Summary
PKR is an interferon-induced kinase that plays a pivotal role in the innate immunity pathway for defense against viral infection. PKR is activated to undergo autophosphorylation upon binding to RNAs that contain duplex regions. Activated PKR phosphorylates the alpha subunit of eukaryotic initiation factor 2, thereby inhibiting protein synthesis in viral infected cells. Viruses have evolved diverse PKR inhibitory strategies to evade the antiviral response. Adenovirus encodes VAI, a highly-structured RNA inhibitor that binds PKR but fails to activate. We have characterized the stoichiometry and affinity of PKR binding to define the mechanism of PKR inhibition by VAI. Sedimentation velocity and isothermal titration calorimetry measurements indicate that PKR interactions with VAI are modulated by Mg2+. Two PKR monomers bind in the absence of Mg2+ but a single monomer binds in the presence of divalent ion. Known RNA activators of PKR are capable of binding multiple PKR monomers to allow the kinase domains to come into close proximity and thus enhance dimerization. We propose that VAI acts as an inhibitor of PKR because it binds and sequesters a single PKR in the presence of divalent cation.
Keywords: Analytical ultracentrifugation, innate immunity, protein-nucleic acid interactions, protein kinase
Viral infection activates the host innate immunity response, leading to synthesis of type 1 interferons and subsequent induction of a large number of antiviral genes.1 Among these, the double-stranded RNA activated protein kinase R (PKR) plays a dominant role.2 The enzyme is induced in a latent form but binding to dsRNA or to RNAs containing duplex regions to undergo autophosphorylation activates it. Activated PKR blocks cap-dependent translational initiation by phosphorylating the alpha subunit of eukaryotic initiation factor 2 (eIF2α) at serine 51. Thus, production of dsRNA during viral infection3 results in PKR activation and inhibition of protein synthesis.
PKR contains an N-terminal dsRNA binding domain (dsRBD), consisting of two tandem copies of the dsRNA binding motif (dsRBM),4 and a C-terminal kinase, with a ~90 amino acid inter-domain linker lying between these domains. The structures of the isolated dsRBD5 and a complex of the kinase domain with eIF2α have been solved.6 The linker is flexible and PKR adopts multiple compact and extended conformations in solution.7
Dimerization plays a key role in PKR activation.8;9 PKR dimerizes weakly in solution and dimerization is sufficient to activate PKR in the absence of RNA.10 Activation of PKR by dsRNA follows a “bell-shaped” curve where low RNA concentrations activate but higher concentrations are inhibitory.11;12 These results can be rationalized in a model where low concentrations of dsRNA favor assembly of multiple PKRs on a single dsRNA whereas high dsRNA concentrations dilute PKR monomers onto separate molecules of dsRNA.13 Consistent with the dimerization model, a minimum of 30 bp of dsRNA are required to bind two PKRs and to activate autophosphorylation, supporting a model where the role of the dsRNA is to bring two or more PKR monomers in close proximity to enhance dimerization via the kinase domain.14;15
The importance of PKR in antiviral defense is underscored by the large number of viruses that produce PKR inhibitors and the variety of mechanisms that are used.16 Adenovirus and Epstein Barr virus encode RNA decoys that bind PKR but do not activate, thereby serving to block the antiviral response. Adenovirus VAI is a ~160 nucleotide RNA that accumulates to high concentration late in viral infection. VAI contains three major domains: the terminal stem, a complex central domain and an apical stem-loop (Figure 1).17;18 There is evidence that the central domain is stabilized by tertiary interactions and possibly forms a pseudoknot.19 Early enzymatic probing measurements20 suggested that Mg2+ alters VAI conformation, but it was recently reported that the melting of VAI is insensitive to divalent ion.21;22 The binding sites for PKR have been mapped to the apical stem and central domain.17;18;20;23–28 PKR binding and inhibitory potency are unaffected by deletion of the entire terminal stem.21 ITC21;29;30 and gel filtration measurements21 indicate that VAI can bind multiple PKR monomers. However, the mechanism of inhibition by VAI remains unclear. It was reported that isolated apical stem functions as a PKR activator and that interactions with other regions of VAI mediate inhibition.30 Alternatively, it was proposed that VAI functions by blocking PKR dimerization.29 Here, we characterize PKR binding to VAI RNA by sedimentation velocity measurements performed under the same conditions employed for enzymatic activity assays.
Figure 1.
Secondary structures of RNAs used in this study. A, Wild type VAI; B, ΔTS mutant lacking the terminal stem. The secondary structures are based on enzymatic structure probing measurements.27 RNAs were prepared by in vitro transcription from plasmid templates as previously described21;35 and were purified on preparative denaturing gels followed by electroelution. The RNAs were annealed by heating to 90°C followed by snap cooling.
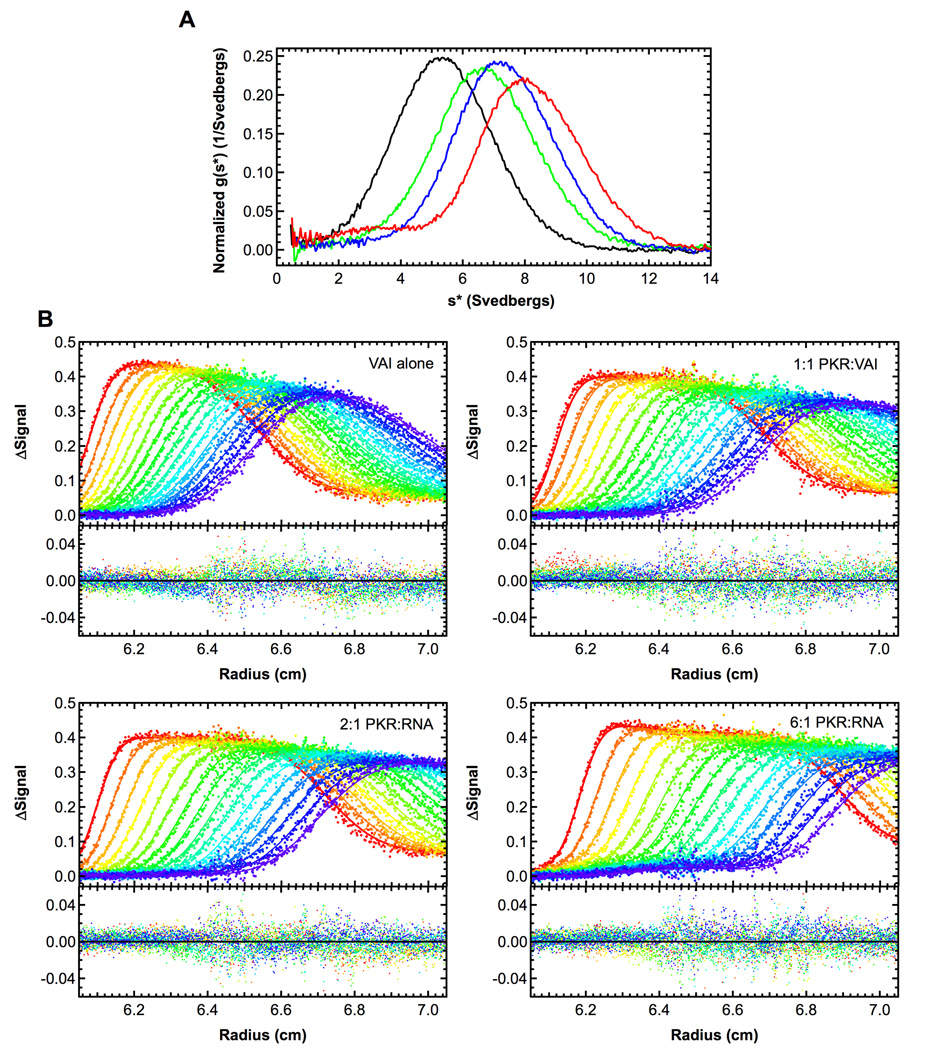
We have used sedimentation velocity analytical ultracentrifugation to define the stoichiometries and affinities for PKR binding to VAI and ΔTS, a VAI mutant that lacks the terminal stem (Figure 1). The data were initially analyzed by the time derivative method to verify binding and to define the association model. Figure 2A shows that VAI has a sedimentation coefficient near 5 S and the peak shifts to the right upon binding PKR. At the highest PKR concentration (6 eq.) the main peak is located near 8.5 S with a shoulder near 3.5 S, corresponding to free PKR. Assuming a model of a single PKR binding leads to an estimate of a frictional ratio (f/f0) of 1.28, which is much lower than the RNA alone or other PKR-RNA complexes (Supplementary materials, Table S1). Therefore, we considered a model of two PKR binding sequentially to VAI. The data fit well to this model in global analysis of the sedimentation velocity difference curves using SEDANAL (Figure 2).31 More reasonable values of f/f0 for the VAI-PKR complexes are obtained from this fit. As expected, a poor fit is obtained for a model of a single PKR binding, with RMS = 0.00946 and systematic deviations in the residuals. However, using a model of two PKR molecules binding results in RMS = 0.00841 without significant systematic deviations (Figure 2). The first PKR binds with high affinity (Kd = 14 nM) and the second binds with lower affinity (Kd = 600 nM) (Table 1). In contrast to wild type VAI, only one PKR binds to the ΔTS mutant that lacks the terminal stem. The dissociation constant of 158 nM (Table 1) lies between Kd1 and Kd2 for VAI. The fit quality is not improved using a model that incorporates binding of a second PKR and the fitted sedimentation coefficients for the PKR- ΔTS complexes are unreasonable.
Figure 2.
Sedimentation velocity analysis of PKR binding to VAI. A) Normalized g(s*) distributions of 0.4 µM VAI (black), VAI + 1 eq. PKR (green), VAI + 2 eq. PKR (blue) and VAI + 6 eq. PKR (red). Distributions were calculated using DCDT+.36 B) Global analysis of difference curves. The data were subtracted in pairs to remove systematic noise and the four data sets at the indicated ratios of PKR: VAI were fit to 2:1 binding model using SEDANAL.31 Fitted parameters were the sedimentation coefficients of the complexes, the VAI loading concentrations and the dissociation constants The top panels show the data (points) and fit (solid lines) and the bottom panels show the residuals (points). The best fit parameters are shown in Table 1. For clarity, only every 2nd difference curve is shown. Samples were prepared in a buffer consisting of 20 mM HEPES pH 7.5, 200 mM NaCl, 0.1 mM EDTA and 0.01 mM TCEP (AU 200) and loaded into two channel aluminum-epon double-sector cells equipped with quartz windows. Data were collected using absorbance optics in a Beckman-Coulter XL-I analytical ultracentrifuge. Conditions: rotor speed, 35,000 RPM; temperature, 20°C; wavelength, 260 nm.
Table 1.
PKR-RNA interaction parameters derived from sedimentation velocity analysis.
| RNA | [Mg2+] mM | Model | Kd1 (nM) | Kd2 (nM) | s(RNA)a | s(RP)a | s(RP2)a | RMSb |
|---|---|---|---|---|---|---|---|---|
| VAI | 0 | R + P ↔ RP RP + P ↔ RP2 |
14 (3,41) | 601 (359, 1200) | 5.34 | 6.78(6.63,7.04) | 8.54 (8.30, 9.00) | 0.00841 |
| ΔTS | 0 | R + P ↔ RP | 158 (136, 182) | -- | 4.33 | 6.43 (6.38, 6.48) | -- | 0.00854 |
| AS | 0 | R + P ↔ RP | 1.04 µM (0.92 µM, 1.17 µM) |
-- | 2.99 | 4.94 (4.88, 5.00) | -- | 0.00757 |
| WT | 5 | R + P ↔ RP | 334 (278, 401) | -- | 5.29 | 7.18(7.07,7.29) | -- | 0.00691 |
| ΔTS | 5 | R + P ↔ RP | 367 (302, 440) | -- | 4.41 | 6.19(6.10,6.29) | -- | 0.0104 |
| AS | 5 | R + P ↔ RP | 3.90 µM (3.39 µM, 4.56 µM) |
-- | 2.98 | 5.04 (4.89, 5.23) | -- | 0.00944 |
Parameters obtained by global nonlinear least squares analysis using SEDANAL (ref). The values in parenthesis correspond to the 95% joint confidence intervals.
Uncorrected sedimentation coefficicient (Svedbergs).
RMS deviation of the fit in absorbance units.
Because Mg2+ has been reported to affect VAI conformation20 we have analyzed the effects of divalent ion on the hydrodynamic properties of VAI and the binding of PKR. The uncorrected sedimentation coefficient of VAI decreases very slightly in the presence of Mg2+ (Table 1). However, this effect is due to the change in buffer density and viscosity and in both cases f/f0 =1.61 (Supplementary materials, Table S1). Thus, Mg2+ does not induce a large-scale change in VAI conformation which would affect the hydrodynamic properties of the RNA. The sedimentation velocity results are supported by small angle X-ray scattering studies, where the radius of gyration (Rg) of VAI increases very slightly from 45.48 ± 0.15 Å to 47.72 ± 0.14 Å upon addition of 5 mM Mg2+ (Wong, C.J., Launer-Felty, K. and Cole, J., unpublished observations). Although Mg2+ does not induce significant structural changes, the affinity of PKR binding to VAI is strongly reduced by about 20-fold in the presence of 5 mM Mg2+ and the second PKR binding event is not even detected (Table 1).*
We have confirmed the sedimentation velocity results using ITC. In the absence of Mg2+, a distinctly bimodal isotherm is observed, indicating that two PKR bind to VAI under these conditions (Figure 3). The dissociation constants obtained by fitting these data are in good agreement with those derived from sedimentation velocity. The enthalpy change for binding of the first PKR is about twice that of the second PKR. In the presence of Mg2+ only one PKR binds (Figure S1), and again, the Kd values agree well with the previous results in Table 2. The large enthalpy change correlates with the first, high-affinity PKR binding event observed in the absence of Mg2+.
Figure 3.

ITC analysis of PKR binding to VAI. Measurements were performing using a VP-ITC (Microcal, Inc.) at 20°C. Both RNA and protein were exchanged in AU 200 buffer by exhaustive dialysis. The calorimeter cell contained 4 µM VAI and the syringe contained 75 µM PKR. A single 2-µL injection was performed followed by 29 10-µL injections. The top panel shows the background-corrected ITC data and the bottom panel shows the integrated data (points) and fit of the data to a model of two independent binding sites (solid line) performed using ORIGIN (Microcal, Inc.) The best fit parameters are shown in Table 2.
Table 2.
PKR-VAI interaction parameters derived from ITC analysis.
| [Mg2+] mM | Model | Kd1 (nM) |
N1 | ΔH1 (kcal/mol) |
ΔS1 (cal/mol) |
ΔG1 (kcal/mol) |
Kd2 (nM) |
N2 | ΔH2 (kcal/mol) |
ΔS2 (cal/mol) |
ΔG2 (kcal/mol) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | R + P ↔ RP RP + P ↔ RP2 |
10±7 | 0.68±0.02 | −15.9±0.5 | −17.7 | −10.8 | 794±367 | 1.34±0.10 | −7.6±1.1 | 2.1 | −8.2 |
| 5 | R + P ↔ RP | 667±125 | 1.12±0.04 | −13.2±0.5 | −16.9 | −8.3 | -- | -- | -- | -- | -- |
Parameters obtained by global nonlinear least squares analysis using ORIGIN version 5.0 software.
To validate the functional relevance of the sedimentation velocity and ITC experiments performed in the presence of Mg2+, enzymatic activity assays were performed under the same conditions used for the biophysical measurements. Figure 4 shows the inhibition of dsRNA-induced PKR autophosphorylation by wild type VAI RNA and ΔTS. Both RNAs effectively inhibit this reaction and within error, their apparent potencies are about the same. Consistent with the present data, it has previously been reported that the central domain and apical stem comprise the PKR binding regions of VAI17;18;20;23–28 and deletion of the terminal stem does not affect PKR inhibitory potency.21
Figure 4.
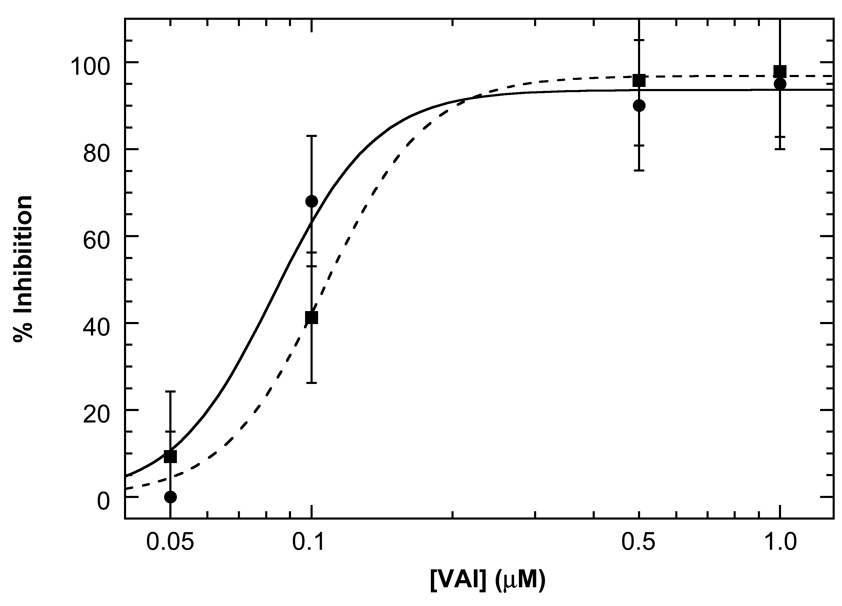
Inhibition of dsRNA activation of PKR by VAI and ΔTS. PKR autophosphorylation assays were performed as previously described10;15 in AU 200 + 5 mM MgCl2. 100 nM PKR and variable concentrations of VAI were pre-incubated for 10 minutes at 30°C, followed by addition of 10 ug/ml poly(rI:rC). After 20 minutes, 0.4 mM ATP containing 4µCi γ-32P[ATP] was added and the reaction was incubated for another 40 minutes before quenching with sample loading buffer. PKR autophosphorylation was quantified by phosphorimager analysis and was referenced to samples containing no inhibitor. Each point represents the average of three measurements and the error bars indicate the estimated standard error.
Although it is difficult to quantitatively analyze PKR activation kinetic data due to the existence of multiple phosphorylation sites and the complexity of autocatalytic reactions,10 the fact that VAI and ΔTS completely inhibit autophosphorylation at concentrations above 0.5 µM is consistent with the PKR binding affinities determined for these RNAs in the presence of Mg2+ (Table 1). Note that divalent ion is required for PKR activity so it is not possible to assess inhibition in the absence of Mg2+. The inhibitory potency of VAI reported here is also consistent with previous studies where PKR inhibition is only detected at VAI concentrations above 100–300 nM.21;24;29;33 Neither VAI nor ΔTS is capable of activating PKR in the absence of the dsRNA poly (rI:rC) (data not shown).
It is notable that Mg2+ strongly modulates the affinity of PKR binding to VAI RNA. Typically, PKR binding is not dependent on divalent ion34 and the ~ 20-fold increase in Kd upon addition of Mg2+ is larger than expected from the slight increase in ionic strength. For example, the Kd for PKR binding to an unrelated 20 bp dsRNA increases only about 3-fold upon addition of 5 mM Mg2+ in the presence of 200 mM NaCl (Supplementary materials, Table S2). Similarly, divalent ion only slightly reduces PKR binding affinity for ΔTS (Table 1). Thus, our data suggest that Mg2+ induces a change in the mechanism of PKR binding to VAI. This change is not reflected in the hydrodynamic properties of VAI (Table 1 and Table S1), ruling out large-scale rearrangement in VAI conformation. In thermal denaturation experiments, addition of Mg2+ induces an overall stabilization of VAI but there is no evidence for specific interactions.21;22 However, Mg2+ does alter the pattern of enzymatic cleavage and chemical modification within the VAI central domain.20 Divalent ion is often required for stabilization of RNA tertiary structure and we propose that Mg2+ is required for correct folding of the VAI central domain. However, the nature of the structural change in VAI induced by Mg2+ is not clear and further experiments are underway to probe the structure of the central domain and to define the role of divalent ion.
It is well established that dimerization plays a key role in the activation of PKR by RNA. Binding of multiple PKR monomers is required for activation of PKR by both homogeneous duplex RNAs15 and natural RNA hairpins that contain secondary structure defects.14 Thus, we propose that VAI acts as an in vivo inhibitor of PKR because it binds only a single PKR under physiological conditions where magnesium is present. It was previously suggested that viral RNA inhibitors are capable of binding only one PKR monomer.8 The high affinity of PKR binding to VAI or ΔTS is noteworthy. Typically, PKR binds most strongly to regions of homogeneous duplex RNA and secondary structure defects impede interaction. The apical stem consists of a stem-loop with about 21 basepairs of duplex interrupted by two one-base bulges and three noncanonical basepairs, and the central domain contains a short stem loop with only 7 basepairs. PKR binds weakly to the isolated apical stem, with Kd > 3 µM in the presence of Mg2+ (Launer-Felty, K. and Cole, J.L., unpublished observations). Thus, the highly structured central domain is likely responsible for mediating the high affinity of PKR binding to the VAI and thus comprises a novel PKR binding motif.
Supplementary Material
Acknowledgements
This work was supported by grant number AI-53615 from the NIH to J.L.C. We thank Arlene Albert for providing access to the VP-ITC calorimeter.
Footnotes
The Mg2+ concentration of 5 mM was chosen to match conditions for PKR activity assays and is somewhat higher that the typical intracellular concentration (~ 0.5 mM Mg2+).32 We also detect strong inhibition of the second PKR binding event by 0.5 mM Mg2+ (data not shown).
References
- 1.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:369–434. doi: 10.1016/S0079-6603(06)81010-X. [DOI] [PubMed] [Google Scholar]
- 3.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded RNA binding motif: Interference and much more. Nature Rev. Mol. Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 5.Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. Structure of the double-stranded RNA binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 7.VanOudenhove J, Anderson E, Krueger S, Cole JL. Analysis of PKR structure by small-angle scattering. J Mol Biol. 2009;387:910–920. doi: 10.1016/j.jmb.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson HD, Mathews MB. The regulation of the protein kinase PKR by RNA. Biochimie. 1996;78:909–914. doi: 10.1016/s0300-9084(97)86712-0. [DOI] [PubMed] [Google Scholar]
- 9.Cole JL. Activation of PKR: an open and shut case? Trends Biochem. Sci. 2007;32:57–62. doi: 10.1016/j.tibs.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemaire PA, Lary J, Cole JL. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. J. Mol. Biol. 2005;345:81–90. doi: 10.1016/j.jmb.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 11.Hunter T, Hunt T, Jackson RJ, Robertson HD. The characteristics of inhibition of protein synthesis by double-stranded ribonucleic acid in reticulocyte lysates. J. Biol. Chem. 1975;250:409–417. [PubMed] [Google Scholar]
- 12.Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kostura M, Mathews MB. Purification and activation of the double-stranded RNA-dependent eIF-2 kinase DAI. Mol. Cell. Biol. 1989;9:1576–1586. doi: 10.1128/mcb.9.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinicke LA, Wong CJ, Lary J, Nallagatla SR, Diegelman-Parente A, Zheng X, Cole JL, Bevilacqua PC. RNA dimerization promotes PKR dimerization and activation. J Mol Biol. 2009;390:319–338. doi: 10.1016/j.jmb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemaire PA, Anderson E, Lary J, Cole JL. Mechanism of PKR Activation by dsRNA. J. Mol. Biol. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. Inhibition of PKR by RNA and DNA viruses. Virus Res. 2006;119:100–110. doi: 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Furtado MR, Subramanian S, Bhat RA, Fowlkes DM, Safer B, Thimmappaya B. Functional dissection of adenovirus VAI RNA. J. Virol. 1989;63:3423–3434. doi: 10.1128/jvi.63.8.3423-3434.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mellits KH, Mathews MB. Effects of mutations in stem and loop regions on the structure and function of adenovirus VA RNAI. EMBO J. 1988;7:2849–2859. doi: 10.1002/j.1460-2075.1988.tb03141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y, Mathews MB. Secondary and tertiary structure in the central domain of adenovirus type 2 VA RNA I. RNA. 1996;2:937–951. [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke PA, Mathews MB. Interactions between the double-stranded RNA binding motif and RNA: definition of the binding site for the interferon-induced protein kinase DAI (PKR) on adenovirus VA RNA. RNA. 1995;1:7–20. [PMC free article] [PubMed] [Google Scholar]
- 21.Wahid AM, Coventry VK, Conn GL. Systematic deletion of the adenovirus-associated RNAI terminal stem reveals a surprisingly active RNA inhibitor of double-stranded RNA-activated protein kinase. J. Biol. Chem. 2008;283:17485–17493. doi: 10.1074/jbc.M802300200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coventry VK, Conn GL. Analysis of adenovirus VA RNAI structure and stability using compensatory base pair modifications. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellits KH, Kostura M, Mathews MB. Interaction of adenovirus VA RNAI with the protein kinase DAI: nonequivalence of binding and function. Cell. 1990;61:843–852. doi: 10.1016/0092-8674(90)90194-j. [DOI] [PubMed] [Google Scholar]
- 24.Mellits KH, Pe'ery T, Mathews MB. Role of the apical stem in maintaining the structure and function of adenovirus virus-associated RNA. J. Virol. 1992;66:2369–2377. doi: 10.1128/jvi.66.4.2369-2377.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghadge GD, Malhotra P, Furtado MR, Dhar R, Thimmapaya B. In vitro analysis of virus-associated RNA I (VAI RNA): inhibition of the double-stranded RNA-activated protein kinase PKR by VAI RNA mutants correlates with the in vivo phenotype and the structural integrity of the central domain. J. Virol. 1994;68:4137–4151. doi: 10.1128/jvi.68.7.4137-4151.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahman A, Malhotra P, Dhar R, Kewalramani T, Thimmapaya B. Effect of single-base substitutions in the central domain of virus-associated RNA I on its function. J. Virol. 1995;69:4299–4307. doi: 10.1128/jvi.69.7.4299-4307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clarke PA, Pe'ery T, Ma Y, Mathews MB. Structural features of adenovirus 2 virus-associated RNA required for binding to the protein kinase DAI. Nucleic Acids Res. 1994;22:4364–4374. doi: 10.1093/nar/22.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spanggord RJ, Beal PA. Selective binding by the RNA binding domain of PKR revealed by affinity cleavage. Biochemistry. 2001;40:4272–4280. doi: 10.1021/bi002512w. [DOI] [PubMed] [Google Scholar]
- 29.McKenna SA, Lindhout DA, Shimoike T, Aitken CE, Puglisi JD. Viral dsRNA inhibitors prevent self-association and autophosphorylation of PKR. J. Mol. Biol. 2007;372:103–113. doi: 10.1016/j.jmb.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna SA, Kim I, Liu CW, Puglisi JD. Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs. J. Mol. Biol. 2006;358:1270–1285. doi: 10.1016/j.jmb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Stafford WF, Sherwood PJ. Analysis of heterologous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys. Chem. 2004;108:231–243. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 32.Gunther T. Concentration, compartmentation and metabolic function of intracellular free Mg2+ Magnes Res. 2006;19:225–236. [PubMed] [Google Scholar]
- 33.Pe'ery T, Mellits KH, Mathews MB. Mutational analysis of the central domain of adenovirus virus-associated RNA mandates a revision of the proposed secondary structure. J. Virol. 1993;67:3534–3543. doi: 10.1128/jvi.67.6.3534-3543.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bevilacqua PC, Cech TR. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain of the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 35.Coventry VK, Conn GL. Analysis of adenovirus VA RNAI structure and stability using compensatory base pair modifications. Nucleic Acids Res. 2008;36:1645–1653. doi: 10.1093/nar/gkn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal. Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.