Abstract
Hybridization and polyploidization are now recognized as major phenomena in the evolution of plants, promoting genetic diversity, adaptive radiation and speciation. Modern molecular techniques have recently provided evidence that allopolyploidy can induce several types of genetic and epigenetic events that are of critical importance for the evolutionary success of hybrids: (1) chromosomal rearrangements within one or both parental genomes contribute toward proper meiotic pairing and isolation of the hybrid from its progenitors; (2) demethylation and activation of dormant transposable elements may trigger insertional mutagenesis and changes in local patterns of gene expression, facilitating rapid genomic reorganisation; (3) rapid and reproducible loss of low copy DNA sequence appears to result in further differentiation of homoeologous chromosomes; and (4) organ-specific up- or down-regulation of one of the duplicated genes, resulting in unequal expression or silencing one copy. All these alterations also have the potential, while stabilizing allopolyploid genomes, to produce novel expression patterns and new phenotypes, which together with increased heterozygosity and gene redundancy might confer on hybrids an elevated evolutionary potential, with effects at scales ranging from molecular to ecological. Although important advances have been made in understanding genomic responses to allopolyploidization, further insights are still expected to be gained in the near future, such as the direction and nature of the diploidization process, functional relevance of gene expression alterations, molecular mechanisms that result in adaptation to different ecologies/habitats, and ecological and evolutionary implications of recurrent polyploidization.
Keywords: adaptation, cDNA-AFLP, epigenetic changes, evolution, gene expression, hybridization, microarrays, MSAP, polyploidy, transcriptome
Introduction
One of the most important insights derived from whole-genome sequencing projects of Arabidopsis thaliana is that angiosperms have undergone multiple rounds of polyploidization events during their evolution (e.g., Wendel, 2000; Bowers & al., 2003; Blanc & Wolfe, 2004; De Bodt & al., 2005; Cui & al., 2006), an idea that Stebbins (1966) partly predicted forty years ago. Despite their current cytological behavior, most typical diploid plant genomes still harbour evidence of multiple rounds of ancient genomic doubling events (see reviews by Wolfe, 2001; Adams & Wendel, 2005a, b; Wendel & Doyle, 2005). Palaeopolyploids most likely recover stability and fertility within a few generations via chromosome rearrangements and genetic diploidization. Recent studies have indicated that ancient genomic duplications were crucial for the creation of many important developmental and regulatory genes found in extant angiosperm genomes (Wendel, 2000) and might even have had an important role in the origin and initial diversification of the flowering plants (De Bodt & al., 2005). Indeed, genome duplication events double the amount of genetic material for evolutionary experimentation; further, functional divergence of duplicated genes (neo- and subfunctionalization, see below) allows an increase in biological complexity over evolutionary timescales (Ohno, 1970).
Another widespread phenomenon, presently recognized as having extensive evolutionary and ecological implications for promoting adaptation and rapid speciation---in combination or not with polyploidy---is natural hybridization (see, e.g., Barton, 2001; Rieseberg & al., 2003; Seehausen, 2004; Hegarty & Hiscock, 2005). Hybridization is an important source of genetic variation and functional novelty; new combinations of favorable genes may provide starting points for evolution of new lineages. Hybrids often exhibit novel or extreme characters when compared with parental taxa, referred to as “transgressive segregation” (Anderson, 1949; Arnold, 1997; Rieseberg & al., 1999; Seehausen, 2004; Salmon & al., 2005). Complementary action of different expressed loci, with additive effects when recombined in hybrids, might be the trigger for such new adaptive phenotypes that can, for example, move into previously unexploitable habitats.
Hybrid speciation can occur by one of two routes: without a change in ploidy after hybridization (homoploid speciation) or with a change in ploidy relative to those in the parents (allopolyploid speciation). Newly formed hybrids are not true species unless they maintain their taxonomic identity and are reproductively isolated from both their parental taxa, either genetically or ecologically (Rieseberg & al., 2003). Allopolyploidy generally results in instantaneous speciation because backcrossing to the diploid parents produces a high proportion of inviable or mostly sterile offspring (Seehausen, 2004). However, in taxa producing large numbers of small seeds, such as orchids and some Solanaceae (e.g., Nicotiana) triploids perhaps could produce enough viable gametes to act as bridges between allotetraploids and their lower ploidy parents (e.g., Aagaard & al., 2005). The newly mixed genomes may more easily be stabilized in polyploid taxa, and polyploidy confers on hybrids a selective advantage in overcoming initial hybrid sterility by providing each chromosome with a precise pairing partner (Hegarty & Hiscock, 2005). Allopolyploids can also exhibit “transgressive” characters, most probably through gene deletion (Wu & al., 2006; see below). Finally, permanent coexistance of favorable traits may be effectively preserved in allopolyploids as fixed, non-segregating heterozygosity.
Due to increased heterozygosity (potentially resulting in heterosis; Lippman & Zamir, 2007) and buffering of disadvantageous alleles (gene redundancy), allopolyploids can be effective competitors with their diploid relatives, and polyploid complexes may have high adaptive potential (Soltis & Soltis, 1993; Seehausen, 2004; Bell & Travis, 2005; Comai, 2005). Polyploids are fit and well adapted, and they may even out-compete their diploid relatives, not only in abundance and distribution, but also by exhibiting broader ecological amplitude. Although ecological divergence seems usually difficult because it may require simultaneous changes at multiple traits (genes), hybridization may overcome this difficulty because it provides genetic variation at many genes in a single generation (see, e.g., Rieseberg & al., 2003). Moreover, probably because of their higher tolerance of increased selfing, many polyploid taxa have superior colonizing abilities relative to their diploid parents (Soltis & Soltis, 2000). On the other hand, heterosis is expected to be transitory (Barton, 2001) and restricted to the first few generations after hybridization in diploid hybrids, but it may be maintained over longer periods in allopolyploids (Otto, 2003; Comai, 2005).
In the first generations following chromosome doubling, newly formed allopolyploids might still experience reduced fertility because initial formation of an allopolyploid must be a shock (Pikaard, 2001; Comai, 2005; Hegarty & Hiscock, 2005), despite their obvious success in nature. They face a number of genomic challenges, such as potential conflict due to different instructions being given for the same tasks and activation of transposable elements that can damage the newly formed compound genomes. A broad range of techniques (e.g., AFLP/SSR mapping, cDNA-AFLP differential display, genomic in situ hybridization - GISH, fluorescent in situ hybridization - FISH, anonymous cDNA microarrays etc.) has recently provided evidence that new allopolyploids might deal with these challenges by rapid genetic and epigenetic adjustments, e.g., by silencing some redundant genes and reducing the incidence of chromosomal infidelity (e.g., Soltis & al., 2004; Adams & Wendel, 2005a, b; Salmon & al., 2005).
Molecular markers for analyzing genetic and epigenetic consequences of polyploidy and/or hybridization
Although expressed DNA segments (coding genes together with promoter regions and other regulatory elements) make up only a limited fraction of the genome, they are exactly the elements producing differences between phenotypes. Even genetically similar organisms may exhibit different levels of gene expression (see, e.g., Le Quere & al., 2004). The state-of-the-art approach to analyze variation of gene expression is based on the microarray technique, and the future is represented by whole transcriptome sequencing via the exciting 454 (Margulies & al., 2005). Unfortunately, these methods are still too expensive for use on a regular base. Handy alternatives are represented, for example, by single-strand conformation polymorphism (SSCP) analysis of cDNA or modifications of standard AFLP protocols for measuring expression variation of multiple genes (cDNA-AFLP) or variation in the distribution of DNA methylation (MSAP).
Methods to analyse the transcriptome1 start from mRNA (or from its complement - cDNA) derived from different tissues---often subjected to specific stimuli/stress---and developmental stages. To ensure comparability of transcriptome/methylation patterns among accessions, prior to analysis plants are usually grown in controlled environments and the techniques are performed on different organs/tissues separately. For total or mRNA isolation and cDNA synthesis, commercially available kits may be used. Modifications of standard protocols for significant improvements in the quantity and quality of the first-strand cDNA synthesis in plants have been recently published (e.g., Xiao & al., 2005).
cDNA microarrays
This high-throughput technology (Schena & al., 1995) has shown enormous potential in almost every aspect of molecular biology (Kammenga & al., 2007) but has been especially used in whole transcriptome analysis. Microarrays are theoretically able to (quantitatively) assess activity of tens of thousands to---in the case of nanoarrays---a million genes at the same time. Usually, known molecules (DNA fragments, full-length cDNAs or cDNA fragments, oligonucleotides, genes or gene fragments, open reading frames, proteins etc.) are printed, in-house or commercially, using a computer-controlled, high-speed robot on a microscale solid support (e.g., glass slide, nylon membrane etc.) at extremely high density in indexed positions. The next experimental step is hybridization of labeled probes to the array created as above. Hybridized arrays are then imaged to measure fluorescent intensities for each spot on the slide. Recent examples of microarray technology applied to questions relating to hybrid and/or polyploid evolution in plants include studies in Arabidopsis (Chen & al., 2004; Madlung & al., 2005) and Senecio (using “anonymous” cDNA microarrays, Hegarty & al., 2005). The work of Madlung & al. (2005), as the first microarray study on allopolyploidy, has mainly shown widespread activation of transposable elements after hybridization and ploidy change. Although microarray techniques are able to monitor sensitively thousands of genes simultaneously, several problems are still expected to be resolved in the near future, such as high cost and requirement for sequence knowledge (but see Hegarty & al., 2005), which results in restricted availability and difficulties encountered in distinguishing transcripts from homoeologous genes (for reviews see, e.g., Dunwell & al., 2001; Gibson, 2002; Ranz & Machado, 2006).
cDNA-AFLP
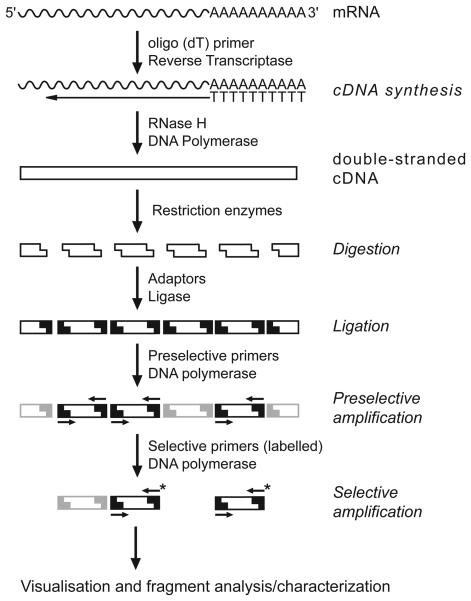
A comprehensive transcript-profiling method that has been used for quantitatively measuring gene expression variation between individuals and groups of phenotypes is the cDNA-amplified fragment length polymorphism (e.g., Kuhn, 2001; Donson & al., 2002; Breyne & al., 2003). The standard AFLP procedure (Vos & al., 1995) is performed on a pool of cDNAs generated from a given tissue (Fig. 1; Bachem & al., 1996, 1998). It is based on ligation of adaptors to restriction fragments of cDNA and use of a set of specific primers for subsequent polymerase chain reaction (PCR) amplification of these fragments under stringent conditions. By combining different restriction enzymes (even just frequently cutting enzymes) and using primers with 1--3 selective nucleotides, the number of amplified fragments can be optimised. The molecular basis of transcript polymorphism (i.e., presence vs. absence of a given cDNA amplified fragment) can be caused by sequence polymorphism (deletions, insertions and rearrangements) or by gene silencing (epigenetically controlled). For example, using cDNA-AFLP Soltis & al. (2004) were successful in revealing differences in transcripts between the neopolyploid Tragopogon miscellus, its diploid parents and populations of T. miscellus of reciprocal origin. This method is fast, requires only a small quantity of mRNA, can be applied in the absence of any prior sequence data, is reproducible and sensitive and exhibits a good correlation with northern analysis or RT-PCR (Bachem & al., 1996, 1998; Donson & al., 2002; He & al., 2003). A shortcoming of this method is that the source of fragment variation cannot be directly identified; however, the fragments of interest can be recovered for further characterization via e.g., cDNA cleaved amplified polymorphic sequence analysis---CAPS---see Konieczny & Ausubel, 1993). Finally cDNA-AFLP can differentiate---in certain circumstances---between homoeologous gene copies, an advantage over microarrays, which rely upon a hybridization-based approach.
Fig. 1.
The steps involved in the cDNA-AFLP technique. Filled elements symbol adaptors ligated to restricted fragments. Small arrows represent (pre-)selective primers, the ones with an asterisk are labeled. Grey fragments will not be amplified in the respective step. Visualisation can be achieved using denaturing gels or capillary sequencers.
MSAP
In addition to rapid and dramatic structural changes in allopolyploids, some systems show mainly epigenetic changes via repatterning of DNA methylation (e.g., Spartina anglica, Ainouche & al., 2003). Methylation patterns of DNA are relatively stable over cell generations but can be altered rapidly and dramatically by hybridization and/or polyploidy. They are essential for regulating plant development through their influence on gene transcription; they are also involved in a number of specific biological processes, such as gene silencing and mobile element control. Epigenetic alterations that might be encountered in allopolyploids can be investigated using methylation sensitive AFLP (MSAP, e.g., Ainouche & al., 2003; Salmon & al., 2005). The standard AFLP technique is applied to genomic DNA by making use of two methylation sensitive/insensitive isoschizomers (e.g., MspI and HpaII) as frequent cutters in parallel restriction reactions. These enzymes recognize the same DNA sequence motif but display differential sensitivity to DNA methylation. A difference in banding patterns indicates methylation variation. For example, in Spartina Salmon & al. (2005) found as much as 30% of the parental methylation patterns to be altered in two hybrids and the allopolyploid, S. anglica. Most of the polymorphic MSAP fragments are probably made up of non-coding regions; nevertheless low percentages of methylated fragments are expected to correspond with epigenetic gene silencing. Sequence analysis of polymorphic cDNA-AFLP and MSAP fragments may allow identification of the genes involved in such alteration.
Genetic consequences of allopolyploid speciation
Recent studies (see below) have documented that allopolyploidy has occurred frequently in the evolutionary history of plants, and such events can induce extensive genomic and genetic alterations that are of critical importance for the evolutionary success of allopolyploids: chromosomal rearrangements, transposable element activation, DNA sequence elimination, gene silencing and novel patterns of expression through epigenetic regulation.
Chromosomal rearrangements
Because recently formed allopolyploids usually possess a complete copy of each parental genome, often displaying colinearity of genes, miss-pairing of homoeologous chromosomes may occur during meiosis. Recombination within one or both parental genomes makes the two combined genomes non-homologous---resulting in proper meiotic pairing (for a discussion on the genetic control of chromosome pairing in allopolyploids see, e.g., Ji & Langridge, 1990)---and represents an additional source of genetic novelty in polyploids. Although there is minimal evidence that rearrangements in genomic DNA (i.e., inversions, translocations and duplications, see Coghlan & al., 2005) cause speciation, recombination between different parental linkage groups also intensifies reproductive isolation of the hybrid from its progenitors, limiting potential for introgressive backcrossing (Rieseberg & al., 2003; Coghlan & al., 2005; Hegarty & Hiscock, 2005). Rapid and extensive chromosomal rearrangement has been recently documented in some allopolyploids (e.g., Triticum, Levy & Feldman, 2004; Nicotiana, Lim & al., 2004; Brassica, Pires & al., 2004; Arabidopsis, Pontes & al., 2004), but not others (e.g., Gossypium, Liu & al., 2001; Spartina anglica, Ainouche & al., 2003; Tragopogon natural allopolyploids, Soltis & al., 2004). Therefore, although it is believed that genome rearrangements may be necessary for restoring nuclear-cytoplasmic compatibility (Soltis & Soltis, 1999), it is clear that certain species may be able to respond to allopolyploidy and adapt to a novel genomic environment without a corresponding reorganisation of the genome. In the allopolyploid Spartina anglica, Ainouche & al. (2003) failed to find significant genomic reorganisation; the phenotypic divergence between the hybrid and parental taxa in this system appears to be mostly of epigenetic---rather than physical---in nature, and it seems to be triggered by hybridization rather than genome duplication (see also Salmon & al., 2005). In Nicotiana (Lim & al., 2004) the degree of “genomic shock” in allopolyploids---i.e., how divergent were the parental taxa at hybridization---seems to influence the presence of chromosome translocations. Indeed, whereas N. tobacum, which originated from widely diverged parental species, shows such genomic reorganisation, no translocations were observed in N. arentsii and N. rustica.
DNA sequence elimination (including gene deletion)
Another consequence of allopolyploid formation, possibly associated with chromosomal rearrangements, is the rapid and reproducible genome-wide removal of some, but not all redundant DNA sequences from hybrid genomes (e.g. in wheat, Shaked & al., 2001; but see He & al., 2003). This process appears to represent a mainly non-random mechanism (Paterson & al., 2006) resulting in differentiation of homoeologous chromosomes, leading to correct meiotic pairing in hybrids. In Tragopogon miscellus (Tate & al., 2006), allopolyploids recurrently formed within the last 80 years, and most of the genomic changes seem to be represented by stochastic sequence loss, rather than gene silencing. Recent studies in maize (e.g., Lai & al., 2004) have also reported contractions in the allopolyploid genome via physical or epigenetic loss of ca. half of all duplicated genes in the roughly 11 million years following genome duplication. Furthermore, it has been demonstrated that, like homoploid hybrids, allopolyploids can achieve results similar to “transgressive segregation” through deletion of less-adaptive or maladaptive loci as the polyploids become progressively more diploidized (Wu & al., 2006), permitting them to occupy habitats unavailable to either parent.
Transposable element activation
Some studies have also shown polyploidy-induced demethylation and activation of dormant mobile elements in newly formed allopolyploids (e.g., Kashkush & al., 2003; Liu & Wendel, 2003; Madlung & al., 2005) as a consequence of genomic instability or “genomic shock” (McClintock, 1984). For example, activation of transposons appears to have occurred in some allopolyploid lines of Arabidopsis (Madlung & al., 2005). In newly synthesized wheat allopolyploids, Kashkush & al. (2003) showed that activation of certain transposable elements can result in silencing of the adjacent downstream genes. Active transposable elements have the potential for insertional mutagenesis and changes in phenotype while altering local patterns of gene expression, but they may also facilitate rapid genomic reorganisation in new polyploids. Because polyploids contain duplicate copies of all genes, transposon insertions into “single” copy genes are less deleterious; thus, transposable elements can multiply and persist for far longer in polyploids than in diploids (Hegarty & Hiscock, 2005).
Gene expression alterations of epigenetic nature (including gene silencing)
The classical model of genome evolution (Ohno, 1970) predicts that in polyploids homoeologous gene copies shared between parental genomes may lead to gene silencing and loss over time. The spectrum of duplicated gene fates seems to range between buffering of vital functions (perhaps via homogenization, Chapman & al., 2006) and functional divergence (see below). Indeed, transcriptional analyses using techniques such as cDNA-AFLP or cDNA-SSCP have confirmed that, in practice, duplicated genes are only rarely found to be expressed at equal levels; instead, there is an up- or down-regulation of one of the duplicated genes, resulting in unequal expression or even epigenetic silencing of one copy. Silencing arising immediately after polyploid formation, in the absence of gene deletion, is probably epigenetically induced via cytosine methylation, a relatively easy, reversible process. Differential expression of genes in allopolyploid species has been demonstrated, for example, in Arabidopsis suecica (Comai & al., 2000), Triticum aestivum (He & al., 2003), multiple Gossypium hybrids (Adams & al., 2003), but seems to happen at a smaller scale than gene loss in Tragopogon mirus and T. miscellus (Soltis & al., 2004; Tate & al., 2006). These studies have also indicated that duplicated genes can be silenced immediately or in a period of only a few generations following genome doubling (Adams & al., 2003, 2004; Kashkush & al., 2003; Wang & al., 2004; Hegarty & al., 2005). For example, two genes in Arabidopsis polyploids were expressed in diploid A. thaliana, silenced in autotetraploid A. thaliana, and then rapidly reactivated following hybridization with A. arenosa in allopolyploids (Wang & al., 2004). Duplicate gene preservation is differential across gene classes and more likely when one member of such pairs diversifies in function (neofunctionalization) or when the gene pair has partitioned different components of the ancestral function (subfunctionalization, Adams & Wendel, 2005a, b; Comai, 2005), indicating one of the evolutionary roles of allopolyploidy. However, both copies can retain their original function over a long evolutionary time if the genes have allele-dosage effects because polyploidy increases potential variation in expression levels, providing selective advantages (De Bodt & al., 2005).
Many manifestations of parental “non-additivity” in gene expression are organ- and even tissue-specific (Adams & al., 2003, 2004; Otto, 2003), additionally indicating differential regulation of the two homoeologous combined genomes during plant development (Wang & al., 2004). For example, in Gossypium allopolyploids Adams & al. (2003) showed that some genes have been reciprocally silenced in different organs (i.e., one copy has been silenced in some organs and the other copy in other organs). Silencing of some specific duplicated genes in polyploids can be a directed, repeatable process in synthetic or natural allopolyploids with multiple origins (from the same parental taxa), potentially reflecting parental effects, but this might also arise from parental polymorphism or a more stochastic process for other genes (Adams & al., 2003, 2004; He & al., 2003; Wang & al., 2004). Soltis & al. (2004) observed, using cDNA-AFLP, expression variation in gene-silencing patterns between natural allopolyploid Tragopogon populations that have reciprocal parentage. In another case, allotetraploid North American Nicotiana (Wu & al., 2006), a new anti-herbivore defense system evolved after polyploidy, in which the signaling system retained was that of one parent, but the defense system was derived from the other parent (and the other copies of these loci were physically deleted).
Concluding remarks and future perspectives
Important advances have been made in hybridization and polyploidy research, and this has helped us recognize their major implications in evolution of plants, especially by promoting genetic diversity, adaptive radiation and speciation. Further insights are expected to come from investigations of genomic organization and the transcriptome, which will enable a deeper understanding of the processes behind these events. Some issues on which further work is needed include: the direction of genetic diploidization after hybridization and chromosome doubling, including an improved understanding of which parts of this process are stochastic and which directed (repeatable); functional relevance of correlations between gene expression and development of a phenotype; molecular driven adaptation of pairs of related hybrids to different ecologies/habitats; and the ecological and evolutionary implications of recurrent polyploidization.
Recently formed polyploids (neopolyploids, see Soltis & al., 2004) provide a rich opportunity for observing the earliest changes in polyploid genome structure in an ecological context using natural populations. Post-glacially formed hybrids are particularly under-investigated, although they might provide information on how these changes become stabilized over time and what is the influence of historical evolutionary patterns in these processes (e.g., migration, bottleneck effects). Similarly, comparisons of diploids with related polyploid hybrids as well as with non-hybrids would enable assessments of the relative importance of hybridization and polyploidization. Finally, an important tool for studying genome-wide consequences of hybridization and polyploidization is also offered by natural recurrent polyploidy.
It is now recognized that multiple origins are the rule for most polyploids (Soltis & al., 2003), and hybridization has been shown to occur repeatedly between different individuals of the same parental taxa, often in disjunct localities (Soltis & Soltis, 2000; Hegarty & Hiscock, 2005; Hegarty & al., 2005). Therefore, many polyploid species of plants are polytopic, having arisen multiple times from the same parental species. This is the case, for example in Tragopogon and Glycine (Doyle & al., 2004; Soltis & al., 2004), in which hybridization in different diploid populations combined with polyploidization to produce arrays of allopolyploids. Indeed, there are few examples of polyploid plants for which a single origin is likely (but see, e.g., Ainouche & al., 2004; Paun & al., 2006). Recurrent formation can create an array of genetically, ecologically, morphologically and physiologically distinct genotypes/populations, among which subsequent gene flow, independent assortment and recombination may produce additional variation (Soltis & Soltis, 1999, 2000). Moreover, in allopolyploid populations of recurrent origin, “reciprocal silencing” of duplicated genes may lead to reproductive isolation and speciation (Soltis & al., 2004). Differential epigenetic modifications of homoeologous genes in polyploid populations, particularly among populations of separate origin, may contribute to the molecular basis of natural variation and therefore play an important evolutionary role (Soltis & al., 2004; Comai, 2005). Nevertheless, the long-term evolutionary significance of recurrent polyploid formation is unclear. Furthermore, investigating whether genetic and genomic alterations depend on which genome was maternally inherited by the hybrid would indicate whether imprinting and/or cytoplasmic factors are involved.
The analysis of the transcriptome is among the most modern and advanced strategies to provide information on how plasticity of gene expression can respond to hybridization and polyploidization events, and it can specifically identify transcripts that are differentially expressed in and among such taxa compared to their parents. Understanding these factors is of great importance for understanding the role of hybridization and polyploidy in evolution, and it is especially relevant in many plant groups in which hybridization has been widespread over a long evolutionary timeframe.
Acknowledgements
Financial support from the Austrian Science Foundation (FWF) for an Erwin Schrödinger fellowship to OP (project no. J26406-B03) and from NSF to DES (MCB - 0346437 DEB-0089483) is acknowledged.
Footnotes
For an extended review on molecular markers from the transcriptome see Gupta & Rustgi (2004).
Literature cited
- Aagaard SM, Sastad SM, Greilhuber J, Moen A. A secondary hybrid zone between diploid Dactylorhiza incarnata ssp. cruenta and allotetraploid D. lapponica (Orchidaceae) Heredity. 2005;94:488–496. doi: 10.1038/sj.hdy.6800643. [DOI] [PubMed] [Google Scholar]
- Adams KL, Cronn RC, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA. 2003;100:4649–4654. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams KL, Wendel JF. Novel patterns of gene expression in polyploid plants. Trends Genet. 2005a;21:539–543. doi: 10.1016/j.tig.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr. Opin. Pl. Biol. 2005b;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Ainouche ML, Baumel A, Salmon A. Spartina anglica Hubbard: a natural model system for analysing early evolutionary changes that affect allopolyploid genomes. Biol. J. Linn. Soc. 2004;82:475–484. [Google Scholar]
- Ainouche ML, Baumel A, Salmon A, Yannic G. Hybridization, polyploidy and speciation in Spartina (Poaceae) New Phytol. 2003;161:165–172. [Google Scholar]
- Anderson E, editor. Introgressive Hybridization. Wiley; New York: 1949. [Google Scholar]
- Arnold ML, editor. Natural Hybridization and Evolution. Oxford University Press; Oxford: 1997. [Google Scholar]
- Bachem CWB, van der Hoeven RS, de Bruijn SM, Vreugdenhil D, Zabeau M, Visser RGF. Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Pl. J. 1996;9:745–753. doi: 10.1046/j.1365-313x.1996.9050745.x. [DOI] [PubMed] [Google Scholar]
- Bachem CWB, Oomen RJFJ, Visser RGF. Transcript imaging with cDNA-AFLP: a step-by-step protocol. Pl. Molec. Biol. Report. 1998;16:157–173. [Google Scholar]
- Barton NH. The role of hybridization in evolution. Molec. Ecol. 2001;10:551–568. doi: 10.1046/j.1365-294x.2001.01216.x. [DOI] [PubMed] [Google Scholar]
- Bell MA, Travis MP. Hybridization, transgressive segregation, genetic covariation and adaptive radiation. Trends Ecol. Evol. 2005;20:358–361. doi: 10.1016/j.tree.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distribution of duplicated genes. Pl. Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, et al. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Breyne P, Dreesen R, Cannoot B, Rombaut D, Vandepoele K, Rombauts S, Vanderhaeghen R, Inzé D, Zabeau M. Quantitative cDNA-AFLP analysis for genome-wide expression studies. Molec. Genet. Genom. 2003;269:173–179. doi: 10.1007/s00438-003-0830-6. [DOI] [PubMed] [Google Scholar]
- Chapman BA, Bowers JE, Feltus FA, Paterson AH. Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiosperm genome duplication. Proc. Natl. Acad. Sci. USA. 2006;103:2730–2735. doi: 10.1073/pnas.0507782103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Wang J, Tian L, Lee HS, Wang JJ, Chen M, Lee JJ, Josefsson C, Madlung A, Watson B, Lippman Z, Vaughn M, Pires JC, Colot V, Doerge RW, Martienssen RA, Comai L, Osborn TC. The development of Arabidopsis model system for genome-wide analysis of polyploidy effects. Biol. J. Linn. Soc. 2004;82:689–700. doi: 10.1111/j.1095-8312.2004.00351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan A, Eichler EE, Oliver SG, Paterson AH, Stein L. Chromosome evolution in eukaryotes: a multi-kingdom perspective. Trends Ecol. Evol. 2005;21:673–682. doi: 10.1016/j.tig.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens Y, Byers B. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Pl. Cell. 2000;12:1551–1567. doi: 10.1105/tpc.12.9.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Wall PK, Leebens-Mack JH, Lindsay BG, Soltis DE, Doyle JJ, Soltis PS, Carlson JE, Arumuganathan K, Barakat A, Albert VA, Ma H, dePamphilis CW. Widespread genome duplications throughout the history of flowering plants. Genom. Res. 2006;16:738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Maere S, Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol. Evol. 2005;20:591–597. doi: 10.1016/j.tree.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Donson J, Fang Y, Espiritu-Santo G, Xiang W, Salazar A, Miyamoto S, Armendarez V, Volkmuth W. Comprehensive gene expression analysis by transcript profiling. Pl. Molec. Biol. 2002;48:75–97. [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL, Rauscher JT, Brown AHD. Evolution of perennial soybean polyploid complex (Glycine subgenus Glycine): a study of contrasts. Biol. J. Linn. Soc. 2004;82:583–597. [Google Scholar]
- Dunwell JM, Moya-León MA, Herrera R. Transcriptome analysis and crop improvement (a review) Biol. Res. 2001;34:153–164. doi: 10.4067/s0716-97602001000300003. [DOI] [PubMed] [Google Scholar]
- Gibson G. Microarrays in ecology and evolution: a preview. Molec. Ecol. 2002;11:17–24. doi: 10.1046/j.0962-1083.2001.01425.x. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Rustgi S. Molecular markers from the transcribed/expressed region of the genome in higher plants. Funct. Integr. Genom. 2004;4:139–162. doi: 10.1007/s10142-004-0107-0. [DOI] [PubMed] [Google Scholar]
- He P, Friebe BR, Gill BS, Zhou JM. Allopolyploidy alters gene expression in the highly stable hexaploid wheat. Pl. Molec. Biol. 2003;52:401–414. doi: 10.1023/a:1023965400532. [DOI] [PubMed] [Google Scholar]
- Hegarty MJ, Hiscock SJ. Hybrid speciation in plants: new insights from molecular studies. New Phytol. 2005;165:411–423. doi: 10.1111/j.1469-8137.2004.01253.x. [DOI] [PubMed] [Google Scholar]
- Hegarty MJ, Jones JM, Wilson ID, Barker GL, Coghill JA, Sanchez-Baracaldo P, Liu G, Buggs RJA, Abbott RJ, Edwards KJ, Hiscock SJ. Development of anonymous cDNA microarrays to study changes to the Senecio floral transcriptome during hybrid speciation. Molec. Ecol. 2005;14:2493–2510. doi: 10.1111/j.1365-294x.2005.02608.x. [DOI] [PubMed] [Google Scholar]
- Ji LH, Langridge P. The genetic control of chromosome pairing in wheat. Funct. Pl. Biol. 1990;17:239–251. [Google Scholar]
- Kammenga JE, Herman MA, Ouborg NJ, Johnson L, Breitling R. Microarray challenges in ecology. Trends Ecol. Evol. 2007;22:273–279. doi: 10.1016/j.tree.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 2003;33:102–106. doi: 10.1038/ng1063. [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Pl. J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Kuhn E. From library screening to microarray technology: strategies to determine gene expression profiles and to identify differentially regulated genes in plants. Ann. Bot. 2001;87:139–155. doi: 10.1006/anbo.2000.1314. [DOI] [PubMed] [Google Scholar]
- Lai J, Ma J, Swigonová Z, Ramakrishna W, Linton E, Llaca V, Tanyolac B, Park Y-J, Jeong O-Y, Bennetzen JL, Messing J. Gene loss and movement in the maize genome. Genome Res. 2004;14:1924–1931. doi: 10.1101/gr.2701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Quere A, Schutzendubel A, Rajashekar B, et al. Divergence in gene expression related to variation in host specificity of an ectomycorrhizal fungus. Molec. Ecol. 2004;13:3809–3819. doi: 10.1111/j.1365-294X.2004.02369.x. [DOI] [PubMed] [Google Scholar]
- Levy AA, Feldman M. Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biol. J. Linn. Soc. 2004;82:607–613. [Google Scholar]
- Lim KY, Matyasek R, Kovarik A, Leitch AR. Genome evolution in allotetraploid Nicotiana. Biol. J. Linn. Soc. 2004;82:599–606. [Google Scholar]
- Lippman ZB, Zamir D. Heterosis: revisiting the magic. Trends Genet. 2007;23:60–66. doi: 10.1016/j.tig.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Liu B, Brubaker CL, Mergeai G, Cronn RC, Wendel JF. Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome. 2001;44:321–330. [PubMed] [Google Scholar]
- Liu B, Wendel JF. Epigenetic phenomena and the evolution of plant allopolyploids. Molec. Phylog. Evol. 2003;29:365–379. doi: 10.1016/s1055-7903(03)00213-6. [DOI] [PubMed] [Google Scholar]
- Madlung A, Tyagi AP, Watson B, Jiang H, Kagochi T, Doerge R, Martienssen R, Comai L. Genomic changes in synthetic Arabidopsis polyploids. Pl. J. 2005;41:221–230. doi: 10.1111/j.1365-313X.2004.02297.x. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y-J, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Ohno S, editor. Evolution by Gene Duplication. Springer-Verlag; Berlin: 1970. [Google Scholar]
- Otto SP. In polyploids, one plus one does not equal two. Trends Ecol. Evol. 2003;18:431–433. [Google Scholar]
- Paterson AH, Chapman BA, Kissinger JC, Bowers JE, Feltus FA, Estill JC. Many gene and domain families have congruent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet. 2006;12:597–602. doi: 10.1016/j.tig.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Paun O, Stuessy TF, Hoerandl E. The role of hybridization, polyploidization and glaciation in the origin and evolution of the apomictic Ranunculus cassubicus complex. New Phytol. 2006;171:223–236. doi: 10.1111/j.1469-8137.2006.01738.x. [DOI] [PubMed] [Google Scholar]
- Pikaard CS. Genomic change and gene silencing in polyploids. Trends Genet. 2001;17:675–677. doi: 10.1016/s0168-9525(01)02545-8. [DOI] [PubMed] [Google Scholar]
- Pires JC, Zhao J, Schranz ME, Leon EJ, Quijada PA, Lukens L, Osborn TC. Flowering time divergence and genomic rearrangements in resynthesized polyploids (Brassica) Biol. J. Linn. Soc. 2004;82:675–688. [Google Scholar]
- Pontes O, Neves N, Silva M, Lewis MS, Madlung A, Comai L, Viegas W, Pikaard CS. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl. Acad. Sci. USA. 2004;101:18240–18245. doi: 10.1073/pnas.0407258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Machado CM. Uncovering evolutionary patterns of gene expression using microarrays. Trends Ecol. Evol. 2006;21:29–37. doi: 10.1016/j.tree.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Archer AM, Wayne RK. Transgressive segregation, adaptation and speciation. Heredity. 1999;83:363–372. doi: 10.1038/sj.hdy.6886170. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Raymond O, Resenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, Lexer C. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- Salmon A, Ainouche ML, Wendel JF. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae) Molec. Ecol. 2005;14:1163–1175. doi: 10.1111/j.1365-294X.2005.02488.x. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Pl. Cell. 2001;13:1749–1759. doi: 10.1105/TPC.010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. Molecular data and the dynamic nature of polyploidy. 1993;12:243–273. [Google Scholar]
- Soltis DE, Soltis PS. Polyploidy: recurrent formation and genome evolution. Trends Ecol. Evol. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Pires JC, Kovarik A, Tate JA, Mavrodiev E. Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biol. J. Linnean Soc. 2004;82:485–501. [Google Scholar]
- Soltis PS, Soltis DE. The role of genetic and genomic attributes in the success of polyploids. Proc. Natl. Acad. Sci. USA. 2000;97:7051–7057. doi: 10.1073/pnas.97.13.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins G. Chromosome variation and evolution. Science. 1966;152:1463–1469. doi: 10.1126/science.152.3728.1463. [DOI] [PubMed] [Google Scholar]
- Tate JA, Ni Z, Scheen A-C, Koh J, Gilbert CA, Lefkowitz D, Chen ZJ, Soltis PS, Soltis DE. Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocically formed allopolyploid. Genetics. 2006;173:1599–1611. doi: 10.1534/genetics.106.057646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian L, Madlung A, Lee HS, Chen M, Lee JJ, Watson B, Kagochi T, Comai L, Chen ZJ. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics. 2004;167:1961–1973. doi: 10.1534/genetics.104.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Pl. Molec. Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- Wendel JF, Doyle JJ. Polyploidy and evolution in plants. In: Henry RJ, editor. Plant Diversity and Evolution. CABI Publishing; Wallingford, UK: 2005. pp. 97–117. [Google Scholar]
- Wolfe KH. Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Baldwin I. Evolution of proteinase inhibitor defenses in North American allopolyploid species of Nicotiana. Planta. 2006 doi: 10.1007/s00425-006-0256-6. doi: 10.1007/s00425-006-0256-6. [DOI] [PubMed] [Google Scholar]
- Xiao N-Z, Ba L, Holm PB, Wang X-Z, Bowra S. Quantitative transcript analysis in plants: improved first-strand cDNA synthesis. Acto Biochim. Biophis. Sin. 2005;37:429–434. doi: 10.1111/j.1745-7270.2005.00052.x. [DOI] [PubMed] [Google Scholar]