Summary
MET amplification activates ERBB3/PI3K/AKT signaling in EGFR mutant lung cancers, and causes resistance to EGFR kinase inhibitors. We demonstrate that MET activation by its ligand, HGF, also induces drug resistance, but through GAB1 signaling. Using high-throughput FISH analyses in both cell lines and in lung cancer patients, we identify subpopulations of cells with MET amplification prior to drug exposure. Surprisingly, HGF accelerates the development of MET amplification both in vitro and in vivo. EGFR kinase inhibitor resistance, due to either MET amplification or autocrine HGF production, was cured in vivo by combined EGFR and MET inhibition. These findings highlight the potential to prospectively identify treatment naïve EGFR mutant lung cancer patients who will benefit from initial combination therapy.
Introduction
Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) gefitinib and erlotinib are effective clinical therapies for advanced non-small cell lung cancer (NSCLC) patients with EGFR activating mutations (Asahina et al., 2006; Inoue et al., 2006; Paz-Ares et al., 2006; Sequist et al., 2008; Tamura et al., 2008). A recent phase III clinical trial demonstrated that patients with EGFR mutant NSCLC had superior outcomes with gefitinib treatment compared to standard first line cytotoxic chemotherapy (Mok et al., 2008). However, despite these dramatic benefits from EGFR TKIs in this genetically defined cohort, all of these patients ultimately develop resistance (referred to as acquired resistance herein) to gefitinib and erlotinib. Two mechanisms of acquired resistance have been validated in patients. Secondary mutations in EGFR itself, including the EGFR T790M “gatekeeper” mutation is observed in 50% of resistance cases, and amplification of the MET oncogene is observed in 20% of resistance cases (Balak et al., 2006; Bean et al., 2007; Engelman et al., 2007b; Kobayashi et al., 2005; Kosaka et al., 2006; Pao et al., 2005). Both resistance mechanisms lead to maintenance of ERBB3/PI3K/AKT signaling in the presence of gefitinib (reviewed in (Engelman and Janne, 2008)).
In addition to these genetic alterations, activation of IGF-1Rβ/IRS-1 signaling through loss of IGF binding proteins also drives gefitinib resistance in EGFR wild-type cancer cell lines (Guix et al., 2008). Additionally, a recent study suggested that the MET ligand, HGF, can promote short-term resistance in two EGFR mutated cancer cell lines (Yano et al., 2008). Both ligand-dependent resistance mechanisms maintain PI3K/AKT activation despite EGFR inhibition. However, differences between IGF and HGF driven resistance in terms of potency and activation of downstream signaling pathways have yet to be thoroughly examined. Furthermore, the contribution of HGF, if any, to gefitinib resistance mediated by MET amplification is unknown.
Strategies for overcoming acquired resistance to gefitinib are now undergoing clinical evaluation. In preclinical studies, the EGFR T790M mutation can be overcome by second-generation, irreversible EGFR inhibitors (Engelman et al., 2007a; Kobayashi et al., 2005; Riely, 2008). In addition, the growth of EGFR mutant cancers with MET amplification can be inhibited by combined treatment with EGFR and MET kinase inhibitors (Bean et al., 2007; Engelman et al., 2007b). Indeed, there are now clinical trials assessing both irreversible EGFR inhibitors and a combination of MET and EGFR inhibitors in patients with acquired resistance to gefitinib/erlotinib. Further, clinical activity of the irreversible EGFR inhibitor, PF00299804, has been observed in NSCLC patients that have developed acquired resistance to gefitinib/erlotinib (Janne et al., 2008). As an alternative strategy, to delay or avoid the emergence of resistance, there is increased enthusiasm to utilize agents effective against specific resistance mechanisms as initial systemic therapies. For example, PF00299804 is now being assessed in a phase II clinical trial of EGFR TKI naïve patients. However, there are currently no methods to predict the specific resistance mechanism that a cancer will develop.
In the current study, we modeled in vitro resistance to PF00299804 in the TKI sensitive EGFR mutant NSCLC cell line HCC827 (Engelman et al., 2006; Engelman et al., 2007b; Ogino et al., 2007) . In addition, we evaluated the potency of the MET ligand, HGF, to promote resistance to EGFR TKIs and determined whether MET amplification pre-exists in a subpopulation of cells prior to treatment with a TKI.
Results
MET amplification causes resistance to the irreversible EGFR inhibitor PF00299804 by activating ERBB3 signaling
We generated in vitro resistant clones of HCC827 cells to the irreversible pan-ERBB kinase inhibitor, PF00299804, using previously described methods (Engelman et al., 2006; Engelman et al., 2007b). HCC827 cells were exposed to increasing concentrations of PF00299804, starting with 1nM, until they were able to proliferate freely in 1 M PF00299804, which occurred after 6 months of drug selection. This concentration was chosen because it is ~ 1000 fold greater than the IC50 for growth inhibition of HCC827 cells and approximately 5 times greater than the serum concentration of PF00299804 observed in NSCLC patients in the phase I clinical trial (Janne et al., 2008; Schellens et al., 2007). Five independent clones were isolated and expanded for further studies. All five HCC827 PF00299804 resistant (PFR) clones were resistant to PF00299804 in vitro (Figure 1A and data not shown). No secondary EGFR mutations (e.g. T790M) were detected in any of the clones (data not shown).
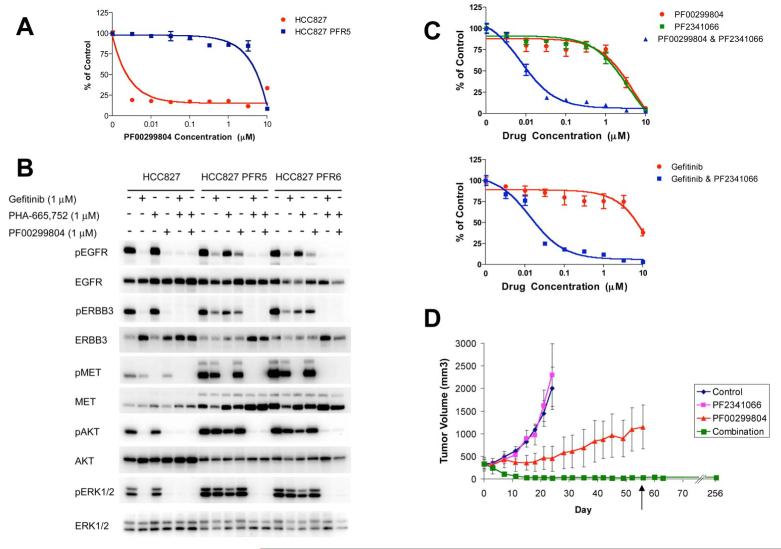
Figure 1. HCC827 PFR cells are resistant to PF00299804, but combined MET and EGFR inhibition blocks PI3K/AKT and ERK signaling and restores sensitivity in vitro and in vivo.
(A) Parental and resistant HCC827 PFR5 cells treated with increasing concentrations of PF00299804. Cell viability relative to untreated controls measured after 72 hours. Each data point represents the mean ±SD of 6 wells. (B) HCC827 and HCC827 PFR5 and PFR6 cells were treated for 6 hours with 1 μM PF00299804 or gefitinib, PHA-665,752, or their combination. Cell lysates were immunoblotted to detect indicated proteins. (C) Upper, HCC827 PFR6 cells treated with increasing concentrations of PF00299804, PF2341066, or their combination. Lower, HCC827 PFR6 cells treated with increasing concentrations of gefitinib alone or in combination with PF2341066. Cell viability relative to untreated controls measured after 72 hours. Each data point represents the mean ±SD of 6 wells. (D) HCC827 PFR xenogafts in nu/nu mice were treated with PF2341066, PF00299804, or their combination. Tumors measured twice weekly. Only combination treatment led to tumor shrinkage and was the most effective treatment in vivo (p < 0.0001). Treatment was stopped after 56 days (arrow) and no tumor re-growth was observed in 35 weeks. Each data point represents the mean ±SD for 5 mice.
We next examined the effects of PF00299804 on EGFR, ERBB3, AKT and ERK phosphorylation in the HCC827 PFR clones. Unlike in parental HCC827 cells, ERBB3 activation as well as downstream PI3K/AKT and ERK signaling is maintained in the presence of PF00299804 in HCC827 PFR cells (Figure 1B). We also observed increased total MET protein in the HCC827 PFR cells, and combined MET and EGFR inhibition down-regulated ERBB3, AKT and ERK phosphorylation as well as the modest EGFR phosphorylation that was maintained in the presence of PF00299804 alone (Figure 1B). This behavior following treatment with PF00299804 alone or in combination with a MET inhibitor is similar to that observed in gefitinib resistant HCC827 cells (HCC827 GR cells), which were generated in an analogous manner and contained a focal amplification in chromosome 7 harboring the MET oncogene (Engelman et al., 2007b).
Given the similarities in the HCC827 PFR and GR cells following treatment with either PF00299804 or gefitinib, respectively, we determined whether the addition of a MET inhibitor would overcome resistance to PF00299804. We used both a tool compound PHA-665,752 and the MET inhibitor PF2341066 currently undergoing clinical development (Figure 1C, upper and data not shown) (Zou et al., 2007). The combination of PF00299804 and a MET inhibitor effectively inhibited the growth of HCC827 PFR cells while neither agent alone led to growth inhibition (Figure 1C, upper and data not shown). In addition, the combination of gefitinib and PF2341066 also effectively inhibited the growth of HCC827 PFR cells (Figure 1C, lower). These findings further suggest that the resistance mechanism in the HCC827 PFR cells is not unique or dependent on the differences between reversible (gefitinib) or irreversible (PF00299804) EGFR inhibitors but rather due solely to MET amplification. We also evaluated the effects of the irreversible EGFR inhibitor PF00299804 and the MET inhibitor PF-2341066 in an HCC827 PFR xenograft model. Treatment with PF00299804 alone was modestly more effective than treatment with PF2341066 alone, but the tumors demonstrated resistance to PF00299804. However, combined MET and EGFR inhibition completely inhibited tumor growth and produced complete responses (p<0.0001; Figure 1D). In fact, the combination treatment was discontinued after 56 days (Figure 1D; arrow) and no tumor re-growth has been observed to date in any of the xenografts (after more than 35 weeks off therapy) (Figure 1D), suggesting that the mice have been cured.
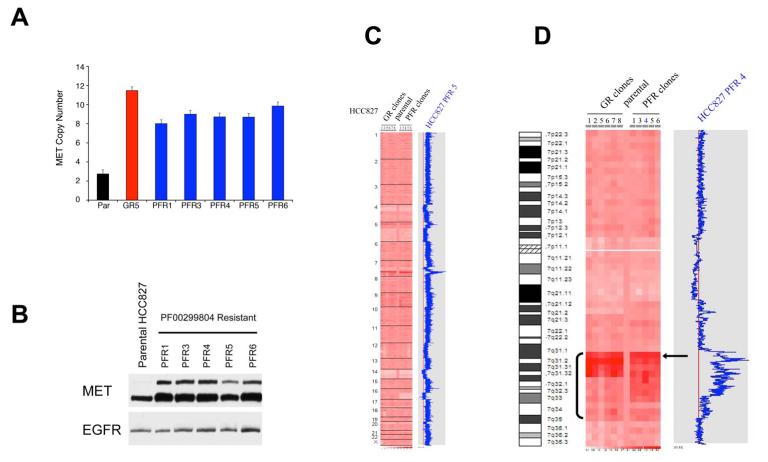
We next determined whether the increase in MET protein expression was due to MET amplification in the HCC827 PFR cells (Figure 2A). All of the PFR clones contained at least a four fold amplification of MET, similar to the amplification previously observed in the gefitinib resistant HCC827 (HCC827 GR) cells ((Engelman et al., 2007b) and Figure 2A). All of the PFR clones also had higher levels of MET protein expression (Figure 2B). Genome-wide SNP analysis revealed that the only area of significant copy number gain in HCC827 PFR cells is on distal chromosome 7, similar to that observed in HCC827 GR cells, and contains the MET oncogene (Figure 2C, D). Furthermore, HCC827 PFR and GR cells share single copy losses of 4p, 5q, 14p, 14q and 19p, but only HCC827 PFR cells have a single copy loss of 16q. Intriguingly, further examination of the region of MET amplification on distal chromosome 7 in both set of clones showed that, although the copy number changes within the amplicons are not identical in the HCC827 GR and PFR cells, the size and the proximal borders of the amplicons are very similar (Figure 2D). Together these findings, along with the multiple shared regions of single copy genomic loss between the HCC827 PFR and GR cells, suggest that the resistant clones may have arisen from a common origin.
Figure 2. HCC827 PFR cells have a focal amplification in MET that is similar to HCC827 GR cells.
(A) MET copy number determined by quantitative PCR. Parental (Par) HCC827 and MET amplified HCC827 GR (GR5) cells were used as negative and positive controls, respectively. Each column represents the mean ±SD for 3 independent experiments. (B) Parental HCC827 cells and PFR clones were immunoblotted to detect indicated proteins. (C) Genome wide view of copy number changes generated using Human Mapping 250K Sty single nucleotide polymorphism (SNP) array and analyzed using the dChip program (see Experimental Procedures). HCC827 GR clones were compared with HCC827 PFR and HCC827 parental clones. Blue curve indicates degree of amplification of each SNP from 0 (left) to 8 (right). (D) Chromosome 7 view of copy number changes in HCC827 parental, GR and PFR cells. Arrow indicates MET oncogene.
HGF activates PI3K/AKT signaling through GAB1 and leads to gefitinib resistance
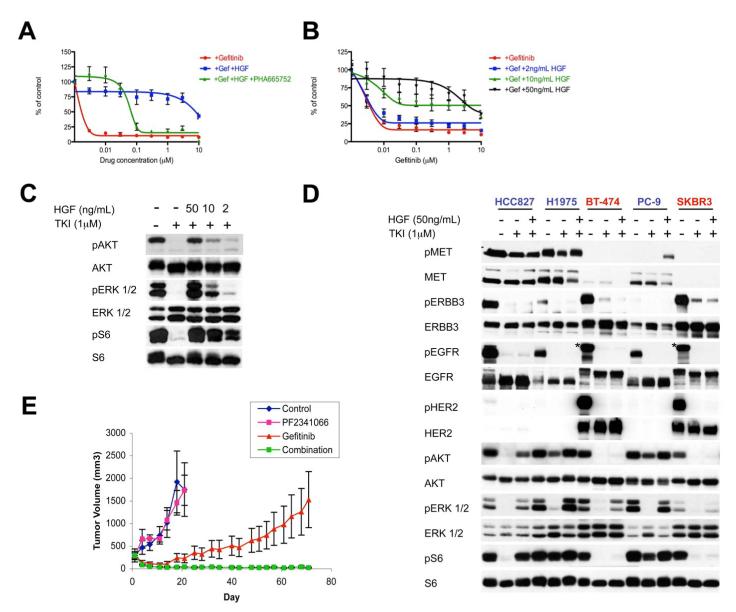
MET amplification was previously shown to cause gefitinib resistance in HCC827 GR cells (Engelman et al., 2007b). We investigated whether activation of MET signaling by its ligand, HGF, could also cause resistance to gefitinib and other ERBB-targeted therapies. In a 72 hour survival assay, HGF induced substantial gefitinib resistance in HCC827 cells that was abolished by the addition of PHA-665,752 (Figure 3A). Furthermore, HGF maintained PI3K/AKT, mTORC1 and ERK activation in the presence of gefitinib in a dose-dependent manner that mirrored its capacity to maintain cell viability (Figures 3B, C).
Figure 3. HGF induces MET dependent resistance only in cell lines in which it activates PI3K/AKT, ERK and mTORC1 signaling.
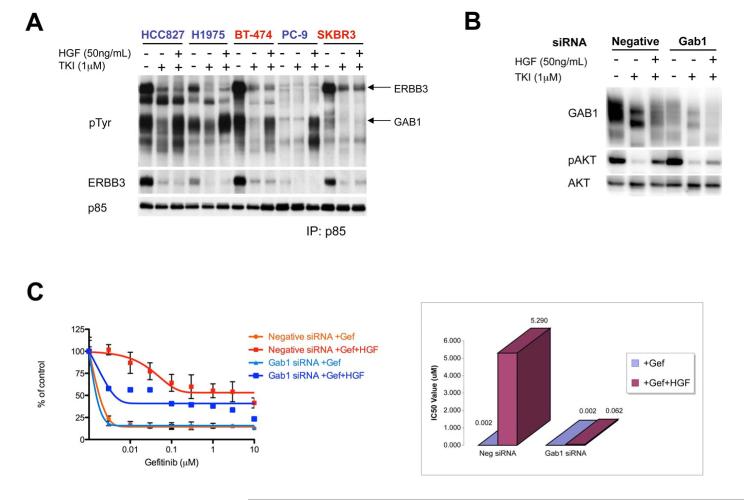
(A, B) HCC827 cells treated with (A) increasing concentrations of gefitinib alone or in combination with PHA-665,752, in the absence or presence of HGF (50ng/mL), or (B) increasing concentrations of gefitinib alone or in combination with the indicated concentrations of HGF. Cell viability relative to untreated controls measured after 72 hours. Each data point represents the mean ±SD of 6 wells. (C) HCC827 cells were treated for 6 hours with 1μM gefitinib alone or in combination with the indicated concentrations of HGF. Cell lysates were immunoblotted to detect indicated proteins. (D) Cells were treated for 6 hours with gefitinib (HCC827, PC-9), PF00299804 (H1975), or lapatinib (BT-474, SKBR3), alone or in combination with HGF (50ng/mL). All drugs were used at 1μM. Cell lysates were immunoblotted to detect indicated proteins. *indicates cross-reaction by the p-EGFR antibody against p-HER2. Cell lines in which HGF rescued viability are labeled in blue, and cell lines in which HGF did not rescue viability are labeled in red. (E) HCC827-HGF xenografts in nu/nu mice treated with PF2341066, gefitinib, or their combination and tumors measured twice weekly. Some growth inhibiton was observed with gefitinib alone, however only combination treatment led to complete tumor shrinkage (p = 0.002). Each data point represents the mean ±SD for 5 mice. See also Figure S2.
We also determined the capacity for HGF to maintain downstream signaling and cell viability in other EGFR and HER2 addicted cancers. In cell lines with EGFR exon 19 deletions (HCC827 and PC-9), and an EGFR-driven lung cancer cell line carrying the T790M resistance mutation (H1975), HGF restored PI3K/AKT, mTORC1 and ERK signaling, despite continued EGFR inhibition in the presence of 1μM gefitinib or PF00299804 (Figure 3D). HGF also rescued each of these cell lines from TKI-induced cell death after 72 hours (Figure 4A and Figure S1A-E). In contrast to the EGFR addicted cancers, HGF did not rescue HER2 amplified breast cancer cell lines from the effects of lapatinib (Figure 4A and Figure S1F, G), nor did it rescue AKT or mTORC1 signaling in either HER2 driven cell line (Figure 3D). Thus, the capacity to rescue cell viability appears to strongly correlate with capacity to restore downstream signaling, especially along the PI3K/AKT pathway. We suspect that HGF had a minimal effect in BT-474 and SKBR3 cells because these cell lines have lower levels of MET expression compared to the other EGFR-driven cell lines that were tested.
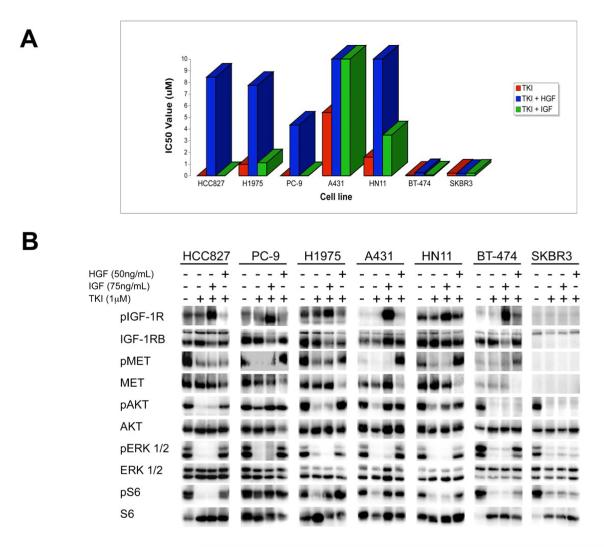
Figure 4. IGF rescues PI3K/AKT and mTORC1 signaling in some cell lines, but fails to activate ERK.
(A) IC50 values for viability curves (Figure S1) in the presence or absence of HGF and IGF. Cells were treated with increasing concentrations of the appropriate TKI alone (red) or in combination with 50ng/mL HGF (blue) or 75ng/mL IGF (green). (B) Cells were treated for 6 hours with gefitinib (HCC827, PC-9, A431, HN11), PF00299804 (H1975) or lapatinib (BT-474, SKBR3) alone or in combination with HGF (50ng/mL) or IGF (75ng/mL). All drugs were used at 1μM. Cell lysates immunoblotted to detect indicated proteins. BT-474 and SKBR3 cell lysates were run on the same gel, and no MET or IGF-1Rβ was detected in SKBR3 cells relative to BT-474 cells. See also Table S1 for quantification.
To confirm the ability of HGF to induce resistance to EGFR TKIs, we introduced the human HGF gene into HCC827 cells (HCC827-HGF). Parental HCC827 cells secrete undetectable levels of HGF; however, HCC827-HGF cells express HGF protein (Figure S2A) and secrete approximately 70ng/mL HGF into the culture medium (data not shown). Further, HCC827-HGF cells are gefitinib resistant (Figure S2B) and maintain PI3K/AKT, ERK and mTOR signaling in the presence of gefitinib (Figure S2A); however gefitinib sensitivity is restored with the addition of a MET inhibitor (Figure S2B). We also evaluated the capacity of HGF to induce gefitinib resistance in vivo using an HCC827-HGF xenograft model. We have previously shown that parental HCC827 cells demonstrate complete responses to gefitinib in vivo (Engelman et al., 2006; Engelman et al., 2007a). However, the HCC827-HGF xenografts demonstrated resistance (Figure 3E). Treatment with gefitinib alone was slightly more effective than no treatment or treatment with PF2341066 alone, but only the combination of gefitinib and PF2341066 completely inhibited tumor growth (p < 0.001; gefitinib vs. gefitinib/PF2341066; Figure 3E). Indeed, 3 out of 4 mice were cured after 70 days of combined treatment with no evidence of re-growth 70 days after stopping treatment.
Since HGF ligand appeared to be a potent inducer of resistance to RTK inhibitors, we compared its efficacy to that of IGF ligand, which we had previously found to cause gefitinib resistance in A431 cells (Guix et al., 2008). Although IGF exposure led to significant rescue from gefitinib-induced cell death in A431 cells, and partial rescue in HN11 EGFR wild-type cells, the other five cell lines tested remained sensitive to ERBB inhibition despite the presence of IGF (Figure 4A and Figure S1). Interestingly, in three of those cell lines (BT-474, HCC827 and H1975), IGF was unable to maintain PI3K/AKT signaling despite potent activation of IGR-1Rβ (Figure 4B and Table S1). Of note, IGF did not restore ERK phosphorylation in any of the six cell lines examined, including those in which it induced IGF-1Rβ and/or PI3K/AKT activation (Figure 4B). Thus, unlike IGF, HGF may be more potent at promoting resistance because it leads to activation of both the PI3K/AKT and ERK pathways. Unexpectedly, IGF restored PI3K/AKT signaling in PC-9 cells, but these cells still remained highly sensitive to EGFR-inhibition after 72 hours (Figure 4A and Figure S1C). This disconnect between maintenance of PI3K/AKT signaling and lack of an effect on cell viability is not due to a brief, transient restoration of downstream signaling, as we observed that IGF maintained PI3K signaling in PC-9 cells for at least 24 hours in the presence of gefitinib (data not shown).
MET amplified gefitinib resistant HCC827 GR cells utilize ERBB3 as the primary adaptor to activate PI3K/AKT signaling (Engelman et al., 2007b). Although HGF treatment was sufficient to rescue AKT phosphorylation in several EGFR-driven cell lines in the presence of TKIs, ERBB3 phosphorylation was not restored (Figure 3D). This suggests that HGF-induced MET activation utilizes an adaptor other than ERBB3 to activate PI3K signaling. To determine which PI3K adaptors were being utilized to maintain HGF-mediated PI3K signaling, we immunoprecipitated the p85 regulatory subunit of PI3K and examined co-precipitating phosphotyrosine proteins (Engelman et al., 2005; Engelman et al., 2007b; Guix et al., 2008). As expected, treatment with a TKI disrupted the association of ERBB3 (and other phosphotyrosine proteins) with p85, and the addition of HGF did not restore the interaction (Figure 5A). However, we observed that HGF potently induced the association between p85 and Grb2 associated binder 1 (GAB1), which runs as a broad, highly tyrosine-phophorylated band at approximately 110kDa.
Figure 5. HGF rescue of PI3K/AKT signaling is mediated though GAB1 instead of ERBB3.
(A) Cells treated for 6 hours with gefitinib (HCC827, PC-9), PF00299804 (H1975), or lapatinib (BT-474, SKBR3), alone or in combination with HGF (50ng/mL). All drugs were used at 1μM. Cell extracts were immunoprecipitated with an anti-p85 antibody followed by Western blot with anti-p-Tyr, anti-ERBB3 and anti-p85 antibodies. (B) HCC827 cells were transfected with a negative control or GAB1 siRNA for 48 hours. Transfected cells were treated for 6 hours with gefitinib (1μM) alone or in combination with HGF (50ng/mL). Cell lysates were immunoblotted to detect indicated proteins. See also Figure S3. (C) HCC827 cells were transfected with GAB1 siRNA or a negative control siRNA for 48 hours, then treated with increasing concentrations of gefitinib, alone or in combination with 50ng/mL HGF. Left, cell viability relative to untreated controls measured after 72 hours. Each data point represents the mean ±SD of 6 wells. Right, plot of IC50 values corresponding to cell viability curves.
To more directly assess if GAB1 mediates HGF-mediated activation of PI3K/AKT signaling and cell viability, we used small interfering RNA (siRNA) to knockdown GAB1 expression in the HCC827 cells. Knockdown of GAB1 reduced HGF-mediated rescue of PI3K/AKT signaling (Figure 5B), and inhibited the ability of HGF to rescue HCC827 cells from gefitinib induced cell death (Figure 5C). Of note, although the addition of HGF leads to substantial loss of GAB1 protein (Figure 5B), the amount of tyrosine phosphorylated GAB1 is dramatically increased (Figure S3), and this facilitates the efficient coupling to PI3K (Figure 5A). Thus, activation of HGF/MET signaling can lead to gefitinib resistance in EGFR mutant cancers by activating PI3K/AKT signaling through two different adaptors: ERBB3 when MET is activated by genomic amplification or GAB1 when MET is activated by HGF.
Transient HGF exposure leads to stable ligand-independent gefitinib resistance in HCC827-50GR cells through selection of a pre-exisiting MET amplified clone
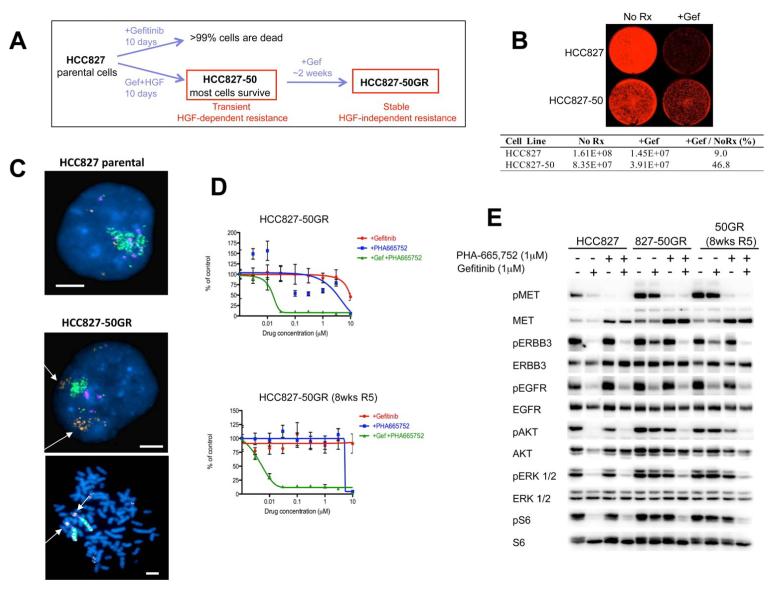
Because HGF-induced resistance to EGFR TKIs appears intimately linked to ligand-induced activation of downstream signaling, we hypothesized that long-term resistance would require continuous exposure to HGF. We observed that by replenishing cells with HGF in combination with the EGFR TKI every 3 days, cells continue to be highly resistant indefinitely (data not shown). Thus, we treated each cell line with HGF in the presence of an EGFR inhibitor for 14 days, and then removed HGF, but maintained the cells in the EGFR TKI. Surprisingly, HCC827 cells treated transiently with HGF remained permanently resistant to gefitinib after HGF withdrawal (Figure 6A, B). These stably resistant cells were termed HCC827-50GR (50ng HGF Gefitinib Resistant) cells (Figure 6A). In contrast, HCC827 cells that are not pretreated with HGF, develop gefitinib resistance only after 6 months of gradually increasing concentrations of drug exposure (Engelman et al., 2007b). In addition, when HCC827-50GR cells were grown in media alone (without gefitinib) for eight weeks, these cells (HCC827-50GR (8wksR5)) maintained their resistance (Figure S4A). Treatment with HGF alone (without gefitinib) for 14 days did not yield stably resistant cells (Figure S4C and Table S2). Thus, lasting resistance conferred by transient HGF requires the selective pressure of gefitinib during ligand exposure.
Figure 6. Transient HGF exposure leads to MET amplification and stable ligand-independent gefitinib resistance in HCC827 cells.
(A) HCC827 cells treated with HGF (50ng/mL) and 1μM gefitinib are resistant to gefitinib (HCC827-50 cells). After the removal of HGF, stably resistant HGF-independent HCC827-50GR cells survive in 1μM gefitinib alone. In contrast, parental HCC827 cells do not survive when treated with 1μM gefitinib. (B) Parental HCC827 cells and HCC827-50 cells (pre-treated with gefitinib in combination with HGF (50ng/mL) for 14 days) were grown in media alone (No Rx) or media treated with 1μM gefitinib (+Gef) for 7 days. Viable cells were visualized and quantified using Syto60 staining. (C) Fluorescence in situ hybridization (FISH) of MET/EGFR/CEP7 probe set with HCC827 and HCC827-50GR cells. MET (orange) EGFR (green) CEP7 (aqua). Metaphase spread (bottom) shows multiple copies of EGFR and MET (arrow) on individual chromosomes. Scale bars represent 10μm. (D) HCC827-50GR cells (upper) and HCC827-50GR cells grown in media alone (without gefitinib) for 8 weeks, 50GR 8wks R5 (lower), were treated with increasing concentrations of gefitinib or PHA-665,752 or their combination for 72 hours. Cell viability was measured relative to untreated controls. Each data point represents the mean ±SD of 6 wells. (E) HCC827 cells and stably resistant HCC827-50GR cells were treated for 6 hours with gefitinib, PHA-665,752, or their combination. All drugs were used at 1μM. Cell lysates were immunoblotted to detect indicated proteins. See also Figure S4 and Table S2 and S3.
Stably resistant HCC827-50GR cells maintained PI3K/AKT, mTORC1 and ERK activation in the presence of gefitinib. Surprisingly, ERBB3 also remained phosphorylated in HCC827-50GR cells treated with gefitinib (Figure S4B), which suggests that although initial HGF-mediated resistance mechanisms utilized GAB1 to activate PI3K/AKT signaling, the ligand-independent HCC827-50GR cells utilize ERBB3 to activate PI3K/AKT signaling. This observation suggests that short-term exposure to HGF may lead HCC827 cells to develop or select the same mechanism of stable resistance, through activation of ERBB3/PI3K signaling, as was observed in MET amplified HCC827 GR cells (Engelman et al., 2007b). Unlike the HCC827 cells, several other EGFR-driven cancer cell lines that were made resistant to EGFR TKIs by HGF treatment did not maintain stable ligand-independent resistance after the withdrawal of HGF (Figure S4D-F and Table S3). These findings suggest that HCC827 cells are uniquely poised to develop stable ligand-independent resistance.
Stably-resistant HCC827-50GR cells had increased total MET protein levels compared to parental cells and maintained MET phosphorylation in the presence of gefitinib (Figure S4B), mimicking MET amplified HCC827 GR cells. Therefore, we examined MET copy number using fluorescent in situ hybridization (FISH), and found significant MET copy number gains in HCC827-50GR cells compared to parental cells (Figure 6C). Quantitative PCR demonstrated a three to four fold amplification of MET, similar to the HCC827 GR and PFR cells (data not shown). These results suggest that MET amplification may be driving ERBB3/PI3K/AKT signaling and gefitinib resistance in HCC827-50GR cells.
To examine this hypothesis, we exposed HCC827-50GR cells to PHA-665,752 alone or in combination with gefitinib. Only the combination of gefitinib and PHA-665,752 resulted in a substantial reduction in the number of viable cells (Figure 6D, upper). In addition, the HCC827-50GR (8wks R5) cells (grown in media without gefitinib for eight weeks) also remained sensitive only to the combination of MET and EGFR inhibition (Figure 6D, lower). Further, treatment with gefitinib in combination with PHA-665,752 completely blocked ERBB3 phosphorylation as well as downstream PI3K/AKT, mTORC1 and ERK signaling in HCC827-50GR and HCC827-50GR(8wks R5) cells (Figure 6E). Taken together, these results suggest that MET inhibition restores EGFR dependence and gefitinib sensitivity in HCC827-50GR cells.
These results led us to examine tissue sections from HCC827-HGF xenograft models treated with gefitinib (Figure 3E). Of three tumors that developed gefitinib resistance, one exhibited significant MET amplification (Figure 7A). Thus, MET amplification is also facilitated by HGF in vivo.
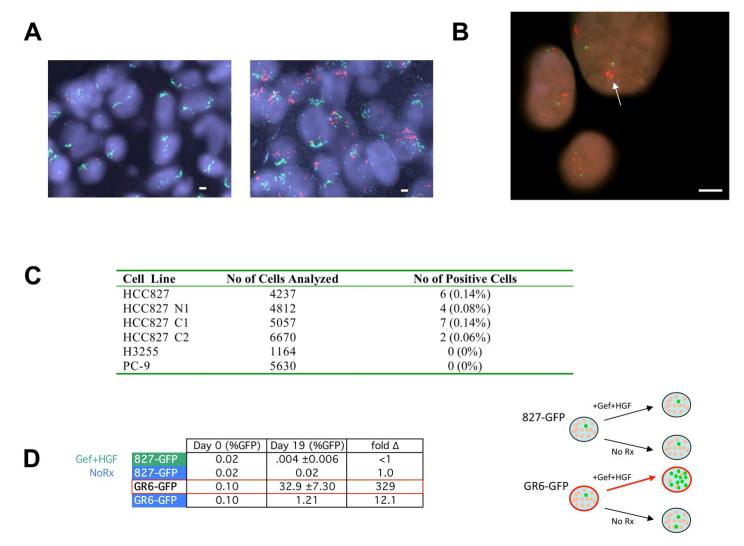
Figure 7. HGF treatment selects out a small pre-existing population of MET amplified HCC827 cells from the parental population in vitro and in vivo.
(A) Fluorescence in situ hybridization (FISH) of MET/EGFR/CEP7 probe set. MET (red) EGFR (green) CEP7 (aqua). Left, tumor sections from control HCC827 xenograft models that do not express HGF showed normal MET copy number. Right, tumor sections from one of three HCC827-HGF xenografts treated with gefitinib (Figure 3E) showed significant MET amplification (arrow). (B) High-throughput FISH analysis of HCC827 cells identifies a subpopulation harboring MET amplification (arrow). MET (RP-11-95I120; red); 7qter (RP-11-6903; green). All scale bars represent 10μm. (C) Parental HCC827 cells and three independent clones harbor a small percentage of MET amplified cells. No pre-existing MET amplification was detected in H3255 or PC-9 cell populations. (D) Left, HCC827 cells were spiked with approximately 0.1% of GFP labeled HCC827 cells or GFP labeled MET amplified HCC827 GR6 cells. Each population was grown in either media alone or media treated with gefitinib (1μM) with HGF (50ng/mL). Cells were collected after 19 days and GFP levels were quantified using FACS. Each data point for cells treated with gefitinib+HGF represents the mean ±SD for 3 independent wells. Fold change is the ratio of Day 19 to Day 0 (%GFP). Right, diagrammatic depiction of results. See also Figure S5.
Because HCC827 GR, PFR and 50GR cells all eventually develop focal MET amplification as a resistance mechanism, we hypothesized that parental HCC827 cells may harbor a pre-existing MET amplified clone. We analyzed 4237 individual HCC827 cell nuclei using high-throughput fluorescence in situ hybridization (FISH) (Experimental Procedures) and identified 6 cells (0.14%; 6/4237) that harbored significant MET copy number gains (Figure 7B, C). These results were confirmed in an independent experiment using a second gefitinib sensitive parental HCC827 cell line (HCC827 N1; Figure 7C). We also generated two subclones derived from single cells from the gefitinib sensitive parental HCC827 cell line (HCC827 C1 and C2). Both subclones were sensitive to gefitinib in vitro (data not shown), and each also contained a low frequency population of MET amplified cells (Figure 7C). We further examined the gefitinib sensitive H3255 and PC-9 cells using FISH. Gefitinib resistant clones of both H3255 and PC-9 have been isolated and reported to contain the EGFR secondary resistance mutation T790M but not MET amplification (Engelman et al., 2006; Ogino et al., 2007). We did not detect a subpopulation of MET amplified cells in the H3255 or the PC-9 cells (Figure 7C).
We hypothesized that the mechanism by which transient treatment with HGF and gefitinib leads to the generation of MET amplified HCC827-50GR cells is by selecting out this small population of pre-existing MET amplified cells from the parental HCC827 cell population. To test this hypothesis, we spiked unlabeled HCC827 parental cells with 0.1% of either GFP labeled HCC827 cells or GFP labeled MET amplified HCC827 GR6 cells. We treated these two populations with either media alone (no selection) or with gefitinib in combination with HGF. Media was changed and fresh HGF was added every 72 hours, and cells were collected after 19 days for FACS to quantify the percent of cells with GFP expression (Figure S5A). As expected, there was no significant change in the percentage of GFP labeled HCC827 cells at the end of 19 days. However, the percentage of GFP labeled MET amplified HCC827 GR6 cells increased over 300 fold to almost 33% in just over two weeks (Figure 7D). Taken together, these results suggest that HGF exposure in the presence of an EGFR inhibitor leads to the rapid selection of a pre-existing MET amplified clone in the HCC827 cells (Figure S5B).
Analyses of tumors with acquired resistance to gefitinib/erlotinib reveal evidence of pre-treatment MET amplification and increased HGF expression in resistant cancers
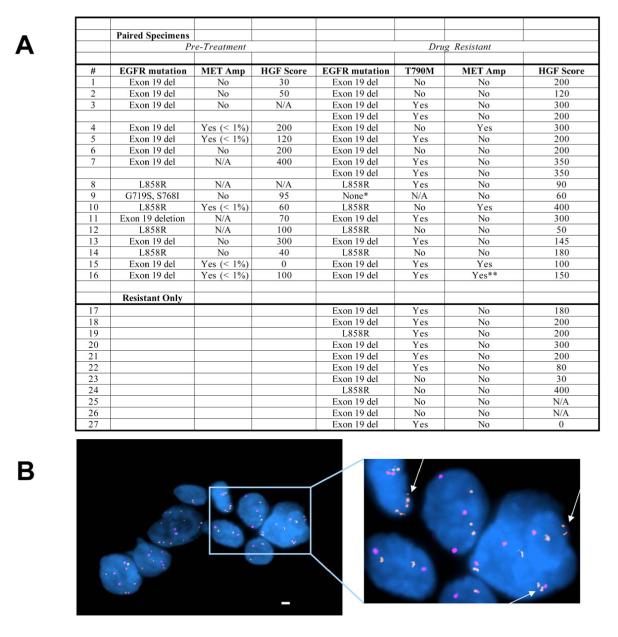
To determine the clinical implications of these in vitro and in vivo observations, we examined tumor specimens from gefitinib or erlotinib treated EGFR mutant NSCLC patients (Figure 8). All patients had a clinical partial tumor response to gefitinib or erlotinib treatment and subsequently developed clinical drug resistance. We evaluated 27 patients, 16 with paired pre and post geftinib/erlotinib treatment specimens and 11 with drug resistance specimens alone. All specimens, when feasible, were evaluated for MET amplification, HGF expression by immunohistochemistry (IHC), and presence of EGFR T790M (Figure 8 and Figure S6). We observed EGFR T790M in 55 % (15/27) and MET amplification in 4/27 (15%) of resistant tumor specimens. In patients with paired tumor specimens, HGF expression was higher in the drug resistant specimens compared to pre-treatment specimens (p = 0.025; Wilcoxon signed-rank test). In patients with drug resistant specimens alone, HGF expression was similar to that of drug resistant specimens in patients with paired tumor specimens. Together these findings support our in vitro and in vivo studies on HGF mediating resistance to EGFR TKIs.
Figure 8. HGF expression and pre-existing MET amplification can be detected in tumor specimens from NSCLC patients.
(A) Summary of tumors from geftinib/erlotinib treated patients, including 16 paired, and 11 drug resistant samples only. Samples were evaluated for EGFR mutational status, MET amplification and HGF expression *Specimen contained less < 30% tumor cells. **MET amplification defined by qPCR as previously described (Engelman et al., 2007b). Data on EGFR T790M and MET amplification in resistant specimens only from patients 1-4 and 17-19 has been previously published (Engelman et al., 2007b). N/A; not available. (B) FISH analysis of pre-treatment sample from patient 10 shows evidence of a subset of MET amplified cells (arrow) before exposure to an EGFR TKI. MET (RP-11-95I120; orange); CEP 7 (aqua). Scale bars represent 10μm. See also Figure S6.
We further evaluated the pre-treatment specimens for evidence of MET amplification. In all 4 patients with MET amplification in the drug resistant specimens, we observed rare (< 1%) tumor cells with MET amplification from the corresponding pre-treatment specimens (Figure 8A, B). In contrast, of 8 cases that had resistant cancers without MET amplification, we observed rare MET amplified tumor cells in only 1 of the corresponding pre-treatment tumor specimen. These findings are consistent with cell line data (Figure 7B, C) where we observed evidence of pre-existing MET amplification only in the cell line that subsequently develops MET amplification as its resistance mechanism.
Discussion
Kinase inhibitors have emerged as effective clinical therapies for cancers that exhibit oncogene addiction to a particular kinase. (Demetri et al., 2002; Druker et al., 2001; Inoue et al., 2006; Mok et al., 2008; Sequist et al., 2008). However, the clinical success of treatment with kinase inhibitors is uniformly limited by the development of drug resistance. To date, resistance mechanisms have predominately involved secondary genomic alterations in the target kinase that alter either the physical (such as steric hindrance) or biochemical (change in ATP affinity) properties of the receptor and result in drug resistance (Gorre et al., 2001; Shah et al., 2002; Yun et al., 2008). We have previously described MET amplification as a mechanism of gefitinib resistance in EGFR mutant cancers (Engelman et al., 2007b), leading to persistent activation of both PI3K/AKT and ERK signaling in the presence of the EGFR TKI (Engelman et al., 2007b).
A critical question for all resistance mechanisms to kinase inhibitors is whether they occur as a result of treatment or whether they pre-exist prior to treatment and are selected out during the course of therapy. At least some imatinib resistant CML clones are thought to be present at low levels prior to treatment and undergo clonal selection during imatinib exposure (Hofmann et al., 2003; Roche-Lestienne et al., 2003; Roche-Lestienne et al., 2002; Shah et al., 2002). Similarly, EGFR T790M can be detected at low levels in EGFR mutant NSCLC patients prior to gefitinib or erlotinib treatment (Maheswaran et al., 2008). Our current findings provide support that this may also be the case for MET amplification both in HCC827 cells (Figure S5B) and in NSCLC patients that subsequently develop MET amplification at the time of clinical gefitinib or erlotinib resistance (Figure 8). The identification of a drug resistance mechanism from a pre-treatment tumor specimen provides the opportunity to specifically target that resistance mechanism prior to its emergence. This approach is clinically appealing as combined treatment with an EGFR and MET inhibitor, specifically in patients with evidence of MET amplification at baseline, may lead to a longer time to progression than is currently observed with gefitinib or erlotinib alone (Asahina et al., 2006; Inoue et al., 2006; Mok et al., 2008; Paz-Ares et al., 2006; Sequist et al., 2008; Tamura et al., 2008). In fact, combined EGFR and MET inhibition in HCC827 cells extinguishes the emergence of MET amplified drug resistant clones (data not shown). However, it will be critical to learn whether upfront treatment with combination therapy is tolerable (toxicity) and/or will provide more clinical benefit than treatment at the time of relapse.
Intriguingly, HCC827 cells appear to be pre-disposed to the development of low level MET amplification as subclones of cells expanded from single cell clones derived from parental HCC827 cells (HCC827 N1 and N2) also are found to contain low levels of MET amplification (Figure 7C). MET is located at a fragile site in chromosome 7, which facilitates its amplification, and subsequently a selection for clones harboring MET amplification can occur under drug pressure (Hellman et al., 2002). Why this occurs only in the HCC827 cells and a subset of lung cancers, and not in other EGFR mutant cell lines and cancers, is currently unknown. Collectively, these studies suggest, but do not prove that the specific mechanisms of resistance that will develop as a result of drug exposure may be pre-determined and occur as a result of drug selection. Understanding why some EGFR mutant cancers are pre-disposed to develop MET amplification will help further refine the clinical development of EGFR and MET inhibitor combinations.
In this study, we also demonstrate two different and distinct roles for HGF in mediating EGFR TKI resistance. First, HGF can independently rescue both PI3K/AKT and ERK signaling in the presence of gefitinib and lead to drug resistance both in vitro and in vivo. Unlike in MET amplified resistant cancers, HGF mediated resistance occurs through GAB1, not ERBB3, signaling. Higher levels HGF can be detected in tumor specimens from NSCLC patients that are clinically resistant to gefitinib or erlotinib compared to pre-treatment tumor specimens (Figure 8A). Notably in some patients without evidence of EGFR T790M or MET amplification, HGF expression is greater in the resistant specimen (patients 1 (Figure S6C) and 14) than in the pre-treatment specimen, supporting a role for HGF alone in promoting drug resistance. This is consistent with prior observations (Yano et al., 2008). Ligand mediated drug resistance is unique to HGF as IGF does not rescue TKI-induced cell death in the majority of cell lines tested. Surprisingly, IGF did not restore P13K/AKT signaling in most EGFR mutant cancers, despite substantial levels of IGF-1Rβ expression and tyrosine phosphorylation. Furthermore, unlike HGF, IGF did not restore ERK signaling even in cell lines in which it restored PI3K/AKT signaling in the presence of a TKI. These signaling differences between HGF and IGF may underlie the lack of drug resistance induced by IGF. In its second role, HGF accelerates the emergence of MET amplification in HCC827 cells both in vitro and in vivo. Intriguingly, this process requires concomitant EGFR inhibition, as HGF exposure alone does not lead to emergence of MET amplified clones. It is possible that in the presence of EGFR inhibition, HGF provides a unique proliferative advantage to a subset of cells with high MET expression (those with amplification) thus facilitating their rapid clonal expansion. Activation of MET signaling is a unique resistance mechanism to kinase inhibitors as it can occur through multiple independent mechanisms, amplification and/or ligand mediated, and when combined can lead to rapid evolution of drug resistance.
Our current findings provide insight into future therapeutic strategies for the treatment of EGFR mutant NSCLC. Although MET amplification has been detected in up to 20% of EGFR mutant patients that develop acquired resistance to gefitinib or erlotinib, activation of MET signaling (by both amplification and mediated by HGF) may in fact account for a larger fraction of gefitinib/erlotinib resistant tumors. It is tempting to speculate that HGF production by the stroma may also partially explain why clinical resistance emerges discordantly in some tissues like the liver, bone and brain, while pulmonary disease continues to respond to erlotinib treatment (personal observation). Our study further implies that the therapeutic combination of an irreversible EGFR inhibitor (effective against EGFR T790M) and a MET inhibitor is an attractive treatment combination for a significant portion of gefitinib/erlotinib resistant EGFR mutant NSCLC patients. In addition, these findings highlight the potential to prospectively identify treatment naïve EGFR mutant lung cancer patients who are likely to develop MET amplification and may benefit from initial combination therapy with a MET inhibitor.
Experimental Procedures
Cell culture reagents, viability studies and Western analyses
Cell lines and growth conditions are described in Supplemental Experimental Procedures. Gefitinib and lapatinib were obtained from commercial sources (American Custom Chemical Corporation and LC Laboratories Woburn, MA). PF00299804, PHA-665,752 and PF2341066 were provided by Pfizer (La Jolla, CA). Cell viability was assessed 72 hours following drug exposure by Syto60 staining (Invitrogen) or by MTS assay (Promega). Cells were lysed in an NP-40 containing lysis buffer, separated by SDS/PAGE electrophoresis and transferred to PVDF membranes. Immunoblotting was performed according to the antibody manufacturer’s recommendations. Antibody binding was detected using enhanced chemiluminescence (PerkinElmer, Waltham, MA).
Generation of in vitro drug resistant HCC827 cells
To generate a resistant cell line, HCC827 cells were exposed to increasing concentrations of PF00299804 similar to our previously described methods (Engelman et al., 2006; Engelman et al., 2007b). PF00299804 concentrations were increased stepwise from 1 nM to 1 μM when the cells resumed growth kinetics similar to untreated parental cells. To confirm the emergence of a resistant clone, MTS assays were performed following growth at each concentration.
In vivo treatment studies
All xenograft studies were performed in accordance with the standards of the Institutional Animal Care and Use Committee (IACUC) under a protocol approved by the Animal Care and Use Committee of Massachusetts General Hospital. Generation and treatment of xenograft models were performed as previously described and detailed in Supplementary Experimental Procedures (Engelman et al., 2007a).
SNP analyses
SNP analyses to evaluate genome wide copy number changes were performed as previously described (Engelman et al., 2007b). Comparison of gene copy number between HCC827 and the PFR clones was performed using dChip software according to previously established methods (Engelman et al., 2007b; Zhao and Vogt, 2008). SNP data is available from the ncbi gene expression omnibus database (accession number: GSE18797).
FISH probes and hybridization
Bacterial artificial chromosome (BAC) clones CTD-2257H21 (EGFR (7p11.2 )) and RP11-95I20 (MET (7q31.2)) were purchased from Children’s Hospital Oakland Research Institute (CHORI; Oakland, CA). DNA was extracted using a Qiagen kit (Valencia, CA) and labeled with Spectrum Green- or Spectrum Orange-conjugated dUTP by nick translation (Vysis/Abbott Molecular, Des Plaines, IL). The CEP7 probe (Vysis/Abbott Molecular, Des Plaines, Il) was used according to manufacturer’s instructions. Chromosomal mapping and hybridization efficiency for each probe set were verified in normal metaphase spreads (data not shown). Three color FISH assays were performed as previously described (Engelman et al., 2007b).
High throughput fluorescence in situ hybridization
A Bioview work station with Duet™ software (Bioview Ltd, Rehovot, Israel) was used to screen for rare MET amplified cells. Automatic scans were performed according to manufacturer’s suggested guidelines after setting classification criteria for each FISH probe. Images were captured and classified in an automated fashion and manually reviewed to ensure accuracy. Any unclassified images were manually reviewed and scored. Any cells that could not be scored were excluded from the analysis. Paraffin embedded specimens derived from NSCLC patients or from xenografts were manually scanned for evidence of MET amplification.
NSCLC patients
Tumor specimens from gefitinib or erlotinib treated patients were obtained from the Dana Farber Cancer Institute/Brigham and Women’s Hospital (Boston, MA), Massachusetts General Hospital (Boston, MA), the Chinese University (Hong Kong, China) and from Guangdong Provincial People’s Hospital (Guangzhou, China) under Institutional Review Board approved studies. All patients provided written informed consent. The presence of an EGFR mutation in each specimen was confirmed by exonspecific amplification (exons 18-21), followed by direct sequencing, or using the Surveyor™ endonuclease coupled with denaturing HPLC (DHPLC), fractionation and sequencing (Janne et al., 2006). The EGFR T790M mutation was detected using Surveyor™ endonuclease coupled with DHPLC or an allele specific PCR (Janne et al., 2006; Maheswaran et al., 2008). Both methods are capable of detecting the EGFR T790M mutation at an allele frequency of 1-5%. HGF immunohistochemistry was performed as using an anti-HGF 7.2 antibody kindly provided by Dr. George Vander Woude at the Van Andel Institute (see Supplemental Experimental Procedures).
Highlights.
Rare MET amplified cells exist in some EGFR mutant lung cancers prior to treatment.
HGF induces resistance to tyrosine kinase inhibitors in EGFR addicted cancers.
HGF accelerates MET amplification by expanding pre-existing MET amplified cells.
Analysis of pre-treatment cancers identifies those poised to become MET amplified.
Significance.
The therapeutic success of EGFR tyrosine kinase inhibitors (TKIs) in EGFR mutant lung cancers is limited by the development of drug resistance, mediated by MET amplification in a subset of patients. Here we observe that MET amplification is present in a small fraction of cells prior to drug exposure and its emergence is dramatically accelerated by its ligand, HGF. These findings provide insight into the origins of drug resistance in EGFR mutant cancers, and support a rationale for combination treatment strategies as initial therapies, specifically in a molecularly defined cohort of patients with evidence of pre-existing MET amplification.
Supplementary Material
Acknowledgements
This study is supported by grants from the National Institutes of Health RO1CA114465 (P.A.J., B.Y.Y.), R01CA135257 (P.A.J., B.Y.Y., J.A.E.), NIH K08 grant CA120060 (JAE), R01CA137008 (J.A.E., P.A.J.), R01CA140594 (J.A.E), National Cancer Institute Lung SPORE P50CA090578 (P.A.J., B.Y.Y., J.A.E.), DF/HCC Gastrointestinal Cancer SPORE P50 CA127003 (J.A.E.), the American Association for Cancer Research (J.A.E.), the V Foundation (J.A.E.), American Cancer Society RSG-06-102-01-CCE (P.A.J., B.Y.Y., J.A.E.), Hazel and Samuel Bellin research fund (P.A.J.), and the Ellison Foundation Scholar (J.A.E.). We also thank Mike Warring and Andrew Cosgrove (MGH) for FACS analysis, and Tatiana Zolotarev for technical histology assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asahina H, Yamazaki K, Kinoshita I, Sukoh N, Harada M, Yokouchi H, Ishida T, Ogura S, Kojima T, Okamoto Y, et al. A phase II trial of gefitinib as first-line therapy for advanced non-small cell lung cancer with epidermal growth factor receptor mutations. Br J Cancer. 2006;95:998–1004. doi: 10.1038/sj.bjc.6603393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, Chiang A, Yang G, Ouerfelli O, Kris MG, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, Heinrich MC, Tuveson DA, Singer S, Janicek M, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, Capdeville R, Talpaz M. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, Cichowski K, Johnson BE, Cantley LC. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc Natl Acad Sci U S A. 2005;102:3788–3793. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borras AM, Gale CM, Naumov GN, Yeap BY, Jarrell E, Sun J, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Gale CM, Lifshits E, Gonzales AJ, Shimamura T, Zhao F, Vincent PW, Naumov GN, Bradner JE, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007a;67:11924–11932. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007b;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- Guix M, Faber AC, Wang SE, Olivares MG, Song Y, Qu S, Rinehart C, Seidel B, Yee D, Arteaga CL, Engelman JA. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman A, Zlotorynski E, Scherer SW, Cheung J, Vincent JB, Smith DI, Trakhtenbrot L, Kerem B. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 2002;1:89–97. doi: 10.1016/s1535-6108(02)00017-x. [DOI] [PubMed] [Google Scholar]
- Hofmann WK, Komor M, Wassmann B, Jones LC, Gschaidmeier H, Hoelzer D, Koeffler HP, Ottmann OG. Presence of the BCR-ABL mutation Glu255Lys prior to STI571 (imatinib) treatment in patients with Ph+ acute lymphoblastic leukemia. Blood. 2003;102:659–661. doi: 10.1182/blood-2002-06-1756. [DOI] [PubMed] [Google Scholar]
- Inoue A, Suzuki T, Fukuhara T, Maemondo M, Kimura Y, Morikawa N, Watanabe H, Saijo Y, Nukiwa T. Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol. 2006;24:3340–3346. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- Janne PA, Borras AM, Kuang Y, Rogers AM, Joshi VA, Liyanage H, Lindeman N, Lee JC, Halmos B, Maher EA, et al. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res. 2006;12:751–758. doi: 10.1158/1078-0432.CCR-05-2047. [DOI] [PubMed] [Google Scholar]
- Janne PA, Schellens JH, Engelman JA, Eckhardt SG, Millham R, Denis LJ, Britten CD, Wong SG, Boss DS, Camidge DR. Preliminary activity and safety results from a phase I clinical trial of PF-00299804, an irreversible pan-HER inhibitor, in patients (pts) with NSCLC. Journal of Clinical Oncology. 2008;26 ASCO #8027. [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Yatabe Y, Endoh H, Yoshida K, Hida T, Tsuboi M, Tada H, Kuwano H, Mitsudomi T. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res. 2006;12:5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok T, Wu Y, Thongprasert S. Phase III, randomized, open label, first-line study of gefitinib vs. carboplatin/paclitaxel in clinically selected patients with advanced non-small cell lung cancer (IPASS) Annals of Oncology. 2008;19 [Google Scholar]
- Ogino A, Kitao H, Hirano S, Uchida A, Ishiai M, Kozuki T, Takigawa N, Takata M, Kiura K, Tanimoto M. Emergence of epidermal growth factor receptor T790M mutation during chronic exposure to gefitinib in a non small cell lung cancer cell line. Cancer Res. 2007;67:7807–7814. doi: 10.1158/0008-5472.CAN-07-0681. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares L, Sanchez J, Garcia-Velasco A, Massuti B, Lopez-Vivanco G, Provencio M, Montes A, Isla D, Amador ML, Rossel R. A prospective phase II trial of erlotinib in advanced non-small cell lung cancer (NSCLC) patients (p) with mutations in the tyrosine kinase (TK) domain of the epidermal growth factor receptor (EGFR) Journal of Clinical Oncology. 2006;24 ASCO #7020. [Google Scholar]
- Riely GJ. Second-generation epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. J Thorac Oncol. 2008;3:S146–149. doi: 10.1097/JTO.0b013e318174e96e. [DOI] [PubMed] [Google Scholar]
- Roche-Lestienne C, Lai JL, Darre S, Facon T, Preudhomme C. A mutation conferring resistance to imatinib at the time of diagnosis of chronic myelogenous leukemia. N Engl J Med. 2003;348:2265–2266. doi: 10.1056/NEJMc035089. [DOI] [PubMed] [Google Scholar]
- Roche-Lestienne C, Soenen-Cornu V, Grardel-Duflos N, Lai JL, Philippe N, Facon T, Fenaux P, Preudhomme C. Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood. 2002;100:1014–1018. doi: 10.1182/blood.v100.3.1014. [DOI] [PubMed] [Google Scholar]
- Schellens JH, Britten CD, Camidge DR, Boss D, Wong S, Diab S, Guo F, Maguire RP, Letrent SP, Eckhardt SG. First-in-human study of the safety, tolerability, pharmacokinetics (PK), and pharmacodynamics (PD) of PF-00299804, a small molecule irreversible pan-HER inhibitor in patients with advanced cancer. Journal of Clinical Oncology. 2007;25 ASCO #3599. [Google Scholar]
- Sequist LV, Martins RG, Spigel D, Grunberg SM, Spira A, Janne PA, Joshi VA, McCollum D, Evans TL, Muzikansky A, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Okamoto I, Kashii T, Negoro S, Hirashima T, Kudoh S, Ichinose Y, Ebi N, Shibata K, Nishimura T, et al. Multicentre prospective phase II trial of gefitinib for advanced non-small cell lung cancer with epidermal growth factor receptor mutations: results of the West Japan Thoracic Oncology Group trial (WJTOG0403) Br J Cancer. 2008;98:907–914. doi: 10.1038/sj.bjc.6604249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano S, Wang W, Li Q, Matsumoto K, Sakurama H, Nakamura T, Ogino H, Kakiuchi S, Hanibuchi M, Nishioka Y, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res. 2008;68:9479–9487. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou HY, Li Q, Lee JH, Arango ME, McDonnell SR, Yamazaki S, Koudriakova TB, Alton G, Cui JJ, Kung PP, et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 2007;67:4408–4417. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.