Data are reviewed for consistency with ZFIN curated data and questions about incorrect or ambiguous nomenclature are resolved with the submitting laboratories. Update dates are displayed for each panel. Panel data are accessible through a query form (Fig. 4) and a map viewer.
Figure 4.
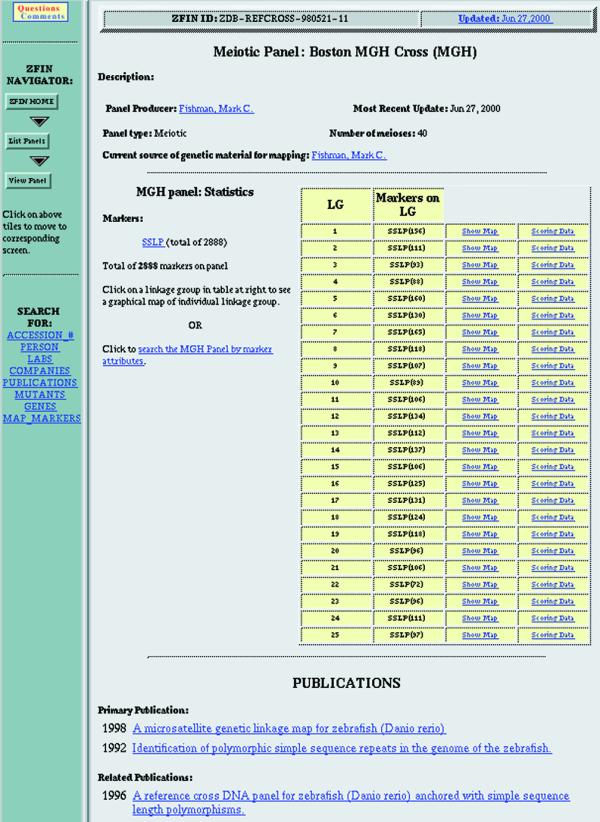
ZFIN mapping panel page. Links to map information in the middle and to publications at the bottom. Navigation bar along the left side.
Map viewer
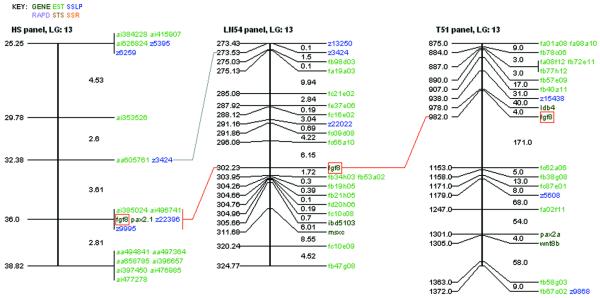
ZFIN provides a unique integrated view of the zebrafish mapping panels that serves as a useful tool for genetic and genomic analyses. Genes, anonymous markers and ESTs are displayed. Framework markers link the individual panels. Detailed information about a marker may be displayed by clicking on the marker on the map. Researchers may generate integrated maps (Fig. 5) by specifying the desired panels, the desired marker types, a marker or gene name, or a location on a linkage group.
Figure 5.
ZFIN map viewer. Allows simultaneous comparisons of several mapping panels. Framework markers (present on each panel) are linked by dark lines. Different marker types are represented by different colors.
NOMENCLATURE
Nomenclature is another important and complex aspect of ZFIN data curation. Each genetic marker is provided with a unique symbol that can be used for identifying sources of information about the marker. The zebrafish nomenclature committee continuously reviews zebrafish gene names to consolidate multiple names for the same gene, to assign uniform names to all members of gene families and to clarify the relationships between genes in zebrafish and other vertebrates. The committee provides guidelines and advice for naming zebrafish genes and mutants and may be contacted at nomenclature@zfin.org. ZFIN users may review current nomenclature guidelines online at http://ZFIN.org/zf_info/nomen.html. Automatic approval for new mutant locus names may be obtained by registering new locus or allele designations with ZFIN’s online locus/allele registration form.
UPDATING DATA
ZFIN data may be updated online at any time by the original submitter. If a user other than the original submitter requests a change, the database staff must perform the update. Questions regarding ZFIN should be addressed to: zfinadmin@zfin.org.
IMPLEMENTATION
ZFIN is implemented in the Informix relational database management system (version 9.2). The database consists of ∼60 tables. Entities are linked to related information using unique identifiers. A web interface of HTML-based forms combined with JavaScript, Java, Perl and CGI scripts provides access to the database. ZFIN curators make use of an advanced set of these forms for entering and modifying data.
FUTURE PLANS
Current development and enhancement of ZFIN include the display of syntenic information on the map viewer, a more complete anatomical parts dictionary and the inclusion of gene expression data.
MIRROR SITES
ZFIN maintains three mirror sites to provide faster worldwide access. The locations are:
Melbourne, Australia: http://zdb.wehi.edu.au/zdb/; Mishima, Japan: http://www.grs.nig.ac.jp:6070/index.html; Strasbourg, France: http://www-igbmc.u-strasbg.fr/index.html
Acknowledgments
ACKNOWLEDGEMENTS
The development of the Zebrafish Information Network is generously supported by the W.M. Keck Foundation. Funds are also provided by the NIH (RR/HD12546).
References
- 1.Streisinger G., Walker,C., Dower,N., Knauber,D. and Singer,F. (1981) Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature, 291, 293–296. [DOI] [PubMed] [Google Scholar]
- 2.Driever W., Solnica-Krezel,L., Schier,A.F., Neuhauss,S.C., Malicki,J., Stemple,D.L., Stainier,D.Y., Zwartkruis,F., Abdelilah,S., Rangini,Z., Belak,J. and Boggs,C. (1996) A genetic screen for mutations affecting embryogenesis in zebrafish. Development, 123, 37–46. [DOI] [PubMed] [Google Scholar]
- 3.Haffter P., Granato,M., Brand,M., Mullins,M.C., Hammerschmidt,M., Kane,D.A., Odenthal,J., van-Eeden,F.J., Jiang,Y.J., Heisenberg,C.P., Kelsh,R.N., Furutani-Seiki,M., Vogelsang,E., Beuchle,D., Schach,U., Fabian,C. and Nüsslein-Volhard,C. (1996) The identification of genes with unique and essential functions in the development of zebrafish, Danio rerio. Development, 123, 1–36. [DOI] [PubMed] [Google Scholar]
- 4.Halpern M.E., Ho,R.K., Walker,C. and Kimmel,C.B. (1993) Induction of muscle pioneers and floor plate is distinguished by the zebrafish no tail mutation. Cell, 75, 99–111. [PubMed] [Google Scholar]
- 5.Weinstein B.M., Stemple,D.L., Driever,W. and Fishman,M.C. (1995) Gridlock, a localized heritable vascular patterning defect in the zebrafish. Nature Med., 1, 1143–1147. [DOI] [PubMed] [Google Scholar]
- 6.Ransom D.G., Haffter,P., Odenthal,J., Brownlie,A., Vogelsang,E., Kelsh,R.N., Brand,M., van-Eeden,F.J., Furutani-Seiki,M., Granato,M., Hammerschmidt,M., Heisenberg,C.P., Jiang,Y.J., Kane,D.A., Mullins,M.C. and Nüsslein-Volhard,C. (1996) Characterization of zebrafish mutants with defects in embryonic hematopoiesis. Development, 123, 311–319. [DOI] [PubMed] [Google Scholar]
- 7.Weinstein B.M., Schier,A.F., Abdelilah,S., Malicki,J., Solnica-Krezel,L., Stemple,D.L., Stainier,D.Y., Zwartkruis,F., Driever,W. and Fishman,M.C. (1996) Hematopoietic mutations in the zebrafish. Development, 123, 303–309. [DOI] [PubMed] [Google Scholar]
- 8.Newman W.M. and Lamming,M.G. (1995) Interactive System Design. Addison-Wesley, Reading, MA.
- 9.Schuler D. and Namioka,A. (1993) Participatory Design: Principles and Practices. Lawrence Erlbaum Associates, Hillsdale, NJ.