Abstract
There has been considerable excitement over the ability to construct linkage maps based only on genome-wide genotype data for single nucleotide polymorphic sites (SNPs) in a population sample. These maps, which are derived from estimates of linkage disequilibrium (LD), rely on population genetics theory to relate the decay of LD to the local rate of recombination, but other population processes also come into play. Here we contrast these LD maps to the classically derived, pedigree-based human recombination maps. The LD maps have a level of resolution greatly exceeding that of the pedigree maps, and at this fine scale, sperm typing allows a means of validation. While at a gross level both the pedigree maps and the sperm typing methods generally agree with LD maps, there are significant local differences between them, and the fact that these maps measure different genetic features should be remembered when using them for other genetic inferences.
Keywords: linkage disequilibrium, genetic linkage, hotspot, population recombination rate, recombination intensity
OVERVIEW AND OBJECTIVES
Classically, recombination rates have been quantified by counting the products of meiosis, tallying either progeny or gametes into recombinant and nonrecombinant classes. In humans, there is an extensive literature on the statistical approaches for obtaining maximum-likelihood and other recombination rate estimators from pedigree-based data (76). By scoring markers across the entire genome, it has been possible to construct whole-genome genetic maps based on such pedigree data. A key limitation of this approach is that the number of sampled individuals is relatively small, so rare events are not adequately ascertained and map resolution is consequently low. This limitation has motivated the development of methods that indirectly sample a much larger number of meiotic events. The leading such approach has been to apply population genetics theory relating local rates of recombination to local levels of linkage disequilibrium (LD) to infer genetic maps from population samples of genotypes. The purpose of this review is to contrast the LD and pedigree-based approaches, highlighting the rich territory at the interface of the two methods.
PEDIGREE-BASED HUMAN GENETIC MAPS
In model organisms, genome-wide linkage maps are constructed by performing controlled crosses and genotyping the progeny in a way that allows the investigator to identify recombination events throughout the genome. The study of human linkage progressed with the cataloging of large pedigrees and the growing availability of molecular markers. The theoretical underpinnings of computational methods for inference of linkage were well understood if the gene or marker order was known (76), and methods of maximum likelihood can be performed with a wide variety of software tools (http://linkage.rockefeller.edu/soft). But when large numbers of genetic markers were involved, entailing a great many possible marker orders, the standard methods became computationally limiting. Lander and Green (60) developed a method for multilocus linkage analysis that applied a hidden Markov chain to efficiently calculate likelihoods in small pedigrees typed for DNA markers. There followed a period of rapid development of computational methods that used a combination of heuristic methods to reduce the search space of gene orders (11, 61–63, 68).
Due to their high heterozygosity, microsatellites became the marker of choice for human linkage studies, and screening repeat libraries resulted in polymerase chain reaction (PCR) primer sets that could score allelic states of thousands of these markers. During the 1990s, James Weber and colleagues at the Marshfield Medical Research Foundation and the Généthon in France established custom-built laboratories for high-throughput genotyping of DNA samples and constructed comprehensive human genetic maps with thousands of microsatellite markers typed on subsets of the extended CEPH (Centre d’Etude du Polymorphisme Humain) family pedigrees (5, 19, 72). It was immediately clear that the intensity of recombination per unit physical length was highly variable across the genome, with consistently low recombination rates in centromeric regions. The total map length on the Marshfield map was 4,400 cM for females, and 2,700 cM for males, and while the sex-specific maps showed generally correlated variation across the genome, there were also sex-specific differences in local recombination rates. There was greater variation among the females in the counts of recombination events than would be expected by chance, and counts were strongly correlated across chromosomes, indicating differences in overall recombination rate among females. These maps subsequently proved highly useful to the human genome sequencing effort, as they contributed independent information on the order and orientation of genes and markers along the chromosome.
A few years later deCODE Genetics, combining extended pedigree information of Icelandic kindreds with the genotyping prowess of their laboratory in Reykjavik, Iceland, produced an even finer resolution map (56) that included 5,136 microsatellite markers genotyped in 869 individuals across 146 Icelandic families. Linkage inference was based on a total of 1,257 meiotic events, approximately 6 times as many as the Marshfield map. The previously published draft human genome sequence and clone physical maps uniquely placed approximately 93% of the markers, and in regions where both genetic and physical maps were available, recombination rates could be correlated with many attributes of the DNA sequence [such as local guanine–cytosine (GC) content, CpG (C—phosphate—G) motifs, and poly(A)/poly(T) stretches (where A and T are adenine and thymine nucleotides)] as well as chromosomal cytological features (telomeres and centromeres). The greatly increased resolution of the deCODE map revealed many local differences between male and female recombination rates.
Pooling genotype data from the CEPH database and from deCODE Genetics, the first Rutgers map, a combined linkage and physical map (59), contained 14,759 markers that had been genotyped in a mixture of CEPH and deCODE (56) families. Version 2 of this map incorporated an additional 13,666 SNP markers genotyped in the CEPH pedigrees for a total of 28,121 mapped polymorphic sites (58, 67). Over 1 million SNP genotypes are now available for these samples, but the genetic map is not much increased in resolution by further addition of markers, because there is already high confidence in the locations of the breakpoints of nearly every meiotic exchange. Further increases in resolution would require additional pedigrees.
Contrasts of the genetic and physical maps have been highly informative. A scatterplot of marker locations, with the physical map along the x-axis and the genetic map along the y-axis, would be expected to fall along a perfect diagonal if gene order and distances were precisely concordant in the two maps. Instead, such plots show some local regions that are flat and others that rise steeply, indicating long physical regions in which little recombination occurs, and short regions where there is intense recombination, respectively (Figure 1). The slope of this plot yields the local rate of recombination per physical distance, or the recombination intensity, typically measured in units of cM/Mbp. Human centromeres have quite low recombination intensity, estimated at well under 1 cM/Mbp, and the steepest regions, often occurring at telomeres, have an average recombination intensity of approximately 5–10 cM/Mbp. This approach led to the discovery of genomic regions with particularly low levels of recombination, which not surprisingly also tend to have much greater levels of linkage disequilibrium (100). Comparisons between the genetic and physical maps also highlighted discrepancies in apparent marker order (18). Roughly 5% of the Marshfield linkage map markers had linkage orders inconsistent with the physical map order in the draft genome sequence. Initially, this was thought to be due to errors in the physical map, but in addition to this possibility, we now know that segmental variation can result in discrepancies between the physical map of any given individual and the reference.
Figure 1.
Genetic versus physical maps. (a) Genetic map location versus physical location for markers in a 20-Mb region of human chromosome 10. The local slope of this relationship provides an estimate of the local recombination intensity in cM/Mbp. (b) Sex-averaged recombination intensity in the same region. Most human chromosomes have very low recombination intensity at centromeres and high recombination intensity at telomeres. Data are from the Rutgers human linkage map version 2 (67).
Linkage maps are affected by genotyping errors, although these are likely fewer than with association-mapping methods because tests of concordance with expected Mendelian transmission provides a strong filter for maintaining data quality. Genotyping errors often result in apparent nearby pairs of recombination events, or close double crossovers. Most mapping efforts delete close doubles from the analysis as likely errors, although some of them may in fact represent gene conversion events or other double exchanges. Similarly, deviations from Mendelian segregation result in marker exclusion; although this is a reasonable and conservative approach, at a genome-wide scale, growing evidence suggests that some markers show genuine exceptions to Mendelian segregation (101). The fact that genetic linkage maps are assembled as an amalgam of meiotic exchange events from many individuals whose local recombination rates almost certainly differ certainly inflates the statistical uncertainty in the averaged map. But perhaps the greatest limitation of genetic maps based on pedigrees is the fact that so few meiotic exchange events can be captured, which limits resolution of the highest-density linkage maps to the order of only 0.5–2 cM. These maps are therefore not useful for understanding the nature of recombination events at a finer scale.
LINKAGE-DISEQUILIBRIUM-BASED GENETIC MAPS
A very different, indirect, means of inferring local rates of recombination, developed over many years, has, with the recent identification of huge numbers of markers made available by high-throughput SNP genotyping methods, shown strong promise for producing genetic maps at a far finer resolution than pedigree maps. The history of this effort is closely tied to the development of tools for genome-wide association mapping in humans. The recognition that it should be feasible to identify genetic variation statistically associated with inflated disease risk, using only case-control samples with sufficiently many typed markers, gave impetus to the quest for large numbers of SNPs across the human genome. It was necessary not only to identify SNPs, but also to determine the degree to which populations exhibited linkage disequilibrium across pairs of SNPs. The International HapMap project identified SNPs from many different sources, and then systematically genotyped them in a panel of 30 CEPH trios, 30 Nigerian Yoruban trios, 45 unrelated Japanese, and 45 unrelated Chinese. This set of 270 people, split equally among European, African, and Asian ancestries, was genotyped and analyzed, yielding a genome-wide map of the haplotype structure in major human populations (1).
SNPs in the first phase of the HapMap project were initially identified by a wide variety of methods, and the ascertainment bias that resulted from detecting SNPs in small samples was felt to compromise the utility of the data for some purposes. Using microarray hybridization, Perlegen Biosciences had fully sequenced a panel of 24 human genomic DNA samples, and the SNPs discovered in this set played an important role in expanding the SNP collection to what became known as HapMap2 (28). Phase II of HapMap characterized more than 3.1 million SNPs, or on average one approximately every 1 kb across the genome (although the distribution was not uniform), in the same 270 individuals. These data yielded a detailed picture of the pattern of linkage disequilibrium across the human genome, and it became abundantly clear that the regions with extensive LD over long spans were regions with low recombination intensity, as inferred from the contrast between genetic and physical maps. In addition, sharply defined regions were found in which LD decayed very rapidly, consistent with local, high levels of recombination. This provided strong impetus to use the LD information to infer which local distribution of recombination events could generate the observed distribution of LD across the genome.
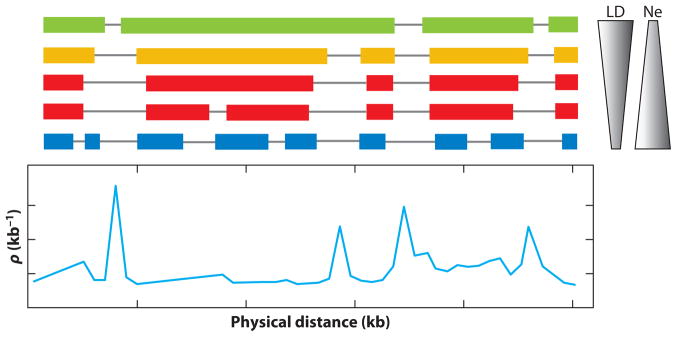
Procedures for estimating recombination rates from population genetic data have been reviewed thoroughly elsewhere (91). Population genetics has a rich history of mathematical theory relating processes that occur in idealized populations to empirically derived metrics. Intuitively, one expects that greater recombination rates will result in more rapid decay of linkage disequilibrium, and population genetics theory provides precise equations for this process. The relationship between local levels of linkage disequilibrium and the local rate of recombination is somewhat involved, with random genetic drift playing a key role. In fact it is through random drift that LD can be maintained at equilibrium in a finite population, since in an infinite population with only neutral variation, all LD eventually decays. So the theory that relates human LD to human recombination rates of necessity includes parameters reflecting population size and demography (Figure 2).
Figure 2.
Relationship between population demography and the linkage disequilibrium (LD) map. Colored boxes indicate the size of recurrently observed haplotypes in the sample. A small population undergoes strong genetic drift, which results in large haplotype blocks, and as the population size increases, these blocks get smaller. We believe that human demography is dominated by an out-of-Africa bottleneck, so that the picture moves from small haplotype blocks in Africa to larger blocks in the out-of-Africa populations due to founder effects and drift. But note that the breaks between the blocks in Africa will often remain breaks outside of Africa, preserving the locations of recombination hotspots. Redrawn from (91). Abbreviations: Ne, effective population size; ρ, the population recombination rate, also written as ρ = 4Neμ, where μ is the mutation rate.
Before we summarize this theory, it is useful to emphasize that the estimates of recombination rate are made in the context of a population genetic model that carries many assumptions. Technically, the model assumes no natural selection, no migration, and constant finite population size with homogeneous rates of recombination. Mutation is ignored. Since these assumptions are clearly violated to some degree in human populations, one might question the validity of the estimates, but in fact it is possible to show that the estimates are robust to many reasonable departures from these idealized conditions. Note however that the parameter estimated in the methods described below is rho = 4Ner, a confounding of recombination with effective population size. This means that factors not included in the model that may distort the local inference of effective population size, such as a selective sweep, will also tend to distort the inference of local recombination. Fortunately, this too is amenable to analysis by simulation.
The formal theory relating the sampling properties of a pair of segregating sites to parameters such as recombination rate was developed by Richard Hudson (37). This work showed that a feasible way to infer a local estimate of rho was to use all the segregating sites in a region and to combine the pairwise inferences in a composite likelihood estimator. The composite estimator does not yield a true maximum likelihood because it combines dependent likelihoods as though they were independent. Fortunately, this approximation works quite well, and its properties have been thoroughly investigated. Along with this important discovery, Hudson developed a means for generating population samples of a region of the genome under a neutral model with recombination (38), and this has been an immensely valuable tool for testing models of LD in finite populations.
Inference of local recombination rates across the human genome using the above population genetic theory remained a steep challenge because calculating the likelihood is computationally difficult. Griffiths and Marjoram (32) developed a Markov chain Monte Carlo (MCMC) approach to approximate the likelihood for estimating rho. This approach could be accelerated by use of importance sampling, but it was still too slow to be feasible for a genome-wide application. Fearnhead and Donnelly (23, 24) finally cracked the problem by introducing a novel importance sampler to calculate the likelihood under a coalescent-based model. This method was several orders of magnitude faster than anything else available and allowed construction of the first genome-wide maps of the population recombination rate, rho. Approximate methods based on the composite likelihood are much faster still. McVean and colleagues (71) implemented and applied the composite likelihood, and using the 1.6 million SNPs from the HapMap project, a full genome-wide LD map was subsequently generated (73). Over the next few sections, we will return to the results of this paper, but the key finding not predicted from the pedigree-based linkage maps is the number and intensity of apparent hotspots of recombination across the human genome.
Before turning to the conclusions drawn from the LD maps, it is important to consider the effects of gene conversion, defined as recombinational exchange events that resolve double-strand breaks without exchange of flanking markers. Such events typically result in the copying of a short tract of sequence from one haplotype onto another, such that if the individual is heterozygous for sites within the conversion tract, the resulting gamete resembles a close double recombinant. Wiuf and Hein (99) developed a model for this process, a geometric length distribution for recombination tracts. Gene conversion alters the pattern of pairwise linkage disequilibrium, and a parameter of Hudson’s (37) model is the ratio of gene conversion to recombination rates. This has been used to infer rates of gene conversion from polymorphism data (29).
Other methods for inferring gene conversion rates from population sample data have been devised (30, 95), for example, using triplets of SNPs. Gene conversion results in reduced LD at a very local scale but has less impact on LD at greater distances; this results in a steeper decline in LD for nearby markers, so inclusion of gene conversion can provide improved empirical fits to the LD decay curves. Models incorporating both crossing over and gene conversion fit the short-range data (0–5 kb) of chromosome 21 much better than do models that include crossing over alone. The estimated ratio of gene-conversion rate to crossing-over rate has a range of 1.6–9.4, depending on the assumed conversion tract length (in the range of 500–50 bp) (78, 79). These methods appear to be reasonably good at estimating an average gene conversion rate over many loci, but they cannot reliably estimate local gene conversion rates for a single region of the genome. One reason for this is that the methods are highly sensitive to genotyping error (83). Several lines of evidence indicate that recombination intensity varies widely across the genome, but evidence for variation in rates of gene conversion is sparser (42, 94). In short, our understanding of determinants of rates of gene conversion remains somewhat unsatisfying.
HETEROGENEITY IN LOCAL RECOMBINATION INTENSITY
Before turning to highly localized spikes of recombination rate, known as hotspots, let us first consider broader-scale variation in recombination rates. As mentioned above, centromeric regions tend to have low rates of recombination, and telomeres tend to have elevated rates. But there are many other regional differences in local recombination rates, including non-centromeric stretches of unusually low recombination. What causes this kind of variation, and what are its consequences for the human genome?
The first efforts to study variation in local recombination rates showed that the genetic and physical maps could not be reconciled unless there was local variation on a megabase pair scale in the rate of recombination. However, determining the scale at which recombination rates vary has been challenging. The first attempts focused on targeted gene regions and asked whether the rate of decay of LD was homogeneous across the regions (16, 86). There does appear to be a signal for within-gene variation in recombination rates, but as we will see below, a combination of variation in density and intensity of recombination hotspots could also be driving this.
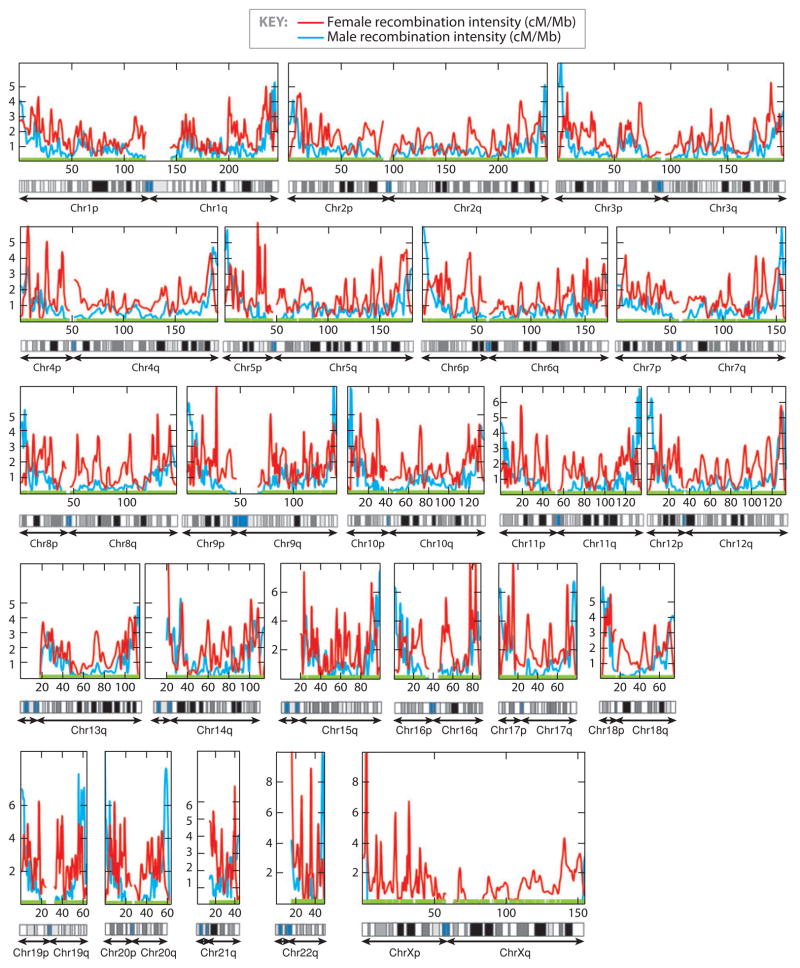
Statistical inference of local recombination intensity by comparison of the genetic and physical maps is not entirely straightforward, because it involves the local slope of the relation plotted in Figure 1. One approach has been to apply locally weighted linear regression (loess) from the best current physical map position to the best current linkage map (20). We updated these calculations using the latest physical and genetic maps (Build 36 and the Rutgers 2007 maps) and applied local regression (66) to obtain smoothed estimates of local recombination intensity (Figure 3). A striking finding is that an increase in recombination in telomeres of males relative to females is now evident for every chromosome.
Figure 3.
Sex-specific recombination intensity across the human genome. The observations that females produce gametes with more recombination than males, centromeres have low recombination, and telomeres have high recombination are all discernable. To obtain the genome-wide local recombination intensity (in cM/Mb), we performed a local regression on the Rutgers human linkage map version 2 (67). The local polynomial degree and weight function parameters were optimized by minimizing the local likelihood within a three-dimensional parameter grid for each chromosome arm (66). The optimal parameter combinations were selected by minimizing local Akaike information criterion (AIC) for male-specific and female-specific linkage maps separately. Computation was done using the Locfit package in the R statistical software v. 2.60 (http://www.r-project.org). Abbreviation: Chr, chromosome.
Large-scale variation in recombination rate across the genome is now widely appreciated, and many efforts have been made to correlate local recombination rate with other properties of the genome. The strongest of these is GC content, which is positively correlated with local recombination rate. The Hill–Robertson effect states that natural selection should be less effective in regions of low recombination, and that this could lead to differences in rates of adaptation among regions of high versus low recombination. Bullaughey and colleagues (6) tested the association between human recombination rate variation and adaptive molecular evolution in primates as scored by the dN/dS ratio (the ratio of rates of substitution at non-synonymous and synonymous nucleotide sites). Using data from human, chimp, and macaque, they found no correlation between rates of recombination and rates of protein evolution, after GC content is taken into account. Genes found in regions of very low recombination, which are expected to show the most pronounced reduction in the efficacy of selection, do not evolve at a different rate than other genes. We will return to the question of the evolutionary consequences of recombination rate variation, but for now it appears that effective population sizes of primates may be small enough that the patterns driven by local selective sweeps and the Hill–Robertson effect are at best weak.
RECOMBINATION HOTSPOTS IN HUMANS
The HapMap project presented human population geneticists with a plethora of genotype frequency data with which to analyze patterns of LD. In particular, the 1.6 million SNPs genotyped in 270 people in HapMap phase 1 enabled scoring local LD patterns at a scale exceeding that available even for model organisms. Initial efforts to estimate the population recombination rate parameter, rho, from human genotype data were limited to local genomic regions. In 2004, Fearnhead and colleagues (25) applied coalescent-based approaches to estimate recombination rates from polymorphism data using full-likelihood methods for the well-characterized hotspot region of the beta-globin gene. A local reduction in LD in this region is clear from resequencing data in population samples. The full-likelihood estimates of local rho in this region provide a satisfying concordance between sperm-typing inference of recombination hotspots and the population recombination rate estimates.
An important means to estimate likelihoods uses simulation of sample data. The approach of Li and Stephens (65) provided a quantum leap in our ability to generate population sample simulations of recombined segments of chromosomes. This approach is not limited to considering only pairwise samples of SNPs, and is computationally extremely fast. The Li and Stephens algorithm could be used to estimate underlying recombination rates from population data, and simulations demonstrated its utility. This algorithm remains a standard tool in many different applications in population genetics, especially for haplotype phasing.
At the time of publication of the first HapMap paper, it was already clear that linkage disequilibrium is organized in a somewhat blockwise fashion, and the human genetics community was eager to catalog the resulting haplotype blocks in order to allow more efficient genome-wide typing of an individual. It is now generally believed that these blocks largely comprise regions delimited by recombination hotspots, but at the time the full extent of hotspots was not fully appreciated, and it was argued that the normal process of random genetic drift, even in the face of homogeneous recombination, can also generate blockwise haplotype structures (80). Although the existence of recombination hotspots is now widely acknowledged, the utility of haplotype blocks has become less critical because of standardized use of commercial SNP genotyping platforms which allow very dense sampling of the genome.
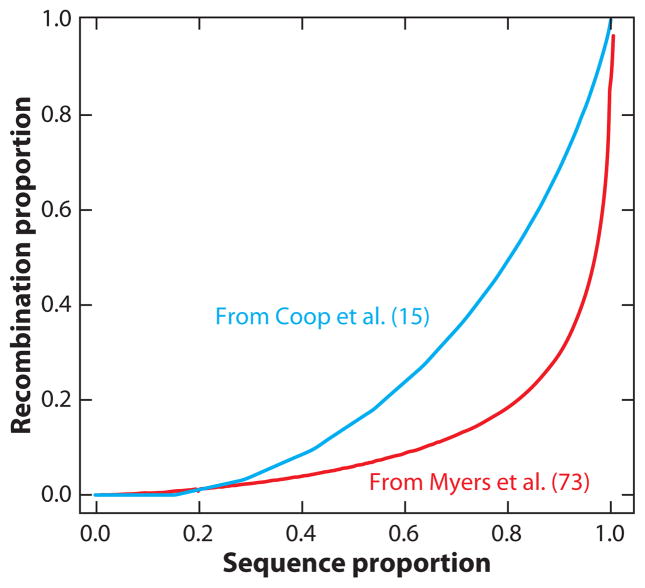
The ability to construct a genome-wide map based on population recombination rate (rho) estimates from population sample data depended on the development of approximate-likelihood methods that provided sufficient computational speed to make the likelihood calculations feasible. Considerable effort was spent in testing these methods by simulations, generating sample genotype data under different scenarios of recombination hotspots and demography, and finding conditions which yielded reasonable estimates of rho (24, 25, 70, 71, 73). Approximate-likelihood methods were then applied to the HapMap sample data, generating the finest-scale resolution recombination map in humans to date (71, 73). The new map showed good correspondence to the pedigree-based maps over large chromosomal scales, and to known hotspots at a very fine scale. The most striking findings were that local rate variation appeared to vary over four orders of magnitude, and that the bulk of recombination in the human genome occurs within hotspots. Initial estimates were that 50% of all recombination occurs in less than 10% of the sequence (Figure 4). There was still reason to pause before throwing out previous maps, since this new map was based on indirect inference from population sample data, but all tests of correspondence with other known attributes of human linkage were encouraging.
Figure 4.
A substantial portion of all human recombination occurs in hotspots. If small windows of the genome are put in rank order of recombination intensity, the lowest 80% of the genome includes only 20% of the recombination events, according to the red curve, adapted from Myers et al. (73). The blue curve presents the result of using pedigree-based recombination data from Coop et al. (15, tbl. S7).
The statistical inference of recombination hotspots from genotype data on samples from a population remains an area of active debate (22, 33, 64, 89, 92). Not only is there a need to improve upon the speed and accuracy of approximate-likelihood methods, but joint inference of gene conversion and integration of imputed genotype data are desirable aims as well. Soon we will have full-genome sequence data from individuals at a scale allowing inference of recombination rates directly from the sequence, and methods will need to accommodate both sequencing error and orders-of-magnitude larger volumes of data. Another problem being tackled with current genotyping data is inferring variation in the background recombination rate. It is common to assume a single background rate, augmented by hotspots of varying density and intensity, but a model allowing variable background recombination rates seems at least as plausible. Auton and McVean (3) developed such a model, and in applying it to data from the human leukocyte antigen (HLA) and minisatellite MS32 regions of the human genome, they find clear differences in background rates in addition to hotspot variability.
Accepting for the moment that recombination hotspots are a genuine feature of the human genome, we can ask which features of the DNA sequence signal the location of a hotspot. Many attempts have been made to identify correlates to hotspot location, including GC content, gene density, and repetitive elements. At best the correlations are weak and offer little predictive power. Myers and colleagues (75) developed an LD map based on the HapMap2 data and found 25,000 recombination hotspots which they then analyzed for predictive features. They applied wavelet-based analysis to determine the scale at which features such as base composition, coding context, and DNA repeats impacted hotspot presence. Word analysis identified a set of statistically significant DNA motifs that are strongly associated with recombination hotspots. They confirmed a rapid turnover of hotspots between humans and chimpanzees, in a manner consistent with the use of the DNA motifs.
The initial success with hotspot motif finding motivated a continued search, both for more refined motifs and for the factors that recognize them. Myers and colleagues (74) found that a 13-bp sequence motif previously associated with the activity of 40% of human hotspots does not function in chimpanzee and is being removed by self-destructive drive in the human lineage. This motif is bound by PRDM9, a rapidly evolving zinc-finger protein that harbors significant human–chimp divergence. PRDM9 catalyzes histone H3 lysine 4 trimethylation in a manner consistent with a role in recombination. Polymorphisms in both mice and humans influence both the binding of PRDM9 and the induction of recombination (4). Initial evidence for a role of PRDM9 in recombination hotspots appears quite strong, but it remains a challenge to imagine how such rapid turnover of recombination landscapes can be adaptive.
DIRECT SCORING OF HOTSPOTS BY SPERM GENOTYPING
As exciting as the inference of hotspots was, the fact that the LD data yielded only indirect evidence of them strongly motivated the direct scoring of local rates of recombination to verify the findings. The laboratories of Norman Arnheim and Alec Jeffreys had earlier developed methods for both single-sperm and pooled-sperm PCR genotyping that are effective in scoring recombinant gametes. In one study (36), sperm-DNA typing was applied to determine the distribution of crossover events within a 1-Mbp region of human chromosome 4p. The investigators typed 602 sperm using PCR and detected 29 recombinants. Genotyping SNPs in the region showed that a 280-kb interval had a six- to ninefold excess of counts of recombination breakpoints compared to flanking regions. This was among the first clues that the human genome may harbor local hotspots of recombination.
Jeffreys et al. (41) attracted strong attention to sperm genotyping technologies and demonstrated clearly at the level of individual recombination events that the human genome has recombination hotspots. Their observation that recombination in humans is “intensely punctate” has held up well. This paper analyzed a 216-kb segment of the class II region of the major histocompatibility complex (MHC) that had been thoroughly studied for LD-map–based inference of recombination hotspots (manifested as discrete points of low LD that also terminate haplotype blocks). The sperm typing confirmed beautifully that the LD-based inferred recombination hotspots were in fact coincident with meiotic crossover hotspots. The six hotspots they defined all share a remarkably similar symmetrical morphology but vary considerably in intensity and are not obviously associated with any primary DNA sequence determinants of hotspot activity. The MHC hotspots occur in clusters and together account for almost all crossovers in this region of the MHC.
Subsequent publications documented the local concentration of meiotic breakpoints in other regions, as assessed by sperm genotyping (2, 40–49, 69, 93). Every region examined was found to have some local inflation of recombination, if not outright hotspots, and this seems to be a widespread characteristic of mammalian genomes (51). The correspondence between the LD-based hotspots and the sperm-typing hotspots remained strong but not perfect. It is not difficult to imagine reasons for the few discrepancies: A recombinational hotspot may fail to show reduced LD if it is relatively young (9); the vagaries of population drift or of natural selection may give rise to a high-frequency haplotype that spans a hotspot, resulting in LD spanning the hotspot.
One means for dissecting the effects of recombination, demographic history, random genetic drift, and natural selection on hotspot intensity is to compare human populations that have highly distinct demographic histories. Kauppi and colleagues (53) examined such populations by genotyping SNPs across a 75-kb span of the MHC region in individuals from northern Europe, northern Finland (Saami), and Zimbabwe. Previous studies had identified three recombination hotspots and a 60-kb long LD block in European samples. Despite the wide variation in demographic histories among these three populations, which is reflected in corresponding variation in haplotype diversity and composition, all three populations showed very similar patterns of LD. The three hotspots were evident in all three populations, and nucleotide diversity in the hotspot regions was also elevated. This early example confirmed predictions from simulations (71) that the ability to detect recombination hotspots from patterns of LD should be robust to demography, and thus good correspondence of hotspots across human populations might be expected (see the section, Among-Population Heterogeneity in Recombination Rates, below).
To date, some 26 recombination hotspots have been characterized using allele-specific PCR to selectively amplify recombined DNA molecules (52). There emerges a consistent picture that meiotic crossover hotspots in humans are highly localized. Interestingly, where there are hotspots, it appears that flanking DNA segments exhibit many fewer exchange events, as though recombination were suppressed. There does appear to be a corresponding increase in noncrossover exchanges (gene conversion), consistent with models in which the relative proportion of exchange and nonexchange resolutions of double-strand breaks remains fairly constant. It is somewhat puzzling that the sperm-typing methods rarely find hotspots with recombination rates greater than 12 cM/Mbp, whereas the LD-based maps suggest that hotspots can approach 100 cM/Mbp. However, rates estimated from both methods have large confidence intervals: Sperm-typing methods are limited by small sample sizes (counts of meiotic exchanges), while LD-based mapping methods rely on the vagaries of sampling from an astronomical number of neutral coalescents, with a resulting inherent uncertainty in the derived estimates (91).
Although sperm typing and population recombination estimates are generally concordant, instances of population and species differences suggest that hotspots are not permanent features of the genome (12, 14, 47, 51). The lack of shared hotspots between humans and chimpanzee (81, 82), in particular, highlights the short evolutionary timeframe in which recombination hotspots operate, which is somewhat surpising given that humans and chimps differ by only slightly more than 1% at the DNA sequence level. The transient nature of hotspots suggests that there exists a mechanism for hotspot turnover, one possibility being gene conversion biased to propagate alleles that locally disrupt hotspots. Coop and Myers (13) demonstrate that gene conversion bias may serve to erase hotspots by shortening their life span. Biased gene conversion may be a sufficiently strong force to produce the observed lack of sharing of intense hotspots between species.
STRUCTURAL VARIATION AND THE LINKAGE MAP
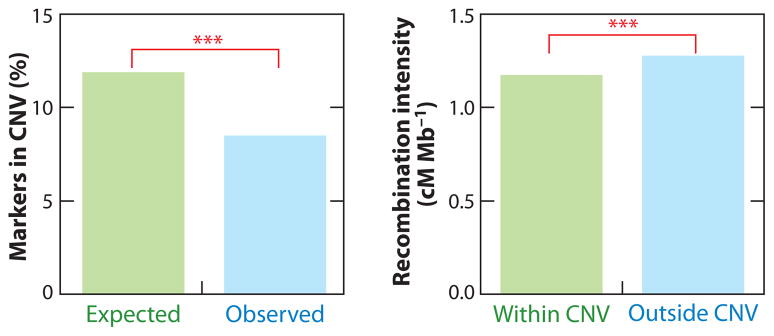
Another potential source of variation in the linkage map arises from variation among individuals in the structure of the physical genome. Copy number variation is now widely recognized to be an important feature of the human population, with any pair of human individuals differing by insertions and deletions amounting to well over 1 Mbp on average (54). Inversions of genetic regions also can attain fairly appreciable frequency (90). The effects of such variation on standard SNP genotyping platforms might be expected to result in regions with an inflated count of SNPs that display typing errors or Mendelian exceptions, or with a reduction of the observed SNP density (Figure 5). Development of commercial SNP genotype arrays often involved extensive empirical testing and elimination of those SNPs that appeared problematic; almost certainly, a significant portion of these dropped SNPs occur in repetitive regions or regions harboring other copy number variation. Regions repeated within the genome may also have unusual recombination characteristics, since they potentially undergo both orthologous and ectopic (paralogous) recombination. In fact, the local average intensity of recombination (in cM/Mbp) is significantly reduced in regions near copy number variants (Figure 5).
Figure 5.
Structural variation and recombination. In copy number variation (CNV) regions, there is a deficit of markers in the Rutgers human linkage map (67) (left). The (HapMap) CNV data are from Redon et al. (27, 85). The CNV regions also have a lower sex-averaged recombination intensity compared with the rest of the genome (right). The asterisks indicate that the two contrasts are significant at P < 0.001. Abbreviation: CNV, copy number variation.
A study of the low copy number repeats containing NF1REPa and NF1REPc on chromosome 17 and a third paralogous copy on chromosome 19 showed that all copies share a recombination hotspot (84). The extra effort in obtaining accurate genotyping data for such repeated regions suggests that it will not be easy to study them at a genome-wide scale and that, even as we progress into whole-genome sequencing of individuals, repeated regions will present challenges. It is also proving difficult to accurately score copy number variants using high-throughput SNP-typing platforms, although the technical problems of typing CNVs do seem surmountable, and the impact of CNVs on local patterns of recombination will hopefully soon be clarified.
LOCAL CONTRASTS OF GENETIC AND LD-BASED RECOMBINATION MAPS
Pedigree-derived linkage maps provide a picture of the actual meiotic exchanges in specific individuals, whereas LD-based maps reflect an integration of population-level processes over many previous generations. LD maps provide an historical average, incorporating the effects of population genetic events such as bottlenecks, population growth, and natural selection. Aside from the different time scales, distortions in local Ne may drive additional discrepancies between pedigree and LD maps. It is worth emphasizing that the LD-based maps are estimates of the parameter rho = 4Ner, which confounds population recombination rate with effective population size so it is somewhat misleading to claim that the LD-based maps estimate recombination intensity (in cM/Mbp). The estimates are model based, and because effective population size weighs into the estimates, any errors in demographic inference will manifest as errors in estimates of recombination intensity. In fact, the HapMap estimates of recombination intensity, besides assuming that all variation is neutral, also assume constant Ne = 10,000 for the European population and Ne = 15,000 for the African population.
Since the pedigree-based genetic map provides direct estimates of recombination rate, and the LD-based map provides an estimate of the compound parameter rho, in principle, discrepancies between the maps can provide evidence for distortion of Ne. This idea goes back at least to Hill (35), who provided a means of estimating Ne from LD data.
IMPACT OF LOCAL RECOMBINATION INTENSITY VARIATION ON GENOME-WIDE ASSOCIATIONS
The recognition that local recombination intensity varies widely gave rise to the notion that for genome-wide association testing, an optimal choice of SNPs would require careful tuning of SNP density to the local recombination rate. In particular, regions of high recombination would shorten the lengths of shared haplotypes such that larger numbers of SNPs would be required to tag those regions. One effective way to choose such tagSNPs was to algorithmically select a maximally informative set of common single-nucleotide polymorphisms that included all known common SNPs for direct assay, and any additional SNPs that fall below a threshold level of r2 (8). With the threshold (r2 = 0.8), the LD-selected tagSNPs resolve >80% of all haplotypes across a set of 100 candidate genes. This method nicely accommodates variation in recombination rate, and in principle provides an empirical solution to the problem. In the end, however, commercial enterprises produced SNP chips with a standardized array of SNPs that were orders of magnitude less expensive for genotyping, and the tagSNP problem became more or less irrelevant. The commercial arrays are heavily weighted toward SNPs that genotype reliably. One lingering issue that the tagSNP problem highlighted is that the optimal selection of SNPs should in principle be tuned for different major population groups; the commercial chips provide us instead with a compromise set of SNPs that is definitely suboptimal for some populations.
AMONG-POPULATION HETEROGENEITY IN RECOMBINATION RATES
The observation of major differences in recombination hotspots between humans and chimpanzees raises the issue of hotspot differences among human populations. This is an important consideration if we are to use the same SNP resources for association scans in different populations. The recognition that linkage disequilibrium patterns can differ in a consistent way came with the observation that LD decays more rapidly in African than in non-African samples (86). However, this most likely reflects long-term effective population size differences rather than recombination differences. Non-African populations, which have experienced one or more bottlenecks, have a lower effective size, resulting in slower decay of LD. Whether on top of this there is heterogeneity in local patterns of LD remains an interesting question.
Several studies tackle the problem of contrasting LD across population samples. One primary lesson is that apart from the shift in overall levels, local regions of high and low LD remain largely consistent across populations (17). To the extent that some of the LD reflects the state of the human population prior to the migration out of Africa, this result is not surprising. But both adaptive and neutral forces have likely acted in population-specific manners over the last 100,000 years, so it is not surprising that within the overall pattern of consistent changes in local LD, there are regions with population-specific changes in LD (21, 87). If patterns of LD are sufficiently heterogeneous, then the tools for inference of association in genome-wide association studies would have to be tuned for each population. Too few populations have been ascertained with sufficient depth to assess this fully, but the major population groups that have been examined appear to have sufficiently similar patterns at least for common SNPs such that genome-wide association study (GWAS) approaches should be applicable across populations. It remains to be seen how the inclusion of increasing numbers of rare SNPs will impact this picture.
Modeling among-population heterogeneity in SNP and haplotype frequencies is feasible with classical population genetics approaches, and methods for generating large simulated samples are also readily available. What is not yet routine, but desirable, is to model heterogeneity in hotspot density and intensity across populations. Calabrese and colleagues (7) developed such a population genetics model, featuring recombination hotspots that are heterogeneous across the population and whose population frequency changes with time. They produced a diffusion approximation to the model, and used simulations to show that hotspot turnover could account for observed patterns of LD.
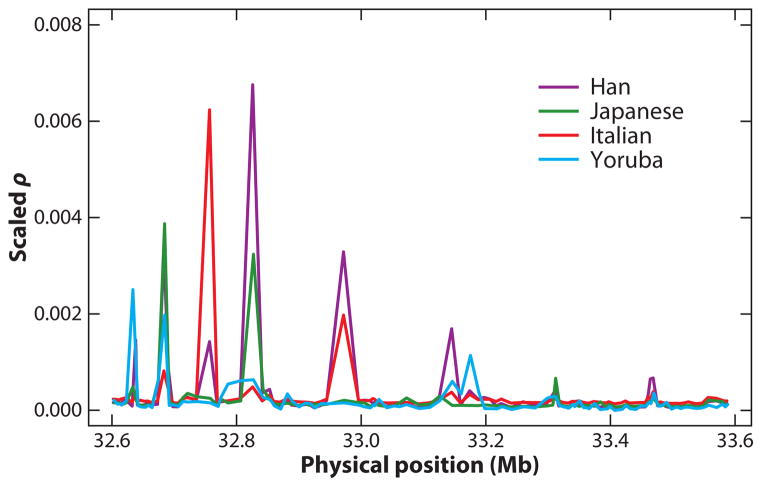
Our understanding of among-population variation in rho is at present limited, and only with the generation of genotype samples from multiple populations can we really address the heterogeneity in rho across populations. When rho is calculated separately based on SNP genotype data for each of several population samples (31), one does indeed find many instances of local differences (Figure 6). It has been argued that the primary causes are a combination of random drift (in the context of particular demographic changes) and natural selection. Identifying which differences are caused by actual population-specific changes in recombination rate is a surmountable challenge. In CAP10, there is at least one instance of a population-specific difference in hotspot activity (10). The HapMap3 data (39) and eventually the 1000 Genomes data will provide an excellent opportunity to explore such local variation in rho at a genome-wide scale.
Figure 6.
Among-population heterogeneity in linkage disequilibrium (LD). A collection of 107 single nucleotide polymorphisms (SNPs) spanning 1 Mbp of chromosome 22q was genotyped by Graffelman et al. (31) in samples from 28 different human populations. Plotted are the estimates of rho across this region in the major population groups studied in the HapMap project. There is an overall similarity across populations but also pronounced local differences.
AMONG-INDIVIDUAL VARIATION IN RECOMBINATION RATES
One might expect that the challenges in identifying among-population differences in recombination rate would become even more daunting for among-individual differences. However, with sufficient density of genotyped markers, it is possible to localize recombination events in each transmitted gamete. The existence of heritable differences in total recombination rate among individuals has been recognized for many years, and was thoroughly documented in the early genome-wide genetic maps (5). Coop and colleagues (15) used genome-wide SNP genotype data collected in Hutterite nuclear families to localize a total of 728 crossovers with high spatial resolution. By tallying which breakpoints were in recombination hotspots, they could show that overall hotspot usage was similar in males and females, with individual hotspots often active in both sexes, and that 60% of crossovers occurred in confirmed recombination hotspots. There were large differences in the number of recombination events among both males and females, and these differences were heritable as assessed by the clustering on the pedigrees. In a similar analysis of the deCODE data, a factor that determined local recombination rate was mapped to RNF212 (57). Baudat and colleagues (4) also identify an association between PRDM9 allelic differences and overall recombination rate in humans. Model organisms such as Drosophila and yeast have many recombination-influencing mutations, some of which influence region-specific recombination rates, so the existence of such variation in humans should not come as a surprise. The association between elevated rates of chromosomal nondisjunction and low rates of recombination show that the differences may not be entirely benign in humans (55).
OUTSTANDING QUESTIONS AND FUTURE OPPORTUNITIES
An open question about the pattern of recombination in the human genome is whether the unstable hotspot organization has any adaptive function. Theory suggests that natural selection may act to tune local rates of recombination, but the strength of such selection is typically quite weak, so mutational events that create and erase hotspots may dominate the process. A worthwhile goal is to develop an evolutionary perspective to understand why the pattern of recombination in the human genome is what we observe.
We have already touched upon some lines of evidence regarding the evolutionary divergence of the primate recombination map. The first demonstration that the map might vary significantly came from a study of the beta-globin recombination hotspot (96). By resequencing a 15-kb segment in a human population sample, and in samples of chimpanzees and rhesus macaques, Wall and colleagues pinpointed the location of the human beta-globin hotspot and tested whether the data were consistent with hotspots in chimpanzee and macaque. The chimp data had population structure complications, but the macaque data clearly showed the absence of a hotspot. This was followed by studies of the hotspots in TAP2 in humans and 24 chimpanzees by Ptak and colleagues (82) who found very little support for recombination rate variation at TAP2 in the western chimpanzee data. Later an additional 14 Mb of sequencing in chimpanzee and human samples confirmed the generality of the instability of recombination hotspots (81).
Perhaps our greatest ignorance of human recombination stems from a failure to understand the evolutionary basis for the fluidity of recombination hotspots. There exists an extensive body of literature on the theory of genetic modifiers of recombination rate, reviewed by Feldman and colleagues (26). In a constant environment, modifiers that reduce recombination rate are generally favored, but since nearly all organisms persist in maintaining some form of recombination, it has been necessary to investigate more complex scenarios. One general conclusion is that increased recombination may be favored under certain forms of negative epistasis among alleles. Hey (34) summarizes this situation as follows: LD among SNPs can be actively maintained by selection, and when this occurs, a modifier allele that raises the recombination rate (thus decreasing LD) can cause selection to act more efficiently. As recombination generates new configurations of beneficial alleles, both the beneficial alleles and the recombination modifiers increase in concert. Otto and Lenormand (77) marshall evidence from model organisms that, as artificial directional selection proceeds, there is often a correlated elevation in recombination rate. There are many differences in hotspot density in and near human protein coding genes stratified by gene function, including a significant elevation of recombination hotspots in brain-expressed genes (50). Whether the expansion by humans into new environments and subsequent adaptive evolution was accelerated by recombination-inflating hotspots remains an intriguing possibility.
Finally, returning to the primary empirical questions about the distribution of recombination events across the human genome, there remains room for many additional advances. Analysis of LD took a big step forward with our understanding of the sampling properties of pairs of loci in a population sample (37), and population genetics theory continues to develop analytical results that are useful and relevant to understanding recombination and LD (88). Incorporation of a full Bayesian approach to the problem has promise, although computational demands of Markov chain Monte Carlo (MCMC) approaches remain a concern. In one successful attempt, Wang and Ranalla (97, 98) developed a full-likelihood MCMC method for estimating recombination rate under a Bayesian framework. This method seems particularly ambitious when it is realized that the genealogies are modeled by using marginal individual SNP genealogies related through an ancestral recombination graph. The method does a credible job locating recombination hotspots, including those that occur in clusters. Further development of approaches like this, incorporating such complications as individual recombination rate distortions due to copy number variants, PRDM9 allelic differences, and richer demographic models all seem within reach.
Acknowledgments
The authors thank Dr. Steve Buyske (Rutgers University) for helpful discussions and analyses. This work was supported by National Institutes of Health (NIH) grants GM065509, HG003229, and MH084685. The Rutgers Maps are supported by NIH grant GM080221 (T.C.M.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Andrew G. Clark, Email: ac347@cornell.edu.
Xu Wang, Email: xw54@cornell.edu.
Tara Matise, Email: matise@biology.rutgers.edu.
LITERATURE CITED
- 1.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, et al. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnheim N, Calabrese P, Tiemann-Boege I. Mammalian meiotic recombination hot spots. Annu Rev Genet. 2007;41:369–99. doi: 10.1146/annurev.genet.41.110306.130301. [DOI] [PubMed] [Google Scholar]
- 3.Auton A, McVean G. Recombination rate estimation in the presence of hotspots. Genome Res. 2007;17:1219–27. doi: 10.1101/gr.6386707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–40. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–69. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bullaughey K, Przeworski M, Coop G. No effect of recombination on the efficacy of natural selection in primates. Genome Res. 2008;18:544–54. doi: 10.1101/gr.071548.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calabrese P. A population genetics model with recombination hotspots that are heterogeneous across the population. Proc Natl Acad Sci USA. 2007;104:4748–52. doi: 10.1073/pnas.0610195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark AG. Hot spots unglued. Nat Genet. 2005;37:563–64. doi: 10.1038/ng0605-563. [DOI] [PubMed] [Google Scholar]
- 10.Clark VJ, Ptak SE, Tiemann I, Qian Y, Coop G, et al. Combining sperm typing and linkage disequilibrium analyses reveals differences in selective pressures or recombination rates across human populations. Genetics. 2007;175:795–804. doi: 10.1534/genetics.106.064964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins A, Teague J, Keats BJ, Morton NE. Linkage map integration. Genomics. 1996;36:157–62. doi: 10.1006/geno.1996.0436. [DOI] [PubMed] [Google Scholar]
- 12.Coop G. Can a genome change its (hot)spots? Trends Ecol Evol. 2005;20:643–45. doi: 10.1016/j.tree.2005.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coop G, Myers SR. Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 2007;3:e35. doi: 10.1371/journal.pgen.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coop G, Przeworski M. An evolutionary view of human recombination. Nat Rev Genet. 2007;8:23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- 15.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–98. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 16.Crawford DC, Bhangale T, Li N, Hellenthal G, Rieder MJ, et al. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat Genet. 2004;36:700–6. doi: 10.1038/ng1376. [DOI] [PubMed] [Google Scholar]
- 17.De La Vega FM, Isaac H, Collins A, Scafe CR, Halldorsson BV, et al. The linkage disequilibrium maps of three human chromosomes across four populations reflect their demographic history and a common underlying recombination pattern. Genome Res. 2005;15:454–62. doi: 10.1101/gr.3241705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeWan AT, Parrado AR, Matise TC, Leal SM. The map problem: a comparison of genetic and sequence-based physical maps. Am J Hum Genet. 2002;70:101–7. doi: 10.1086/324774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dib C, Faure S, Fizames C, Samson D, Drouot N, et al. A comprehensive genetic map of the human genome based on 5264 microsatellites. Nature. 1996;380:152–54. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 20.Duffy DL. An integrated genetic map for linkage analysis. Behav Genet. 2006;36:4–6. doi: 10.1007/s10519-005-9015-x. [DOI] [PubMed] [Google Scholar]
- 21.Evans DM, Cardon LR. A comparison of linkage disequilibrium patterns and estimated population recombination rates across multiple populations. Am J Hum Genet. 2005;76:681–87. doi: 10.1086/429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fearnhead P. SequenceLDhot: detecting recombination hotspots. Bioinformatics. 2006;22:3061–66. doi: 10.1093/bioinformatics/btl540. [DOI] [PubMed] [Google Scholar]
- 23.Fearnhead P, Donnelly P. Estimating recombination rates from population genetic data. Genetics. 2001;159:1299–318. doi: 10.1093/genetics/159.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fearnhead P, Donnelly P. Approximate likelihood methods for estimating local recombination rates. J R Stat Soc Lond B. 2002;64:657–80. [Google Scholar]
- 25.Fearnhead P, Harding RM, Schneider JA, Myers S, Donnelly P. Application of coalescent methods to reveal fine-scale rate variation and recombination hotspots. Genetics. 2004;167:2067–81. doi: 10.1534/genetics.103.021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldman MW, Otto SP, Christiansen FB. Population genetic perspectives on the evolution of recombination. Annu Rev Genet. 1996;30:261–95. doi: 10.1146/annurev.genet.30.1.261. [DOI] [PubMed] [Google Scholar]
- 27.Fiegler H, Redon R, Andrews D, Scott C, Andrews R, et al. Accurate and reliable high-throughput detection of copy number variation in the human genome. Genome Res. 2006;16:1566–74. doi: 10.1101/gr.5630906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frisse L, Hudson RR, Bartoszewicz A, Wall JD, Donfack J, Di Rienzo A. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am J Hum Genet. 2001;69:831–43. doi: 10.1086/323612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gay J, Myers S, McVean G. Estimating meiotic gene conversion rates from population genetic data. Genetics. 2007;177:881–94. doi: 10.1534/genetics.107.078907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graffelman J, Balding DJ, Gonzalez-Neira A, Bertranpetit J. Variation in estimated recombination rates across human populations. Hum Genet. 2007;122:301–10. doi: 10.1007/s00439-007-0391-6. [DOI] [PubMed] [Google Scholar]
- 32.Griffiths RC, Marjoram P. Ancestral inference from samples of DNA sequences with recombination. J Comput Biol. 1996;3:479–502. doi: 10.1089/cmb.1996.3.479. [DOI] [PubMed] [Google Scholar]
- 33.Hellenthal G, Stephens M. Insights into recombination from population genetic variation. Curr Opin Genet Dev. 2006;16:565–72. doi: 10.1016/j.gde.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Hey J. What’s so hot about recombination hotspots? PLoS Biol. 2004;2:e190. doi: 10.1371/journal.pbio.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill WG. Estimation of effective population size from data on linkage disequilibrium. Genet Res. 1981;38:209–16. [Google Scholar]
- 36.Hubert R, MacDonald M, Gusella J, Arnheim N. High resolution localization of recombination hot spots using sperm typing. Nat Genet. 1994;7:420–24. doi: 10.1038/ng0794-420. [DOI] [PubMed] [Google Scholar]
- 37.Hudson RR. Two-locus sampling distributions and their application. Genetics. 2001;159:1805–17. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–38. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 39.International HapMap3 Consortium. An integrated haplotype map of rare and common genetic variation in diverse human populations. Nature. 2010 In press. [Google Scholar]
- 40.Jeffreys AJ, Holloway JK, Kauppi L, May CA, Neumann R, et al. Meiotic recombination hot spots and human DNA diversity. Philos Trans R Soc Lond B. 2004;359:141–52. doi: 10.1098/rstb.2003.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet. 2001;29:217–22. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 42.Jeffreys AJ, May CA. DNA enrichment by allele-specific hybridization (DEASH): a novel method for haplotyping and for detecting low-frequency base substitutional variants and recombinant DNA molecules. Genome Res. 2003;13:2316–24. doi: 10.1101/gr.1214603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36:151–56. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- 44.Jeffreys AJ, Murray J, Neumann R. High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol Cell. 1998;2:267–73. doi: 10.1016/s1097-2765(00)80138-0. [DOI] [PubMed] [Google Scholar]
- 45.Jeffreys AJ, Neumann R. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat Genet. 2002;31:267–71. doi: 10.1038/ng910. [DOI] [PubMed] [Google Scholar]
- 46.Jeffreys AJ, Neumann R. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum Mol Genet. 2005;14:2277–87. doi: 10.1093/hmg/ddi232. [DOI] [PubMed] [Google Scholar]
- 47.Jeffreys AJ, Neumann R. The rise and fall of a human recombination hot spot. Nat Genet. 2009;41:625–29. doi: 10.1038/ng.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeffreys AJ, Neumann R, Panayi M, Myers S, Donnelly P. Human recombination hot spots hidden in regions of strong marker association. Nat Genet. 2005;37:601–6. doi: 10.1038/ng1565. [DOI] [PubMed] [Google Scholar]
- 49.Jeffreys AJ, Ritchie A, Neumann R. High-resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum Mol Genet. 2000;9:725–33. doi: 10.1093/hmg/9.5.725. [DOI] [PubMed] [Google Scholar]
- 50.Kato M, Miya F, Kanemura Y, Tanaka T, Nakamura Y, Tsunoda T. Recombination rates of genes expressed in human tissues. Hum Mol Genet. 2008;17:577–86. doi: 10.1093/hmg/ddm332. [DOI] [PubMed] [Google Scholar]
- 51.Kauppi L, Jeffreys AJ, Keeney S. Where the crossovers are: recombination distributions in mammals. Nat Rev Genet. 2004;5:413–24. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- 52.Kauppi L, May CA, Jeffreys AJ. Analysis of meiotic recombination products from human sperm. Methods Mol Biol. 2009;557:323–55. doi: 10.1007/978-1-59745-527-5_20. [DOI] [PubMed] [Google Scholar]
- 53.Kauppi L, Sajantila A, Jeffreys AJ. Recombination hotspots rather than population history dominate linkage disequilibrium in the MHC class II region. Hum Mol Genet. 2003;12:33–40. doi: 10.1093/hmg/ddg008. [DOI] [PubMed] [Google Scholar]
- 54.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kong A, Barnard J, Gudbjartsson DF, Thorleifsson G, Jonsdottir G, et al. Recombination rate and reproductive success in humans. Nat Genet. 2004;36:1203–6. doi: 10.1038/ng1445. [DOI] [PubMed] [Google Scholar]
- 56.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–47. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 57.Kong A, Thorleifsson G, Stefansson H, Masson G, Helgason A, et al. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science. 2008;319:1398–401. doi: 10.1126/science.1152422. [DOI] [PubMed] [Google Scholar]
- 58.Kong X, Matise TC. MAP-O-MAT: Internet-based linkage mapping. Bioinformatics. 2005;21:557–59. doi: 10.1093/bioinformatics/bti024. [DOI] [PubMed] [Google Scholar]
- 59.Kong X, Murphy K, Raj T, He C, White PS, Matise TC. A combined linkage-physical map of the human genome. Am J Hum Genet. 2004;75:1143–48. doi: 10.1086/426405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA. 1987;84:2363–67. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–81. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 62.Lange K, Cantor R, Horvath S, Perola M, Sabatti C, et al. MENDEL version 4.0: a complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genet. 2001;69:A1886. [Google Scholar]
- 63.Lange K, Weeks D, Boehnke M. Programs for Pedigree Analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol. 1988;5:471–72. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- 64.Li J, Zhang MQ, Zhang X. A new method for detecting human recombination hotspots and its applications to the HapMap ENCODE data. Am J Hum Genet. 2006;79:628–39. doi: 10.1086/508066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li N, Stephens M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 2003;165:2213–33. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loader C. Local Regression and Likelihood. xiii. New York: Springer; 1999. p. 290. [Google Scholar]
- 67.Matise TC, Chen F, Chen W, De La Vega FM, Hansen M, et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–86. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matise TC, Perlin M, Chakravarti A. Automated construction of genetic linkage maps using an expert system (MultiMap): a human genome linkage map. Nat Genet. 1994;6:384–90. doi: 10.1038/ng0494-384. [DOI] [PubMed] [Google Scholar]
- 69.May CA, Shone AC, Kalaydjieva L, Sajantila A, Jeffreys AJ. Crossover clustering and rapid decay of linkage disequilibrium in the Xp/Yp pseudoautosomal gene SHOX. Nat Genet. 2002;31:272–75. doi: 10.1038/ng918. [DOI] [PubMed] [Google Scholar]
- 70.McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–41. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McVean GA, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–84. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 72.Murray JC, Buetow KH, Weber JL, Ludwigsen S, Scherpbier-Heddema T, et al. A comprehensive human linkage map with centimorgan density. Cooperative Human Linkage Center (CHLC) Science. 1994;265:2049–54. doi: 10.1126/science.8091227. [DOI] [PubMed] [Google Scholar]
- 73.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–24. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 74.Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:791–92. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Myers S, Spencer CC, Auton A, Bottolo L, Freeman C, et al. The distribution and causes of meiotic recombination in the human genome. Biochem Soc Trans. 2006;34:526–30. doi: 10.1042/BST0340526. [DOI] [PubMed] [Google Scholar]
- 76.Ott J. Analysis of Human Genetic Linkage. xxiii. Baltimore: Johns Hopkins Univ. Press; 1999. p. 382. [Google Scholar]
- 77.Otto SP, Lenormand T. Resolving the paradox of sex and recombination. Nat Rev Genet. 2002;3:252–61. doi: 10.1038/nrg761. [DOI] [PubMed] [Google Scholar]
- 78.Padhukasahasram B, Marjoram P, Nordborg M. Estimating the rate of gene conversion on human chromosome 21. Am J Hum Genet. 2004;75:386–97. doi: 10.1086/423451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Padhukasahasram B, Wall JD, Marjoram P, Nordborg M. Estimating recombination rates from single-nucleotide polymorphisms using summary statistics. Genetics. 2006;174:1517–28. doi: 10.1534/genetics.106.060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Phillips MS, Lawrence R, Sachidanandam R, Morris AP, Balding DJ, et al. Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nat Genet. 2003;33:382–87. doi: 10.1038/ng1100. [DOI] [PubMed] [Google Scholar]
- 81.Ptak SE, Hinds DA, Koehler K, Nickel B, Patil N, et al. Fine-scale recombination patterns differ between chimpanzees and humans. Nat Genet. 2005;37:429–34. doi: 10.1038/ng1529. [DOI] [PubMed] [Google Scholar]
- 82.Ptak SE, Roeder AD, Stephens M, Gilad Y, Paabo S, Przeworski M. Absence of the TAP2 human recombination hotspot in chimpanzees. PLoS Biol. 2004;2:e155. doi: 10.1371/journal.pbio.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ptak SE, Voelpel K, Przeworski M. Insights into recombination from patterns of linkage disequilibrium in humans. Genetics. 2004;167:387–97. doi: 10.1534/genetics.167.1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raedt TD, Stephens M, Heyns I, Brems H, Thijs D, et al. Conservation of hotspots for recombination in low-copy repeats associated with the NF1 microdeletion. Nat Genet. 2006;38:1419–23. doi: 10.1038/ng1920. [DOI] [PubMed] [Google Scholar]
- 85.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 87.Serre D, Nadon R, Hudson TJ. Large-scale recombination rate patterns are conserved among human populations. Genome Res. 2005;15:1547–52. doi: 10.1101/gr.4211905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Song YS, Song JS. Analytic computation of the expectation of the linkage disequilibrium coefficient r2. Theor Popul Biol. 2007;71:49–60. doi: 10.1016/j.tpb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 89.Spencer CC, Deloukas P, Hunt S, Mullikin J, Myers S, et al. The influence of recombination on human genetic diversity. PLoS Genet. 2006;2:e148. doi: 10.1371/journal.pgen.0020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37:129–37. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 91.Stumpf MP, McVean GA. Estimating recombination rates from population-genetic data. Nat Rev Genet. 2003;4:959–68. doi: 10.1038/nrg1227. [DOI] [PubMed] [Google Scholar]
- 92.Tapper W, Gibson J, Morton NE, Collins A. A comparison of methods to detect recombination hotspots. Hum Hered. 2008;66:157–169. doi: 10.1159/000126050. [DOI] [PubMed] [Google Scholar]
- 93.Tiemann-Boege I, Calabrese P, Cochran DM, Sokol R, Arnheim N. High-resolution recombination patterns in a region of human chromosome 21 measured by sperm typing. PLoS Genet. 2006;2:e70. doi: 10.1371/journal.pgen.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wall JD. Close look at gene conversion hot spots. Nat Genet. 2004;36:114–15. doi: 10.1038/ng0204-114. [DOI] [PubMed] [Google Scholar]
- 95.Wall JD. Estimating recombination rates using three-site likelihoods. Genetics. 2004;167:1461–73. doi: 10.1534/genetics.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wall JD, Frisse LA, Hudson RR, Di Rienzo A. Comparative linkage-disequilibrium analysis of the beta-globin hotspot in primates. Am J Hum Genet. 2003;73:1330–40. doi: 10.1086/380311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Y, Rannala B. Bayesian inference of fine-scale recombination rates using population genomic data. Philos Trans R Soc Lond B. 2008;363:3921–30. doi: 10.1098/rstb.2008.0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y, Rannala B. Population genomic inference of recombination rates and hotspots. Proc Natl Acad Sci USA. 2009;106:6215–19. doi: 10.1073/pnas.0900418106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wiuf C, Hein J. The coalescent with gene conversion. Genetics. 2000;155:451–62. doi: 10.1093/genetics/155.1.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu A, Zhao C, Fan Y, Jang W, Mungall AJ, et al. Comparison of human genetic and sequence-based physical maps. Nature. 2001;409:951–53. doi: 10.1038/35057185. [DOI] [PubMed] [Google Scholar]
- 101.Zöllner S, Wen X, Hanchard NA, Herbert MA, Ober C, Pritchard JK. Evidence for extensive transmission distortion in the human genome. Am J Hum Genet. 2004;74:62–72. doi: 10.1086/381131. [DOI] [PMC free article] [PubMed] [Google Scholar]