Abstract
The Rvb1 and Rvb2 proteins are two members of the AAA+ family that are involved in roles as diverse as chromatin remodeling, transcription, small nucleolar RNA maturation, cellular transformation, signaling of apoptosis and mitosis. These proteins are capable of playing a role in such diverse cellular processes because they are components of different macromolecular assemblies. In the last few years, there has been a number of groups reporting on the structure of purified Rvbs. The reported results have been rather controversial because there are significant differences observed among the published structures in spite of the high degree of homology among these proteins. Surprisingly, contradictions are observed not only between structures representing the Rvb proteins from different species, but also between protein structures from the same species. This review describes the available Rvb structures from the different species and makes also a comparative analysis of them. Finally, we identify some aspects of these structural studies worth pursuing additional investigations to ensure that the reported structures reflect physiologically relevant conformations of the Rvb1/Rvb2 complex.
Keywords: AAA+, Rvb1, Rvb2, Pontin, Reptin
Introduction
Proteins members of the AAA+ family share similarity both in sequence and structure (Lupas and Martin 2002; Ogura and Wilkinson 2001; Snider and Houry 2008; Snider et al. 2008). The large variety of cellular activities in which AAA+ proteins are involved (Hanson and Whiteheart 2005) was in fact the unifying theme used to name this family of proteins. AAA+ stands for ‘ATPases associated with diverse cellular processes’ and although as a family of proteins, they are involved in a large number of cellular functions, it is usually found that the role of each specific protein member is typically confined to a particular cellular process (Ogura and Wilkinson 2001).
The Rvb1 and Rvb2 proteins are two members of the AAA+ family that do not follow this pattern, as they are involved in multiple roles as diverse as chromatin remodeling, transcription, small nucleolar RNA maturation, cellular transformation and signaling of apoptosis (Ikura et al. 2000; Shen et al. 2000). These proteins are capable of playing a role in such diverse cellular processes because they are components of different macromolecular assemblies (see accompanying paper from Huen et al. in this issue). In particular, the two proteins are found in large chromatin remodeling complexes such as INO80 (Bakshi et al. 2006; Jin et al. 2005; Shen et al. 2000), p400 (Fuchs et al. 2001) and SWR1 (Krogan et al. 2003; Mizuguchi et al. 2004). They are also essential components of chromatin modifying enzymes such as the TIP60 complex that display histone acetylase activity. In addition, the identification of Rvb proteins in the RNA polymerase II complex (Qiu et al. 1998) and in small nucleolar ribonucleoprotein complexes (snoRNPs) (King et al. 2001; Newman et al. 2000; Zhao et al. 2008) mediates their role in transcriptional processes and nucleolar localization as well. Finally, the participation of the Rvb proteins in cellular transformation and apoptosis is a consequence of their interaction with c-Myc (Wood et al. 2000) and β-catenin (Bauer et al. 2000). In the last few years, the repertoire of cellular activities has been expanded even further upon the observation that these proteins are also associated to the mitotic spindle and centrosomes through a tubulin-mediated interaction (Gartner et al. 2003). These results indicate a role of these proteins in mitosis.
Given their physiological relevance, the Rvb proteins have been found to be essential in Saccharomyces cerevisiae (Qiu et al. 1998), Drosophila melanogaster (Bauer et al. 2000), Caenorhabditis elegans (Matias et al. 2006) and it is likely that they are also essential in mammalian cells. Probably, the variety of cellular processes in where these proteins are involved explains the multiplicity of names given to the Rvb proteins in the literature. Rvb1 is also known as RuvBL1, Tip49a, Tip48/Tip49, ECP-54, Pontin, Tih1, p50 and Tap54β, while Rvb2 has been named RuvBL1, Tip49b, Tip49/Tip48, ECP-51, Reptin, Tih2, p47 and Tap54α.
In the last few years there has been a number of groups reporting on the structure of purified Rvbs (Table 1). In 2006, the X-ray structure of one of the human Rvb proteins (RuvBL1) assembled as a hexameric ring was reported (Matias et al. 2006). A few months later, an electron microscopy (EM) study showed the structure of a complex containing the two human Rvb proteins (RuvBL1 and RuvBL2) that assembled as a double hexamaric ring (Puri et al. 2007). Finally, in 2008 two independent groups published structural information regarding the complex formed by the yeast proteins (Rvb1 and Rvb2) (Gribun et al. 2008; Torreira et al. 2008). The reported results have been rather controversial because in spite the high degree of homology among these proteins, there are significant differences observed among the published structures. Surprisingly, contradictions are observed not only between structures representing the Rvb proteins from different species, but also between protein structures from the same species. This review is a comparative analysis of the available structural information and proposes a number of research avenues to elucidate whether the observed differences simply reflect multiple physiologically relevant functional states of these proteins or they are caused by differences in the methodology used in the studies.
Table 1.
Structural method used for determination of the Rvb protein structures
| Structure | Method for Structural Determination |
|---|---|
| Human Rvb1 (Matias et al. 2006) | X-ray crystallography |
| Human Rvb1/Rvb2 complex (Puri et al. 2007) | Negative staining electron microscopy and single particle reconstruction methods |
| Yeast Rvb1/Rvb2 complex (Gribun et al. 2008) | Negative staining electron microscopy and single particle reconstruction methods |
| Yeast Rvb1/Rvb2 complex (Torreira et al. 2008) | Cryo-electron microscopy and single particle reconstruction methods |
Structure of the human Rvb proteins
The X-ray structure of the human Rvb1 (Matias et al. 2006) and the structure of the human Rvb1/Rvb2 complex obtained from negatively stained electron micrographs (Puri et al. 2007) were reported only a few months apart at the end of 2006 and the beginning of 2007, respectively. This section describes these two structures.
X-ray structure of the human Rvb1 protein
The X-ray structure of human Rvb1 monomer (Matias et al. 2006) shows a protein folded into three domains named from DI to DIII (Fig. 1A). The structure contains the typical AAA+ module that is made from domains DI and III (Fig. 1A). The first one constitutes the P-loop αβα nucleotide-binding core domain and consists of the typical five-stranded β-sheet sandwiched between α-helices. The folding of DIII is also similar to many other AAA+ proteins and it is formed by a bundle of α-helices. The Rvb1 protein in the structure is complexed with an ADP molecule (Fig. 1A), which provides information about how the predicted elements of the AAA+ module for nucleotide binding and hydrolysis relate to the nucleotide molecule (Fig. 1B). The conserved P-loop (Walker A motif) important for binding and orientation of the nucleotide for hydrolysis, the Walker B motif necessary for nucleotide hydrolysis and the sensor domains (sensor 1 and 2) that also interact with the nucleotide molecule and assist in several aspects of the nucleotide recognition and hydrolysis are all in a functionally compatible conformation. The Walker A and B motifs as well as sensor 1 domain are located in DI while sensor 2 domain is placed in DIII (Fig. 1B). Superimposing the Rvb1 coordinates with those from RuvB (Putnam et al. 2001), NSF-D2 (Lenzen et al. 1998) and SV40 large tumor antigen helicase (Li et al. 2003) allowed the authors to predict the specific residues within each element of the AAA+ module involved in nucleotide binding and hydrolysis. However, a mutational analysis verifying these predictions is still not available.
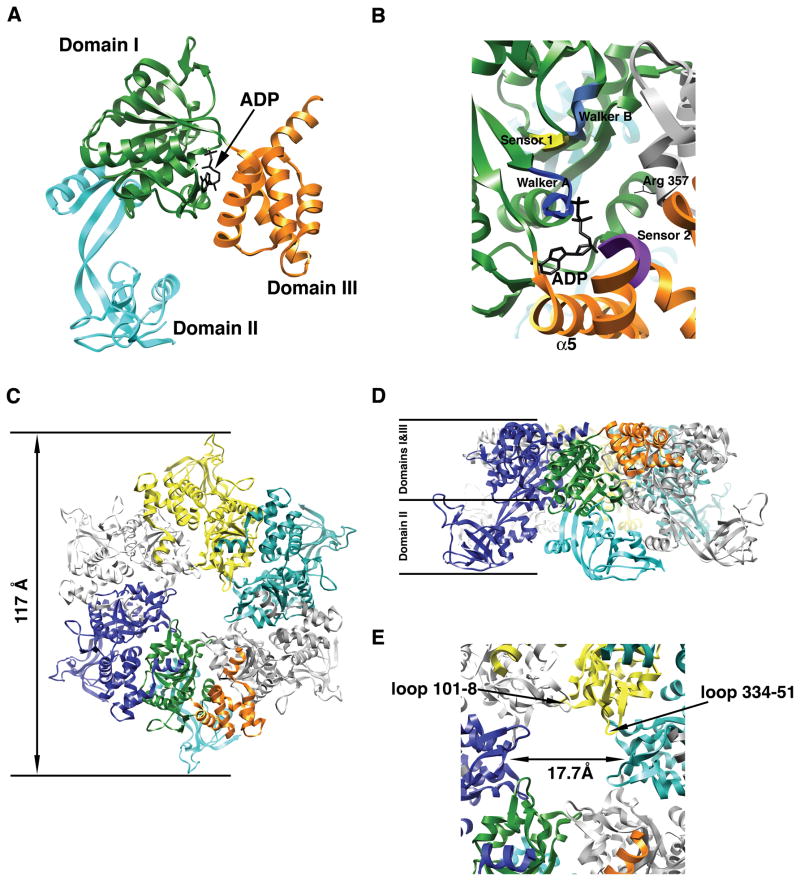
Figure 1. X-ray structure of the human Rvb1 protein.
(A) Ribbon representation of the monomer of human Rvb1 in complex with an ADP molecule. Each domain is represented in a different color. (B) Closed up view of the nucleotide-binding site in the human Rvb1 structure. The elements of the AAA+ module important for nucleotide binding and hydrolysis are represented in different colors and labeled. Domains within the monomer are color coded as in Fig. 1A and the neighboring subunit contributing the “arginine finger” (Arg-357) is colored in grey. (C) Top-view of the ribbon representation of the hexameric ring formed by the human Rvb1 protein in the crystal structure. Each monomer is represented in a different color except the monomer at the bottom that follows the same color code as Fig. 1A for the domains. (D) Side-view of the ribbon representation of the human Rvb1 hexamer. The location of domains I, II and III from the Rvb1 monomers in the hexamer is indicated. Each monomer is represented in a different color and the monomer at the front has its domains colored as in Fig. 1A. (E) Close-up view of the central pore in the human Rvb1 hexamer. The diameter of the pore is indicated and also the two loops within one monomer that are predicted to mediate the binding of ssDNA.
In the crystal structure, the Rvb1 protein oligomerizes as a hexameric ring (Fig. 1C&D), which is also a hallmark of AAA+ proteins (Neuwald et al. 1999). Similarly to other proteins of this family, the ADP molecule was found sandwiched between two of the subunits of the ring. This particular location of the nucleotide molecule in AAA+ proteins provides the opportunity to neighboring subunits in the ring to contribute to the binding and hydrolysis of the ATP usually through an arginine residue that contributes to the nucleotide catalysis (Davey et al. 2003; Johnson and O’Donnell 2003; Karata et al. 1999; Rombel et al. 1999). In the Rvb1 protein, Arg-357 in DI is close enough to the nucleotide binding pocket of the adjacent subunit to act as the “arginine finger” (Fig. 1B).
An important consequence of the oligomerization of Rvb1 into a hexamer is that the ADP molecule bound to the protein becomes completely buried into the structure. One helix in DIII (α5), sensor 2 and the neighboring monomer completely blocks the nucleotide binding pocket making the release of ADP impossible (Fig. 1B), which is consistent with the lack of ATPase activity experimentally observed for human Rvb1 protein in the study from Matias et al. (Matias et al. 2006). These observations support the idea that a conformational change is necessary during the nucleotide hydrolysis cycle for the ADP to ATP exchange. Interestingly, nucleotide induced conformations have been described in the yeast Rvb1/Rvb2 complex (Gribun et al. 2005; Torreira et al. 2008) (see below). The observation that the ATPase activity of the yeast Rvb1/Rvb2 complex (Gribun et al. 2008) showed a significantly higher ATPase activity than human Rvb1 alone (Matias et al. 2006) supports the hypothesis that the presence of Rvb2 in the complex is required to induce the conformational change allowing for nucleotide exchange.
The Walker A and B motifs are usually closely spaced in the sequence of AAA+ proteins. However, in the Rvb proteins there is a long insertion of ~170 amino acids between these two motifs. In the native protein, the Walker A and B motifs still come together to form the nucleotide binding site but the 170 residues insertion domain protrude from DI and constitute the DII domain with an OB fold like structure mostly formed by β–strands (Fig. 1A). The resemblance of this domain in structure and surface charges to the DNA binding domain of other proteins involved in DNA metabolism is remarkable. Matias et al. (Matias et al. 2006) showed that human Rvb1 binding of single (ssDNA) and double (dsDNA) stranded DNA as well as single stranded RNA is mediated by DII. Surprisingly, the protein was found to lack in vitro helicase activity most likely as a consequence of its deficient ATPase activity.
Even so, this crystal structure does not explain how the interaction with nucleic acids may occur in the physiological complex, it certainly provides an initial framework to propose hypothesis for further testing. For instance, it is clear that the diameter of the central channel, which is known to be in many hexameric AAA+ proteins the main nucleic acid interaction site, in the human Rvb1 hexamer (17.7 Å) is too narrow for double stranded nucleic acids to pass through (Fig. 1E). In addition, the negative electrostatic potential of the inner surface of the channel makes it better suited for binding single stranded rather than doubled stranded that binds to wider and positively charged channels (Fletcher et al. 2003; Li et al. 2003). Finally, there are two loops facing the hexameric channel (residues 101–108 & 334–351) (Fig. 1E) that resemble the ones in several hexameric AAA+ proteins, such as the T7 gp4D helicase (Singleton et al. 2000) and that were found to directly mediate the binding of ssDNA. All of these structural characteristics are consistent with the model proposed by Matias et al. (Matias et al. 2006) suggesting that the human Rvb1 hexamer binds to forked DNA substrates. One strand of the fork will pass through the central channel whereas the second strand will bind to the outside of the ring in an interaction mediated by the DII domain. Similarly, binding to ssRNA in transcription events could be also mediated through the central channel in the hexamer. Nonetheless, it is clear that these protein-nucleic acid interactions are nonsequence-specific, which indicates that the protein-nucleic acid interaction may be mediated through the sugar-phosphate backbone of the DNA and RNA. However, it is a possibility that other proteins in the Rvb-containing complexes might dictate certain specificity in these interactions. Additional structures of the Rvb1 protein in complex with single stranded and doubled stranded nucleic acids are required to verify these interaction models.
Three-dimensional structure of the human Rvb1/Rvb2 complex by negative staining electron microscopy
The three-dimensional (3D) reconstruction of the human Rvb1/Rvb2 complex was obtained from negatively stained electron micrographs of an in vitro assembled complex (Puri et al. 2007). Independently purified human Rvb1 and Rvb2 proteins were mixed together and the mixture was further purified and fractionated using metal affinity and size exclusion chromatography. The fraction containing both proteins in equimolar amounts was used for the structural study.
The EM map reveals a double hexameric ring structure with a height of 116 Å (Fig. 2A) and a maximum diameter of 158 Å. The structure encloses a large cavity that opens to the outside through the axial channel that runs through the complex (Fig. 2B) and also through multiple small openings on the sides (Fig. 2A). The central channel is in principle wide enough (26 Å) to accommodate dsDNA (Fig. 2B). The most striking aspect of the structure is its asymmetric character as both rings differ significantly (Fig. 2C&D).
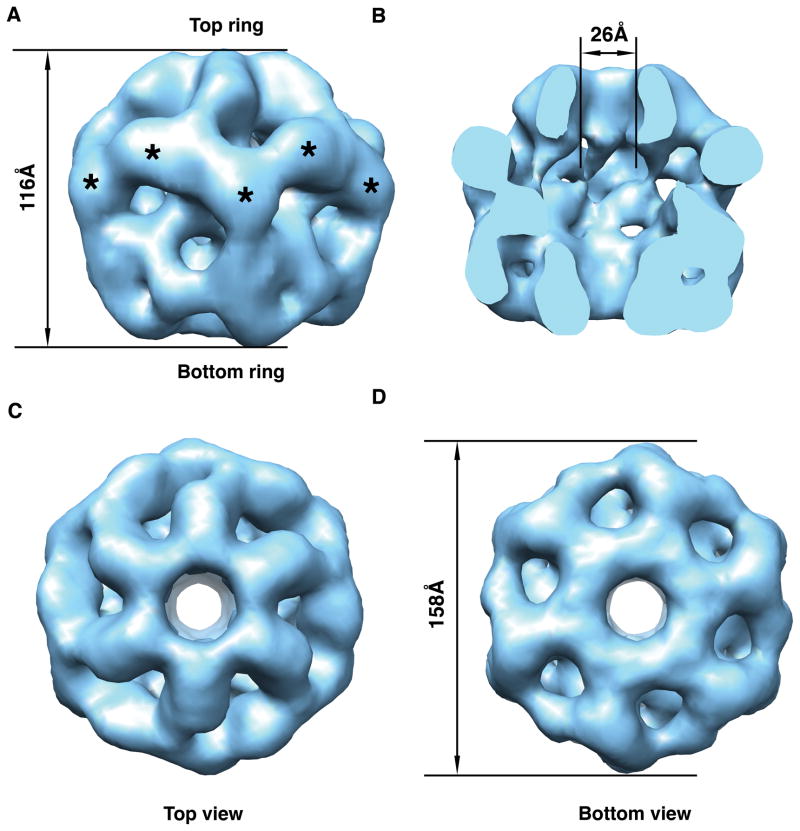
Figure 2. Three-dimensional structure of the human Rvb1/Rvb2 complex obtained from negative staining electron micrographs.
(A) Surface rendering representations of the side view of the structure. Top and bottom rings are defined arbitrarily. Asterisks indicate the putative location of the DII domains. (B) Surface representation of the inside of the complex. The front half of the structure has been removed to appreciate the cavity and the channel going through the structure. (C) & (D) Surface representations of the 3D structure showing the top and bottom view, respectively of the dodecameric structure.
The limited resolution of the structure does not allow to locate the Rvb1 and Rvb2 proteins within the complex. Considering the equimolar content of Rvb1 and Rvb2 in the complex, the different structure of the two rings could indicate that each ring is made exclusively by one of the proteins and thus, two different homomeric rings constitute the complex. Alternatively, the structure is also consistent with a model proposing that the complex is made out of two heteromeric rings of identical composition. In this case, the structural differences between rings would imply that they exist in a different conformation. Size exclusion chromatography experiments performed by Puri et al. (Puri et al. 2007) provide some support for the homomeric rings model. It was found that Rvb2 in the presence of nucleotide (ATP or ADP) and MgCl2 eluted in a peak corresponding to a molecular mass of ~400 kDa, which is slightly higher than the expected size of a Rvb2 hexamer (320 kDa). Conversely, Rvb1 always eluted as monomer regardless of the conditions tested including in the presence of nucleotide and MgCl2. The different ability of Rvb1 and Rvb2 to form stable oligomers by themselves might indicate that in the process of complex formation initially Rvb2 forms a hexameric ring that act as a scaffold promoting the hexamerization of Rvb1 to finally render the described double hexameric ring structure. Additional experiments are required to fully establish the layout of Rvb1 and Rvb2 proteins in the dodecameric complex.
Interestingly, the finding that in the size exclusion chromatography experiments Puri et al. (Puri et al. 2007) were able to assemble stable oligomers with Rvb2 but not with Rvb1 is surprising since the crystal structure of human Rvb1 suggests that this protein is also capable of forming higher order oligomers. The different protein concentration used in the crystallization trials and size exclusion chromatography experiments is probably the cause of this discrepancy.
Structure of the yeast Rvb proteins
Two studies were published in the year 2008 reporting structural information about the structure of the yeast Rvb proteins (Gribun et al. 2008; Torreira et al. 2008). Both studies used electron microscopy and single particle image processing methods. The main highlights of the structures reported are detailed below.
Two-dimensional projection structure of the yeast Rvb1/Rvb2 complex by negative staining electron microscopy
The first study addressing the question of the structural organization of the yeast Rvb1/Rvb2 complex was our work and we used negative staining EM and scanning transmission electron microscopy (STEM) (Gribun et al. 2008). In images obtained by STEM, the intensity at any pixel is directly proportional to the mass, providing quantitative information about the particle mass. This method was used in the study by Gribun et al. (Gribun et al. 2008) to establish that the yeast Rvb1/Rvb2 complex form single hexameric rings. Using this method it was determined that the distribution of masses of a large population of hexamers follows a Gaussian curve centered at the expected mass for a hexamar (~ 300 kDa).
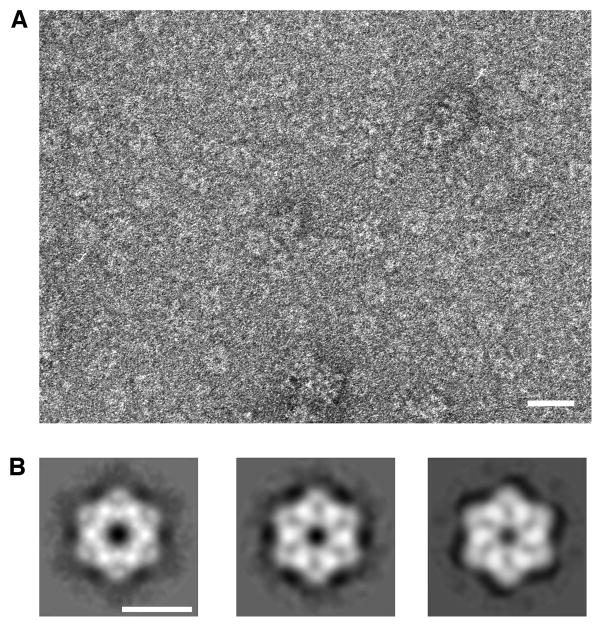
Negative staining images (Fig. 3A) were used to study the two-dimensional structure of the complex and the conformational changes that the hexameric rings undergo in the presence of different nucleotides. Two distinct conformations were identified. In the presence of ADP the ring-shaped particles form a hexagon with straight edges measuring ~142 Å between opposite vertices and a central channel of ~17 Å diameter (Fig. 3B; left panel). The two-dimensional projection structure of the complex in the presence of ATP (Fig. 3B; center panel) or ATPγS (Fig. 3B, right panel) showed dramatic structural differences compared to the ADP structure. In these two cases, the Rvb1/Rvb2 complex showed slightly bigger dimensions measuring ~152 Å between opposite vertices but a smaller (~15 Å diameter) central pore. The ATP and ATPγS hexagon had concave edges and some radial areas of stain penetration instead of the smooth stain-excluded area observed in the ADP form (Fig. 3B).
Figure 3. Two-dimensional structure of the yeast Rvb1/Rvb2 complex.
(A) Electron micrographs of negatively stained ring-shaped particles obtained upon incubation of the Rvb1 and Rvb2 proteins in the presence of ADP. Scale bar represents 200Å. (B) Two-dimensional averages of the yeast Rvb1/Rvb2 complex in the presence of ADP (left panel), ATP (center panel) and ATPγS (right panel). Scale bar represents 100Å.
The limited resolution of the EM averages does not allow differentiating the Rvb1 from the Rvb2 protein in the two-dimensional projection structures, hence whether the single rings are formed by only one (homomeric) or both (heteromeric) Rvb proteins is still unclear. In one hand, our finding that both Rvb1 and Rvb2 alone form large oligomers only at very high concentrations (40 μM) but oligomers are easily formed at low concentrations (5 μM) upon mixing both proteins support the hypothesis that the yeast Rvb1/Rvb2 complex is a single heteromeric hexameric ring (Gribun et al. 2008). Supporting the heteromeric model, we also found that the Rvb1 and Rvb2 mixed together have an enhanced ATPase and DNA helicase activity when compared to the individual proteins (Gribun et al. 2008). Although, an interpretation of these results is that homomeric rings of each protein might enhance the activity of the other protein by short-lived interaction in trans, these finding could also be seen as an indication that both the Rvb1 and Rvb2 proteins are present within the same ring and stimulation occurs in cis. An additional experiment reinforcing the hypothesis that the Rvb1/Rvb2 complex assembles as a heteromeric ring was the immunoprecipitation experiments performed in our study (Gribun et al. 2008). An affinity purified αRvb1 antibody was used to immunocapture the Rvb1 protein from an equimolar mixture of Rvb1 and Rvb2. In this experiment, Rvb2 was co-immunoprecipitated with Rvb1 strongly suggesting that there is a direct interaction between the two proteins within the complex. However, it could still be argued that the interaction detected in the immunoprecipitation experiment is due to the presence of non-specific aggregates between both proteins present in the incubation mixture to assemble the complex. Certainly, further analysis is necessary to clarify this aspect of the structure of yeast Rvb1/Rvb2 complex as well as the layout of the two proteins in the ring.
Three-dimensional structure of the yeast Rvb1/Rvb2 complex by cryo-electron microscopy
The structure of the yeast Rvb1/Rvb2 complex has been also studied by cryo-electron microscopy (Torreira et al. 2008). In this study, untagged Rvb1 was co-expressed with a N-terminal His-Rvb2 construct using recombinant baculovirus. Complexes were then purified using affinity and size exclusion chromatography.
The purified specimen was used to produce a high-resolution 3D reconstruction of the Rvb1/Rvb2 complex (Fig. 4). According to this study, the yeast Rvb1/Rvb2 complex organizes as an asymmetric double ring structure (Fig. 4A) enclosing an internal chamber (Fig. 4B). The asymmetry is produced because the top ring (as defined in Fig. 4A) is slightly taller than the lower (Fig. 4A & B). The total height of the complex is 130Å (Fig. 4A) and the diameter at the widest region is 135Å (Fig. 4D). The top and bottom of the EM map are compact ring structures probably formed by DI and DIII of the Rvb proteins where the AAA+ module is located. Six protein densities project from both the top and bottom ring towards the center of the structure forming two layers of discontinuous density (equatorial domains) that contain lateral openings to the internal chamber of the structure (Fig. 4A). It was proposed that the DII domains that protrude from the top and bottom ring intercalate and interact in these two layers of densities in the middle of the structure to maintain the integrity of the dodecameric structure (Fig. 4A). The internal chamber is also opened to the outside through an axial pore at each end of the structure. In this case, the diameter of the pore is ~25Å which is sufficient to accommodate dsDNA (Fig. 4B).
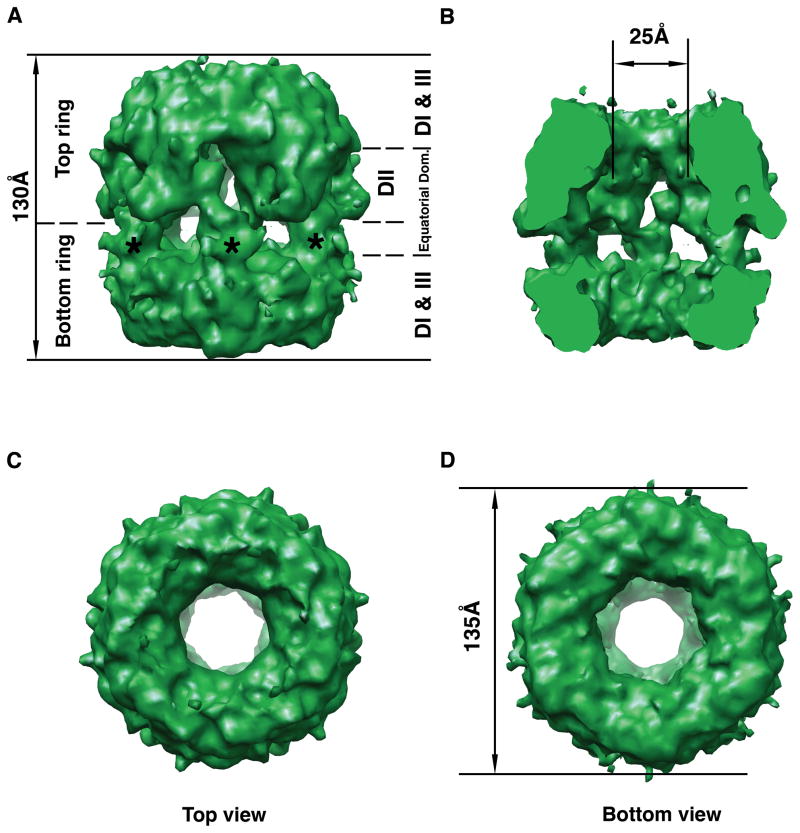
Figure 4. Cryo-electron microscopy 3D structure of the yeast Rvb1/Rvb2 complex.
(A) Side view of the three-dimensional structure of the Rvb1/Rvb2 complex. Top and bottom rings are indicated as well as the location of the DI, DII, DIII and equatorial domains of the monomers assembles within the dodecamer. The asterisks indicate the projected densities from the bottom ring into the equatorial domain. (B) The structure has been cut open to show the internal chamber, and the channel going through the structure. (C) Top view of the cryo-EM map. (D) Bottom view of the 3D EM map of the complex. The diameter of the structure is indicated.
Interestingly, each ring in the complex displays a different conformation mostly because of the disposition of its corresponding equatorial domain rather than the terminal ring. The projected densities from the bottom ring (Fig. 4A; asterisk) extend further than the corresponding ones in the top ring. Thus, the interactions between projections from opposite rings occurs mostly in the top equatorial domain resulting in a slightly wider layer of density with smaller lateral openings than the equatorial domain at the bottom (Fig. 4A).
The complex structurally characterized in the study (Torreira et al. 2008), contained equimolar amounts of Rvb1 and Rvb2. However, similarly to the EM structure of the human Rvb1/Rvb2 complex, it was not possible to establish the layout of each protein within the complex. The 3D reconstruction of the yeast Rvb1/Rvb2 complex is equally consistent with the existence of two homomeric hexameric rings (each one formed by one of the Rvb protein) or with an assembly formed by the stacking of two heteromeric rings containing three monomers of each protein. Antibody labeling experiments presented in the study (Torreira et al. 2008) support the former. However, given the very low efficiency of binding reported for the antibody used in these experiments, we think this question still remains unanswered.
The structure described by Torreira et al. (Torreira et al. 2008) incorporated only the 34% of the particles contained in the purified Rvb1/Rvb2 complex. The remaining particles did not classify and aligned correctly during the reconstruction process producing a double-ring structure with different level of resolution on each ring. Rejection of 66% of the data was necessary to obtain a self-consistent data set able to produce a 3D structure. The behavior of these particles in the reconstruction process was interpreted as an indication that the Rvb1/Rvb2 complex in the sample was present in more than one conformation. A 3D reconstruction from the rejected particles was problematic and structures were not produced in the study (Torreira et al. 2008). Thus, how these other putative conformations of the yeast Rvb1/Rvb2 complex may differ from the described structure or the degree of heterogeneity in the purified complex is still unanswered.
Comparison of the Rvb structures
A problem frequently faced in structural biology is the need to compare structures at very different levels of resolution. This is the case at hand here as some of the available structures contain atomic detail (X-ray model of Rvb1 (Matias et al. 2006)). Some other are three-dimensional structures but at low or moderate resolution (human (Puri et al. 2007) and yeast (Torreira et al. 2008) Rvb1/Rvb2 3D EM maps). Finally, one of the structures is only two-dimensional and solved to a limited resolution (yeast Rvb1/Rvb2 EM average projections (Gribun et al. 2008)). However, there are several appropriate approaches to successfully compare these structures.
Relevant to the structures reviewed here are the methods used to compare X-ray structures with EM density maps. An appropriate method to perform this analysis is to fit the X-ray model into the electron density map (Rossmann 2000; Rossmann et al. 2005). When two structures are in agreement, it is found that after fitting the structures, there is a corresponding electron density in the EM map for every part of the crystallographic model. In those cases where the EM structure is of limited resolution, such as the Rvb1/Rvb2 EM map analyzed here (Puri et al. 2007), the X-ray model should nicely fill the density envelope delineated by the EM structure.
It is also possible to compare atomic detail X-ray structures and high-resolution EM 3D structures to two-dimensional projection structure of limited resolution. In this case, it is required first to low-pass filter the X-ray and 3D EM structure to the resolution of the 2D projection and second the dimensionality of the 3D structures has to be reduced by calculating two-dimensional projections from the 3D structures at the particular view angles represented by the two-dimensional projection structure (Fig. 5).
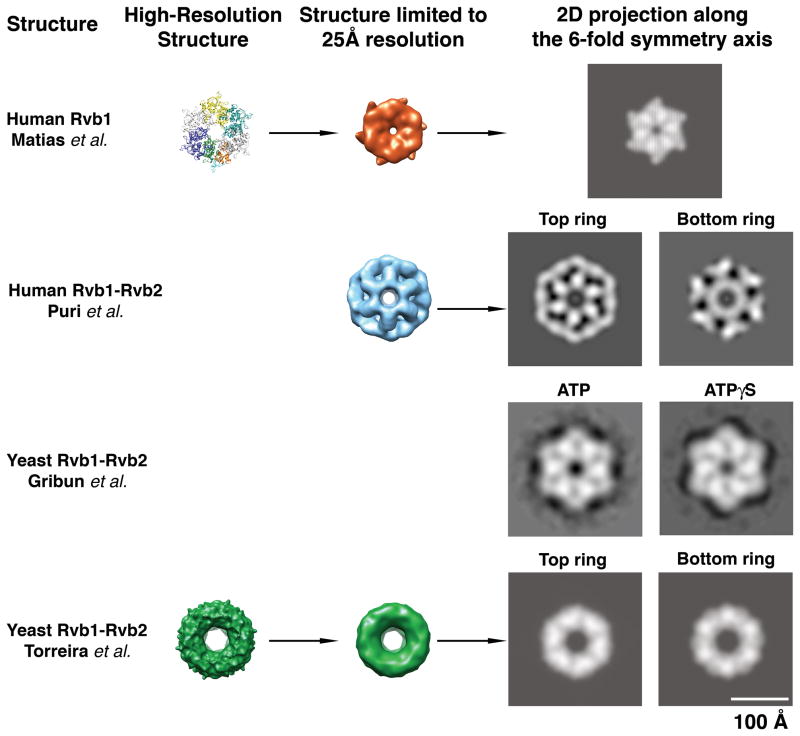
Figure 5. Comparison of the Rvb structures.
To compare the available Rvb structures, high-resolution models such as human Rvb1 and yeast Rvb1/Rvb2 complex were first limited to a resolution of 25Å by applying a Gaussian low-pass filter. A two-dimensional projection along the 6-fold symmetry axis was calculated from the limited resolution 3D structures. In the case of the double-ring EM structure of the human and yeast Rvb1/Rvb2 complexes independent projections were calculated for the top (“Top”) and bottom (“Bottom”) rings (as defined in Fig 2 and 4). The structures are represented at scale thus; their relative size is comparable. In the case of the yeast Rvb1/Rvb2 complex from Gribun et al. (Gribun et al. 2008) the images represent 2D averages of the Rvb1/Rvb2 complex in ATP (left panel) and ATPγS (right panel) obtained upon averaging several hundred 2D projections of these complexes.
Pairwise comparison between the X-ray structure of the human Rvb1 protein and the EM map of the human Rvb1/Rvb2 complex
The X-ray structure of human Rvb1 shows that the Rvb1 protein forms as a single hexameric ring in the crystal (Fig. 1C&D). However, the EM map represents a human Rvb1/Rvb2 complex assembled as a double hexameric ring structure (Fig. 2A). The Rvb1 single-ring X-ray structure and the observation that in the size exclusion chromatography experiments Rvb2 assembled in stable oligomers (Puri et al. 2007) is a strong indication that these proteins can assemble as single homo oligomeric rings suggesting that the human Rvb1/Rvb2 complex might be made of two homomeric rings. However, whether each individual ring in the EM structure is a homomeric or a heteromeric ring is still an open question and it requires further study.
Manual docking of the X-ray structure of the Rvb1 hexamer into the EM map to analyze the agreement between structures revealed that both structures seem incompatible. The elements of the AAA+ module in the crystal structure form a compact ring between subunits and with a flat top surface that contrast with the EM structure where each ring comprises domains separated by large gaps. The observed differences are not explained by the different resolution of the structures.
An additional point of discrepancy between the structures is regarding the diameter of the central channel in the structures and the accessibility of the DII domain in the single and double ring structures. These two characteristics of the assemblies probably have important implications of how these proteins mediate their interaction with nucleic acids. The diameter and the electrostatic character of the central pore in X-ray structure of the Rvb1 ring are consistent with ssDNA binding (Fig. 1E). However, the channel is wide enough to accommodate dsDNA in the EM structure of the Rvb1/Rvb2 complex (Fig. 2B). In the Rvb1/Rvb2 complex, DII domains are sandwiched between the two rings and their mobility is partially restricted through interactions with neighboring subunits (Fig. 2A; asterisks). Certainly a single ring structure, such as the hexamers formed by Rvb1 provides a less constrained and more accessible DII domain for nucleic acid interactions. Additional structures in complex with nuclei acids will clarify how the protein interacts with DNA and RNA.
Pairwise comparison between the X-ray structure of the human Rvb1 protein and the cryo-EM map of the yeast Rvb1/Rvb2 complex
The sequence identity between yeast Rvb1 and Rvb2 with human Rvb1 is of 69% and 43%, respectively. Therefore, it is expected that the X-ray structure of the human Rvb1 protein would fit well in cryo-EM density map of the yeast Rvb1/Rvb2 complex assuming that both structures are in a similar conformation.
Initially, Torreira et al. (Torreira et al. 2008) found that the atomic structure of the human Rvb1 hexamer fit poorly when docked as a rigid body into the ring densities at the ends of the cryo-EM map of the yeast Rvb1/Rvb2 complex. Subsequent rotation and outward translation of the human Rvb1 monomers within the EM density produced a good fitting for DI and DIII domains. DII domains from all the twelve monomers in the complex also reasonably matched the EM densities when fitted independently from the other two domains in the equatorial region closer to the top ring in the structure (Torreira et al. 2008) (Fig. 4A). Overall, these results indicate a good agreement between the X-ray structure of the human Rvb1 protein and the yeast Rvb1/Rvb2 complex. However, the unsuccessful initial rigid body fitting of the human Rvb1 hexamer in the cryo-EM map of the yeast Rvb1/Rvb2 hexamer implies that the cryo-EM map captured a conformational state of the complex that significantly differ from the X-ray structure of the human Rvb1 protein.
As indicated, in order to fit the X-ray structure into the cryo-EM map, the authors had to rotate each monomer in a counter-clock direction by approximately 5–10° and also to displace it outward creating a large central pore of sufficient diameter (~25Å) to allow threading of dsDNA (Fig. 4B). A caveat of such rotational and translational movement of the Rvb monomers is that it produces a displacement of the predicted “arginine finger” that is contributed to the nucleotide binding pocket by a neighboring subunit. Therefore, it follows that the structure of the yeast Rvb1/Rvb2 complex should undergo a conformational change from the described structure in order to properly place all the putative elements of the AAA+ module involved in nucleotide binding and hydrolysis.
How similar are the four available structures from human and yeast Rvb proteins?
As mentioned above, to compare structures of very different level of detail, it is necessary to reduce the resolution and dimensionality of all structures to the lowest value of resolution and dimensions found among the compared structures. Thus, in order to analyze how similar the four structures discussed in this review are, first the resolution of the X-ray structure of human Rvb1 (Matias et al. 2006) and the EM structures of the human (Puri et al. 2007) and yeast (Torreira et al. 2008) Rvb1/Rvb2 complexes was limited to the value in the 2D projection structures of the yeast complex (Gribun et al. 2008). Next, a two dimensional projection was calculated from each 3D structure along their 6-fold symmetry axis. In the case of the double-ring structures (human (Puri et al. 2007) and yeast (Torreira et al. 2008) 3D reconstructions) independent projections were calculated from each ring so the structural features are not cancelled out by adding the projection of the second ring (Fig. 5). These operations reduced the resolution and dimensionality of the 3D structures and it was possible to make a comparison with the 2D average projection of the yeast Rvb1/Rvb2 complex.
The comparison of the 2D projections from human and yeast structures showed certain similarities between the crystal structure of human Rvb1 (Matias et al. 2006) and both yeast structures (Gribun et al. 2008; Torreira et al. 2008) (Fig. 5). This is consistent with the docking experiments described above from Torreira et al. (Torreira et al. 2008) that showed a good agreement between the cryo-EM map of the yeast Rvb1/Rvb2 complex and the X-ray structure of the human Rvb1 hexamer. The differences observed between the 2D projections generated from both structures, mainly the larger central pore in the yeast structure, are not surprising due to the rotation and outward translations applied to the monomers in the human Rvb1 hexamer in order to fit the EM density of the yeast Rvb1/Rvb2 structure (Torreira et al. 2008). We also notice that the projections obtained from the cryo-EM structure of the yeast complex (Torreira et al. 2008) were more rounded than the projection structures obtained by us (Gribun et al. 2008) and the one obtained from the X-ray structure of the human Rvb1 (Matias et al. 2006) that had a hexagonal shape with concave edges. According to the crystal structure of the human Rvb1 hexamer, the DII domains constitute the vertices of the hexamer. These domains in the double-ring structure of the yeast complex (Torreira et al. 2008) extend downward interacting in the equatorial domain and locking their position rather than protruding outward. The different conformation of the DII domains in the cryo-EM map of the yeast complex (Torreira et al. 2008) explains the more rounded shape of the projections obtained from this structure.
As mentioned, the projection from the X-ray structure of human Rvb1(Matias et al. 2006) had commonalities with the negatively stained EM average projections of the yeast Rvb1/Rvb2 complex (Gribun et al. 2008) in the ATP or ATPγS conformation (Fig. 5), but not with the projection of the complex in the presence of ADP (Fig. 3). This is surprising as the X-ray structure of the human Rvb1 reveals that the protein in this structure is complexed with ADP. In addition, we also noticed that the hexameric rings formed by the yeast Rvb1/Rvb2 complex obtained by negative staining electron microscopy (Gribun et al. 2008) are significantly bigger than the ring formed by human Rvb1 (Matias et al. 2006) but they are similar in dimensions to the cryo-EM reconstruction of the yeast complex (Torreira et al. 2008) (Fig. 5). There is no obvious reason at this point to explain the observed difference in size between the X-ray and the EM structures.
Consistent with the incompatibility of the X-ray structure of the human Rvb1 (Matias et al. 2006) and the EM reconstruction of the human complex (Puri et al. 2007) derived from the docking experiment performed with these two structures, the projections obtained from the human Rvb complex were very dissimilar to the projections obtained from all the other structures, including the X-ray structure of the human Rvb1 (Matias et al. 2006) (Fig. 5).
All in all, a conclusion that can be withdrawn from this comparison is that in spite of the double or single-ring structure adopted by these proteins in the different structures, there are a reasonable degree of compatibility between three of the structures (X-ray structure of the human Rvb1 (Matias et al. 2006) and the two EM structures of the yeast complex (Gribun et al. 2008; Torreira et al. 2008). Probably the EM reconstruction of the human Rvb1/Rvb2 complex (Puri et al. 2007), which is incompatible with the other structures, captures the assembly in a somehow a unique conformation.
Roadmap to elucidate the physiologically relevant structures and conformational changes of the Rvb1/Rvb2 complexes
The human Rvb1 and Rvb2 proteins share 40% sequence identity. In addition, there is also a high degree of identity between the yeast and human Rvb proteins. Therefore, it is expected that these proteins share high degree of structural similarity. Consistently, we found a reasonable degree of structural similarity between three of the structures. However, it is surprising that both the human and yeast Rvb proteins were found forming single and double-ring oligomers. Certainly, it is possible that these proteins undergo conformational changes while performing their diverse functions or upon nucleotide hydrolysis. It is also possible that a single-ring structure is required to perform certain aspects of their functionality but a double-ring complex may be necessary for others. Thus, the solved structures may simply represent different functional states. However, careful analysis of the details of these studies during the elaboration of this review red flagged some aspect of these studies worth pursuing additional analysis to ensure that the reported structures reflect relevant physiological conformational states of the Rvb1/Rvb2 complex.
Differences such as the buffer conditions and the presence or absence of nucleotides in the current structures could be triggering these structural differences (Table 2). However, a first point of concern is the usage of differently tagged versions of the Rvb proteins in some of these structural studies (Table 3). Only in the study from Gribun et al. (Gribun et al. 2008), the Rvb1/Rvb2 complex is assembled from untagged proteins. In this study proteins are expressed as N-terminal His tag proteins. However, the tag is removed before the assembly of the complex is performed. Also in the study from Torreira et al. (Torreira et al. 2008), although the gross of the structural characterization was done in a Rvb1/N-terminal His Rvb2, it was reported (Torreira et al. 2008) that the complex still remained as a double ring structure after the His tag was removed right after the complex assembly. Results obtained in our laboratory have shown that an N-terminal His tag attached to either yeast Rvb1 or Rvb2 promotes the assembly of a double rather than a single hexameric structure (manuscript in preparation). Additional structures produced from untagged Rvb proteins are essential at this point to rule out the possibility that the tags may be inducing non-physiological conformational and oligomeric states.
Table 2.
Buffer conditions
| Structure | Buffer Conditions | Nucleotide bound |
|---|---|---|
| Human Rvb1 (Matias et al. 2006) | 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 1mM dithiothreitol | ADP |
| Human Rvb1/Rvb2 complex (Puri et al. 2007) | 20 mM Tris-HCl, pH 8.0, 100mM NaCl, 10% glycerol, 1mM PMSF, 1mM dithiothreitol, 1mM EDTA | No nucleotide added |
| Yeast Rvb1/Rvb2 complex (Gribun et al. 2008) | 25 mM Tris-HCl, pH 7.5, 200 mM KCl, 10% glycerol, 1mM dithiothreitol | ADP, ATP and ATPγS |
| Yeast Rvb1/Rvb2 complex (Torreira et al. 2008) | 25 mM Tris-HCl, pH 8.0, 125 mM NaCl | No nucleotide added |
Table 3.
Protein constructs and expression methods
| Structure | Construct/Assembly | Expression System |
|---|---|---|
| Human Rvb1 (Matias et al. 2006) | N-His-FLAG Rvb1 | E. coli BL21 (DE3) |
| Human Rvb1/Rvb2 complex (Puri et al. 2007) | Rvb1/Rvb2-His-C/in vitro | E. coli BL21 (DE3) |
| Yeast Rvb1/Rvb2 complex (Gribun et al. 2008) | Rvb1/Rvb2/in vitro | E. coli BL21 (DE3) |
| Yeast Rvb1/Rvb2 complex (Torreira et al. 2008) | Rvb1/N-His-Rvb2/in vivo | Baculovirus |
Furthermore, several systems were used in the different structures to overexpress the protein constructs (Table 3). These range from bacterial to insect cells based expression system. Also, some times the complexes were assembled in vitro from purified components and in other cases they were obtained in vivo through co-expression of both the Rvb1 and Rvb2 proteins. It is certainly a concern how human or yeast proteins expressed in bacterial systems as well as the Rvb1/Rvb2 complexes assembled in vitro relate to the native complexes. In this case purification and structural analysis of the endogenous complexes will provide important insights on the effect of the expression and assembly system in the structure of the complex.
Once the effect of protein tags, expression system and in vitro/in vivo assembly in the structure has been study or to some extend rule out, it will be beneficial to perform the structure determination of these complexes at conditions close to physiological or under those conditions where ATPase and helicase activity has been observed. Analysis of the structures in the apo-form but also bound to nucleotides and ultimately to nucleic acids will certainly unveil physiologically relevant structure and conformations of the Rvb1/Rvb2 complex.
In addition, whether the hexameric rings formed by these proteins are homomeric or heteromeric as well as the stoichiometry and layout of Rvb1 and Rvb2 proteins in the complex remain as a major unsolved issue. In the absence of a crystal structure of the human or yeast Rvb1/Rvb2 complex, this is one question where electron microscopy can contribute great insights through protein labeling experiments.
Finally, as our knowledge expands on the protein partners of the Rvb proteins in multiple macromolecular assemblies (see accompanying manuscript from Huen et al.) where these proteins are found, the next challenge will be to determine the structure of these larger assemblies to eventually understand the physiological role of Rvb proteins in the broader context of large macromolecular complexes.
Acknowledgments
Images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081). Our work in this field is supported by a Discovery grant from the National Science and Engineering Research Council of Canada (J.O), the National Cancer Institute of Canada (W.A.H., number 016372), and the Canadian Institutes of Health Research (W.A.H., MOP-93778). J.O. is Canadian Institutes of Health Research New Investigator.
References
- Bakshi R, Mehta AK, Sharma R, Maiti S, Pasha S, Brahmachari V. Characterization of a human SWI2/SNF2 like protein hINO80: demonstration of catalytic and DNA binding activity. Biochem Biophys Res Commun. 2006;339(1):313–320. doi: 10.1016/j.bbrc.2005.10.206. [DOI] [PubMed] [Google Scholar]
- Bauer A, Chauvet S, Huber O, Usseglio F, Rothbacher U, Aragnol D, Kemler R, Pradel J. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J. 2000;19(22):6121–6130. doi: 10.1093/emboj/19.22.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MJ, Indiani C, O’Donnell M. Reconstitution of the Mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J Biol Chem. 2003;278(7):4491–4499. doi: 10.1074/jbc.M210511200. [DOI] [PubMed] [Google Scholar]
- Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat Struct Biol. 2003;10(3):160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane WS, Nakatani Y, Livingston DM. The p400 complex is an essential E1A transformation target. Cell. 2001;106(3):297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- Gartner W, Rossbacher J, Zierhut B, Daneva T, Base W, Weissel M, Waldhausl W, Pasternack MS, Wagner L. The ATP-dependent helicase RUVBL1/TIP49a associates with tubulin during mitosis. Cell Motil Cytoskeleton. 2003;56(2):79–93. doi: 10.1002/cm.10136. [DOI] [PubMed] [Google Scholar]
- Gribun A, Cheung KL, Huen J, Ortega J, Houry WA. Yeast Rvb1 and Rvb2 are ATP-dependent DNA helicases that form a heterohexameric complex. J Mol Biol. 2008;376(5):1320–1333. doi: 10.1016/j.jmb.2007.12.049. [DOI] [PubMed] [Google Scholar]
- Gribun A, Kimber MS, Ching R, Sprangers R, Fiebig KM, Houry WA. The ClpP double ring tetradecameric protease exhibits plastic ring-ring interactions, and the N termini of its subunits form flexible loops that are essential for ClpXP and ClpAP complex formation. J Biol Chem. 2005;280(16):16185–16196. doi: 10.1074/jbc.M414124200. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6(7):519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102(4):463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, Gutierrez JL, Coleman MK, Workman JL, Mushegian A, et al. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem. 2005;280(50):41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- Johnson A, O’Donnell M. Ordered ATP hydrolysis in the gamma complex clamp loader AAA+ machine. J Biol Chem. 2003;278(16):14406–14413. doi: 10.1074/jbc.M212708200. [DOI] [PubMed] [Google Scholar]
- Karata K, Inagawa T, Wilkinson AJ, Tatsuta T, Ogura T. Dissecting the role of a conserved motif (the second region of homology) in the AAA family of ATPases. Site-directed mutagenesis of the ATP-dependent protease FtsH. J Biol Chem. 1999;274(37):26225–26232. doi: 10.1074/jbc.274.37.26225. [DOI] [PubMed] [Google Scholar]
- King TH, Decatur WA, Bertrand E, Maxwell ES, Fournier MJ. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol Cell Biol. 2001;21(22):7731–7746. doi: 10.1128/MCB.21.22.7731-7746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12(6):1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Lenzen CU, Steinmann D, Whiteheart SW, Weis WI. Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell. 1998;94(4):525–536. doi: 10.1016/s0092-8674(00)81593-7. [DOI] [PubMed] [Google Scholar]
- Li D, Zhao R, Lilyestrom W, Gai D, Zhang R, DeCaprio JA, Fanning E, Jochimiak A, Szakonyi G, Chen XS. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature. 2003;423(6939):512–518. doi: 10.1038/nature01691. [DOI] [PubMed] [Google Scholar]
- Lupas AN, Martin J. AAA proteins. Curr Opin Struct Biol. 2002;12(6):746–753. doi: 10.1016/s0959-440x(02)00388-3. [DOI] [PubMed] [Google Scholar]
- Matias PM, Gorynia S, Donner P, Carrondo MA. Crystal structure of the human AAA+ protein RuvBL1. J Biol Chem. 2006;281(50):38918–38929. doi: 10.1074/jbc.M605625200. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303(5656):343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9(1):27–43. [PubMed] [Google Scholar]
- Newman DR, Kuhn JF, Shanab GM, Maxwell ES. Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA. 2000;6(6):861–879. doi: 10.1017/s1355838200992446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure--diverse function. Genes Cells. 2001;6(7):575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- Puri T, Wendler P, Sigala B, Saibil H, Tsaneva IR. Dodecameric structure and ATPase activity of the human TIP48/TIP49 complex. J Mol Biol. 2007;366(1):179–192. doi: 10.1016/j.jmb.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Clancy SB, Tsuruta H, Gonzalez S, Wetmur JG, Tainer JA. Structure and mechanism of the RuvB Holliday junction branch migration motor. J Mol Biol. 2001;311(2):297–310. doi: 10.1006/jmbi.2001.4852. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Lin YL, Thome KC, Pian P, Schlegel BP, Weremowicz S, Parvin JD, Dutta A. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J Biol Chem. 1998;273(43):27786–27793. doi: 10.1074/jbc.273.43.27786. [DOI] [PubMed] [Google Scholar]
- Rombel I, Peters-Wendisch P, Mesecar A, Thorgeirsson T, Shin YK, Kustu S. MgATP binding and hydrolysis determinants of NtrC, a bacterial enhancer-binding protein. J Bacteriol. 1999;181(15):4628–4638. doi: 10.1128/jb.181.15.4628-4638.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG. Fitting atomic models into electron-microscopy maps. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 10):1341–1349. doi: 10.1107/s0907444900009562. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Morais MC, Leiman PG, Zhang W. Combining X-ray crystallography and electron microscopy. Structure. 2005;13(3):355–362. doi: 10.1016/j.str.2005.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406(6795):541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101(6):589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Snider J, Houry WA. AAA+ proteins: diversity in function, similarity in structure. Biochem Soc Trans. 2008;36(Pt 1):72–77. doi: 10.1042/BST0360072. [DOI] [PubMed] [Google Scholar]
- Snider J, Thibault G, Houry WA. The AAA+ superfamily of functionally diverse proteins. Genome Biol. 2008;9(4):216. doi: 10.1186/gb-2008-9-4-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torreira E, Jha S, Lopez-Blanco JR, Arias-Palomo E, Chacon P, Canas C, Ayora S, Dutta A, Llorca O. Architecture of the pontin/reptin complex, essential in the assembly of several macromolecular complexes. Structure. 2008;16(10):1511–1520. doi: 10.1016/j.str.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, McMahon SB, Cole MD. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol Cell. 2000;5(2):321–330. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- Zhao R, Kakihara Y, Gribun A, Huen J, Yang G, Khanna M, Costanzo M, Brost RL, Boone C, Hughes TR, et al. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J Cell Biol. 2008;180(3):563–578. doi: 10.1083/jcb.200709061. [DOI] [PMC free article] [PubMed] [Google Scholar]