Abstract
Alzheimer’s disease (AD) is a complex disorder with a clear genetic component. Three genes have been identified as the cause of early onset familial AD (EOAD). The most common form of the disease is, however, a sporadic one presenting itself in later stages of life (LOAD). The genetic component of this late onset form of AD has been the target of a large number of studies, since only one genetic risk factor (APOE4) has been consistently associated with the disease. However, technological advances allow new approaches in the study of complex disorders. In this review, we discuss the new results produced by genome wide association studies, in light of the current knowledge of the complexity of AD genetics.
INTRODUCTION
Historically, Alzheimer's disease (AD) is the most common cause of dementia. It is a progressive neurodegenerative disorder with an insidious onset, which typically appears in older individuals, but may affect people as early as the third decade of life [Rademakers, et al. 2003, Rogaeva 2002]. The genetics of Alzheimer’s disease is complex and heterogeneous. Most cases are “sporadic” with no apparent familial recurrence of the disease. However, a small percentage of AD cases (1–2% of all cases) have an early onset (EOAD), with symptoms appearing before 65 years of age. In these patients, the disease commonly aggregates within families and typically presents an autosomal dominant pattern of inheritance. Mutations in three genes are known to account for this early onset, familial type of the disease: amyloid precursor protein gene (APP), presenilin 1 gene (PSEN1) and presenilin 2 gene (PSEN2) [Rogaeva 2002]. In fact, early-onset autosomal dominant disease with age of onset younger than sixty years, seems to be completely explained by pathogenic mutations in these three genes.
The most common, late onset and sporadic form of the disease remains mostly a genetic conundrum. The only well established genetic risk factor for late onset AD (LOAD) is the E4 allele of apolipoprotein E although many lifestyle risk factors have been reported including low education, mid-life high blood pressure and cholesterol levels, obesity and diabetes [Rogaeva 2002]. It has been estimated that the four established AD genes account for less than 30% of the genetic variance in EOAD and LOAD, suggesting that numerous additional AD genes may exist [Daw, et al. 2000]. The pursuit of these additional genes has been unproductive until recently. In the last two years however, high throughput technologies able to genotype up to one million single nucleotide polymorphisms (SNPs) have revealed some of the genetic players in different complex disorders. In this review, we will consolidate the current knowledge on the role of APP, PSENs and APOE in AD, we will systematically review the most promising new loci and variants reported to be associated with AD and we will discuss the recent advances in AD genome wide analysis in the context of the known epidemiology of the disorder.
EOAD CAUSATIVE GENES (APP, PSEN1 and PSEN2)
The amyloid precursor protein gene (APP, OMIM 104760, chromosome 21q21) encodes an ubiquitously expressed, integral type I membrane glycoprotein that exists as different alternatively spliced isoforms, with three predominant ones: APP751, APP770 and APP695, the latter being the main isoform found in brain [Yoshikai, et al. 1990]. The proteolytic processing of APP results in the production of different peptides (including Aβ), after a series of secretase cleavages, and occurs through two mutually exclusive pathways: the amyloidogenic pathway (fundamentally considered as the pathogenic pathway) and the non-amyloidogenic or constitutive pathway [Esch, et al. 1990, Haass, et al. 1992, Shoji, et al. 1992]. The identification of Aβ as a metabolic product of APP and the reports of AD families harboring APP causative mutations led to the general concept that Aβ is a key player in the development of AD, and that EOAD mutations are influencing the properties or ratios of the different Aβ isoforms in the brain [Hardy 1997]. Dominant mutations in APP are, however, a rare cause of AD with an estimated frequency of 16% of familial EOAD patients [Raux, et al. 2005]. More recently, two mutations in APP (A673V and E693Δ) have been reported to cause AD only in the homozygous state in families with apparently recessive modes of inheritance [Di Fede, et al. 2009] [Tomiyama, et al. 2008].
In addition to missense variants, copy number mutations have been identified in autosomal dominant early-onset families. Five French families [Cabrejo, et al. 2006, Rovelet-Lecrux, et al. 2006] were first reported to harbor small chromosomal duplications with different break points, but all including the APP locus. Subsequent screens in Finnish and Dutch AD cases revealed additional APP duplications in EOAD cases with prominent cerebral amyloid angiopathy (CAA) [Remes, et al. 2004, Rovelet-Lecrux, et al. 2007, Sleegers, et al. 2006]. The phenotypic spectrum of APP duplications is yet to be fully defined but clearly includes mixed phenotypes of AD and/or CAA. The estimated frequency of duplications also appears to be variable: in the selected Rovelet-Lecrux cohort it was 8% (about half the contribution of missense APP mutations to early onset, autosomal dominant AD) [Raux, et al. 2005]; in the Dutch cohort less than 2% [Sleegers, et al. 2006]; in EOAD familial and sporadic Swedish and Finnish cases there were no duplications in APP identified [Blom, et al. 2008]; and a frequency of 18% was estimated in early onset familial Japanese cases [Kasuga, et al. 2009].
The presenilin 1 (PSEN1, OMIM 104311, chromosome 14q24.3) and presenilin 2 (PSEN2, OMIM 600759, chromosome 1q31-q42) genes have a very similar genetic structure and encode two proteins expressed in a multiplicity of tissues including the brain, with higher levels in the cerebellum and the hippocampus and a primarily neuronal expression [Levy-Lahad, et al. 1996],[Rogaev, et al. 1995]. These are highly homologous, sharing an overall amino acid sequence identity of 67%. Hydrophobicity plots predicted these to be integral membrane proteins [Rogaev, et al. 1995] most likely adopting a transmembrane structure containing nine segments with a hydrophilic intracellular loop region [Henricson, et al. 2005, Laudon, et al. 2005]. PSENs are important components of the multimeric gamma-secretase complex and are predominantly located in the endoplasmic reticulum and Golgi compartments, clearly suggesting their involvement in protein processing [Kovacs, et al. 1996],[De Strooper 2003]. The first disease causing mutations in PSEN1 and PSEN2 were identified in 1995 [Rogaev, et al. 1995, Sherrington, et al. 1995]. At the present time, 175 pathogenic mutations and 7 variants non-pathogenic or with unclear pathogenicity have been identified in PSEN1. PSEN2 harbors fewer mutations: 14 pathogenic mutations and 9 variants non-pathogenic or with unclear pathogenicity (http://www.molgen.ua.ac.be/ADMutations, accessed on August 2009). The PSENs mutation range encompasses mainly missense mutations, thus manifesting in a scattered fashion all over the proteins, with some clustering around transmembrane domains [Guerreiro, et al. 2008, Hardy and Crook 2001].
GENETIC RISK FOR AD (APOE)
The apolipoprotein E gene (APOE, OMIM 107741, chromosome 19q13.2) encodes a glycoprotein synthesized mainly in the liver, brain (primarily by neurons and astrocytes), and also by other cells such as macrophages and monocytes [Siest, et al. 1995]. APOE is involved in the mobilization and redistribution of cholesterol in the periphery and also during neuronal growth and repair [Mahley 1988]; in nerve regeneration, immunoregulation and activation of several lipolytic enzymes [Mahley and Rall 2000]. The three major APOE isoforms (ApoE2, ApoE3 and ApoE4) differ in two sites of the aminoacid sequence (residues 112 and 158) and are encoded by a single genetic locus. The frequencies of the ε2, ε3, and ε4 alleles were estimated at 0.11, 0.72, and 0.17, respectively, but vary widely among populations [Zannis, et al. 1981]. The ε4 allele, (the ancestral allele), is more frequent in populations such as Pygmies (0.407) and Khoi San (0.370). The ε2 allele frequency oscillates with no apparent trend and, for example, is absent in some Native Americans populations [Corbo and Scacchi 1999].
Many studies have demonstrated an association between the ε4 allele and familial and sporadic forms of LOAD. This allele represents an increased risk seen across different ethnic groups of 3 fold for heterozygous carriers and up to 15 fold for individuals who are ε4 homozygotes, when compared to ε3 homozygotes [Ashford 2004]. APOE is known to act in a dose dependent manner in AD: the effect of the ε4 allele in the risk for AD increases from 20 to 90% and the mean age of onset decreases from 84 to 68 years with the increase in the number of ε4 alleles [Corder, et al. 1993]. The ε2 allele has been shown to have an impact on longevity and may confer protection against AD [Corder, et al. 1994]. Distinct binding properties of the different APOE isoforms to the Aβ peptide [Strittmatter, et al. 1993] and tau protein [Strittmatter, et al. 1994] have been suggested to underlie the disparities associated with each genotype. In particular, the ApoE4 isoform binds to the Aβ peptide more rapidly than the ApoE3 isoform, forming novel monofibrils that precipitate into dense structures [Sanan, et al. 1994]. The fact that ApoE4 does not bind to tau protein in vitro, unlike ApoE2 and ApoE3, has suggested to some that this interaction between ApoE3 and tau serves as a protection against tau phosphorylation and consequent neurofibrillary tangle formation [Strittmatter, et al. 1994, Weisgraber 1994].
The ε4 allele appears to be a risk factor and not an invariant cause of AD, indicating that other environmental or genetic factors may need to be concurrently acting with this allele in order to cause AD [Hyman, et al. 1996]. For example, physical activity has been shown to be protective for dementia in non-APOEe4 carriers [Podewils, et al. 2005]. Additionally, while most of the risk at the APOE locus is likely to be encoded at the protein coding polymorphism, it is likely that other genetic variability at this locus, probably altering APOE expression, also contributes to the risk of developing AD [Bekris, et al. 2009, Chartier-Harlin, et al. 1994, Lambert, et al. 2002, Lambert, et al. 1997]. Genetic variability in APOE expression may contribute more to disease risk, rather than independent effects of the adjacent gene TOMM40.
During the past two decades there have been many studies searching for genetic risk factors: hundreds of positive and negative results have been produced, but none apart from APOE have produced clear and reproducible associations.
DIFFERENT TYPES OF STUDIES IN AD GENETICS
Methodologically, two main genetic strategies have governed the field: genetic linkage analysis and case-control association studies. For linkage analysis, researchers have used informative families where a clear heritability of the disease is present and no mutations have been found. In association studies, researchers compare the frequencies of a pre-determined allele between a group of AD cases and a group of healthy individuals. Several considerations are relevant for both strategies, of which we will mention the main ones. A full discussion of all methodological issues involved in these types of studies goes beyond the scope of this review. Here we will briefly describe the known genes and risk factors associated with AD, with a focus on genome wide association studies (GWAS) as a methodology for exploring genetic susceptibility and identifying risk factors and mechanisms for disease. We will also address the previously overlooked role of homozygosity and recessive cases in the major complexity of AD genetics.
LINKAGE STUDIES
Genetic linkage studies aim to identify chromosomal regions associated with disease by measuring the correlated segregation of particular markers with a determined phenotype within a family [Dawn Teare and Barrett 2005]. This type of study usually involves three sequential steps: 1) the identification of the disease causative locus; 2) sequencing the region found in the previous step in a cohort of cases and controls in order to define and characterize the mutation(s) found; 3) uncovering the molecular and biological functions of the genes found [Altshuler, et al. 2008]. Several factors are known to complicate this approach in AD: i) difficulties in getting large, complete and informative multigeneration families; ii) the potential inclusion of phenocopies (individuals with a sporadic indistinguishable form of disease); and iii) genetic heterogeneity, since observing that the pattern of disease in families is consistent with a major gene component does not necessarily imply that only one gene or factor is involved. Additionally, linkage mapping suffers from limitations, such as the low resolution of the results. Usually these studies do not identify one gene or one mutation associated with a disease, but rather a chromosomal region (many times, a very large region) is identified. In addition, the strongest linkage signals tend to come from recessive and highly penetrant, thus very rare, disorders [Dawn Teare and Barrett 2005].
Nonetheless, linkage mapping has been a very important methodology in the study of AD genetics. The four genes undoubtedly associated with AD were identified primarily by linkage analysis. In addition to these genes, several other genomic regions have been implicated using this methodology (Table 1). These loci contain many genes that have been considered candidates and consequently have been studied in order to identify the genetic variation responsible for the development of AD. Until now, no specific genes implicated in AD have been identified in these regions. Most recently, a study by Butler and colleagues used a meta-analysis method to analyze the pooled linkage results from five independent genome scans. These included the results of 2206 affected individuals and 785 families of Caucasian and Caribbean Hispanic descent. This study was able to identify genome-wide suggestive evidence for linkage on chromosomes 1p13.3-q31.1, 7pter-p21.1 and 8p22-p21.1, together with other seven loci presenting nominally significant evidence for linkage [Butler, et al. 2009]. Interestingly, the most significant locus identified in this study (8p22-p21.1) includes the CLU gene, (the top hit from the largest GWAS performed in AD, as discussed below in this review) and previously reported loci by different studies (as 9p, 9q, 10q and 12p) were not identified by Butler and colleagues. Even so, linkage analysis has largely failed to identify risk factors in LOAD, probably due to the low odds ratios associated with the unidentified variants.
Table 1.
Ten most interesting AD linkage regions.
| Chr | Region | Number of studies |
Studies | LOD scores |
Relevant genes | |
|---|---|---|---|---|---|---|
| 1 | 1p13.3-q23.3 | 2 | [Liu, et al. 2007]; [Butler, et al. 2009] | 5.2 | GSTM4; GSTM1; GSTM3; CSF1; NGF; HMGCS2, PRKAB2; APH1A; CTSS; THEM5; FAM63A; CHRNB2; LMNA; PMVK; FDPS; APOA1BP; GBA; NTRK1; CRP; NCSTN |
|
| 1q23.3-q31.1 | 3 | [Liu, et al. 2007]; [Butler, et al. 2009]; [Blacker, et al. 2003] |
2.1 – 4.0 | F11R; USF1; FCER1G; RGS4; APOA2; RXRG, POU2F1; PRDX6; SOAT1; PTGS2 |
||
| 5 | 5p13-p15 | 6 | [Pericak-Vance, et al. 2000]; [Curtis, et al. 2001]; [Olson, et al. 2002]; [Myers, et al. 2002]; [Blacker, et al. 2003]; [Lee, et al. 2006] | 1.4 – 2.8 | SLC6A3; PRKAA1 | |
| 8 | 8p22-p21.1 | 2 | [Butler, et al. 2009]; [Lee, et al. 2008] | >2.0 | NAT1; NAT2; LPL; ADRA1A; CHRNA2; CLU | |
| 9 | 9p21 | 5 | [Pericak-Vance, et al. 2000]; [Curtis, et al. 2001]; [Myers, et al. 2002]; [Scott, et al. 2003]; [Hamshere, et al. 2007] | >1.0 – 4.6 | IFT74 | |
| 9q22-q34 | 8 | [Pericak-Vance, et al. 2000]; [Curtis, et al. 2001]; [Olson, et al. 2002]; [Myers, et al. 2002]; [Blacker, et al. 2003]; [Holmans, et al. 2005]; [Lee, et al. 2006]; [Hamshere, et al. 2007] |
1.6 – 4.2 | FBP1; GOLM1; ABCA1; DFNB31; TLR4; NDUFA8; PSMB7; HSPA5; POMT1; DBH; RXRA; TRAF2; ABCA2 |
||
| 10 | 10q21-q22 | 7 | [Curtis, et al. 2001]; [Olson, et al. 2002]; [Myers, et al. 2002]; [Blacker, et al. 2003]; [Holmans, et al. 2005]; [Liu, et al. 2007]; [Hamshere, et al. 2007] |
1.8 – 4.15 | ZWINT; UBE2D1; TFAM; BICC1; ANK3; CDC2; EGR2; CTNNA3; LRRTM3; DNAJC12; SIRT1; SRGN; SUPV3L1; TSPAN15; VPS26A; HK1; TACR2; NEUROG3; SAR1A; SGPL1; PSAP; CHST3; PPP3CB; SEC24C; NDST2; CAMK2G; PLAU; VCL; AP3M1; MYST4; KCNMA1 |
|
| 12 | 12p11-p13 | 4 | [Pericak-Vance, et al. 1997]; [Curtis, et al. 2001]; [Myers, et al. 2002]; [Holmans, et al. 2005] |
1.4 – 3.9 | TNFRSF1A; CNAP1; GAPDH; GNB3; C1R; APOBEC1; MMP3; A2M; PZP; A2MP; OLR1; LRP6; GRIN2B; GYS2; ABCC9; PKP2P1 |
|
| 19 | 19q12-q13.33 | 9 | [Pericak-Vance, et al. 2000]; [Curtis, et al. 2001]; [Olson, et al. 2002]; [Myers, et al. 2002]; [Li, et al. 2002]; [Blacker, et al. 2003]; [Holmans, et al. 2005]; [Hamshere, et al. 2007]; [Sillen, et al. 2008]; [Butler, et al. 2009] |
1.6 – 7.7 | LRP3; USF2; GAPDHS; PSENEN; AKT2; TGFB1; LIPE; XRCC1; BCL3; APOE; PVRL2; TOMM40; APOC1; APOC2; ERCC2; CARD8; GYS1; LHB; CD33; NR1H2 |
|
| 21 | 21q21-q22 | 4 | [Olson, et al. 2002]; [Myers, et al. 2002]; [Blacker, et al. 2003]; [Holmans, et al. 2005] |
1.6 – 4.5 | PRSS7; NCAM2; APP; C21orf63; C21orf55; RUNX1; C21orf55; DYRK1A; KCNJ6; BACE2 |
|
These regions were selected either from the top 3 regions resulting from the recent meta-analysis performed by Butler and colleagues, or from the overlapping between four or more whole-genome studies assessing linkage with AD risk, based on the AlzGene database of concordant linkage/association regions observed in full genome screens (http://www.alzforum.org/res/com/gen/alzgene/linkage.asp). The relevant genes for each of the represented regions were obtained from crosschecking with the AlzGene database (www.alzgene.org, [Bertram, et al. 2007]).
Note that: results by [Kehoe, et al. 1999] and [Lee, et al. 2004] were not considered as they were significantly extended in the same groups’ follow-up genome screen (Myers, et al, 2002 and Lee et al., 2006, respectively). The results by Hamshere, et al., 2007 consist of an amalgamated of three data sets previously studied by Kehoe, et al., 1999; Myers, et al., 2002; Blacker, et al., 2003 and Holmans, et al., 2005. Other studies have been published that performed re-analyses of existing data either by incorporating other variables (such as evidence for AD with and without psychosis [e.g. [Bacanu, et al. 2002], [Avramopoulos, et al. 2005], [Hollingworth, et al. 2007]), or parent of origin effects e.g. [Bassett, et al. 2002]. Results by [Ashley-Koch, et al. 2005] and [Hahs, et al. 2006] on Amish families were not considered due to the inclusion of MCI cases.
GENE ASSOCIATION STUDIES
The quest for genetic risk factors in clinical genetics has mainly focused on the study of candidate genes (usually focusing only on variants altering the coding sequence of a gene). These types of studies rely on a rather simple principle: to test if a determined allelic or genotypic variant occurs more or less often in a group of people with a particular disease when compared to a similar group of healthy individuals.
The success of this approach relies in an in-depth understanding of the disease and disease pathways, in such a way that the researcher will be able to select, not only the right gene, but also the right variant(s) to be studied. Additionally, the two studied groups need to be homogeneous, well characterized and large enough to allow a statistically powerful analysis [Hattersley and McCarthy 2005].
Most gene association studies in AD have studied a few variants in one or two genes. The large number of genes and even more variants, clearly reduce the chances of true positive findings. Nonetheless, several hundreds of positive associations have been reported. Most of these are certainly false positives resulting mainly from population substructure (i.e. existence of subpopulations in which there was random mating with reduced amount of gene flow, that may lead to spurious associations when the geographical origin/ancestry of cases and controls are not matched) [Tian, et al. 2008], poor statistical analysis and publication bias toward positive results [Choudhry, et al. 2006, Ioannidis, et al. 2001]. Positive associations have been broadly published, while negative results (unless convincingly refuting previous results) would not be reported. When reported, most of these studies pointed to small sample sizes, or specific genetic population backgrounds as main reasons for the negative results. Other issues, particularly in large epidemiologic studies, include case definition or the ability to accurately identify AD versus vascular or other forms of dementia; age of dementia onset estimation; and phenotypic variations in the disease whether cognitive, vascular, psychiatric or metabolic.
In order to address these very large numbers of conflicting reports, a database (the AlzGene database) was created which systematically collects, summarizes and meta-analyzes the results for all the genetic variants studied in association with AD [Bertram, et al. 2007]. As of November 16th 2009 the top ten results in this database included: APOE (E2/3/4), CLU, PICALM, TNK1, ACE, TFAM, CST3, IL1B, CR1 and hCG2039140 [Bertram and Tanzi 2008]. The fifth top hit, ACE, has repeatedly been reported as associated with AD [Webster J, et al. 2009], atherosclerosis [Sayed-Tabatabaei, et al. 2003] and hypertension [Staessen, et al. 1997]. The role of vascular risk factors in AD is further discussed below.
GENOME WIDE ASSOCIATION STUDIES
The development of platforms able to genotype millions of SNPs and of powerful analytical frameworks, able to distinguish true associations; together with the completion of the International Human HapMap project [Consortium 2005], have provided unprecedented tools to the study of the so called “Common Disease-Common Variant” (CD-CV) hypothesis. This theory proposes that common polymorphisms (usually defined as having a minor allele frequency of over 5%) may contribute to the overall susceptibility to common diseases [Cargill, et al. 1999, Chakravarti 1999, Lander 1996]. This type of analysis addresses one of the major pitfalls of gene association studies: the coverage of the study. Instead of studying one or two genetic variants, we are now able to test the majority of common variability in the genome for association with disease, by means of testing tagging SNPs, i.e., polymorphisms in linkage disequilibrium (LD) with each other. This means that if one knows the genotype in one locus, one can predict with a high accuracy (dependent on the strength of the LD and the allele frequencies) the genotype occurring at linked loci. However, these platforms require the analyses of large numbers of samples and some regions of the genome are still not well covered. Thus, the success of genome wide association studies (GWAS) depends on sample size, frequency of risk alleles and individual effect sizes. The smaller the attributable risk associated with any given common variant, the greater the number of samples needed to identify that variant. The risk allele frequency relates to the effect size: a risk variant that has a high odds ratio but is rare in the population is much more difficult to identify with GWAS than a common variant linked to moderate or mild risk within the population. Additionally, population stratification (when cases and non-cases are not genetically similar); population admixture (when several distinct, unrecognized sub-populations comprise the same group of individuals studied) and unreported relatedness in the population may also be problems if the study is not correctly designed [Simon-Sanchez and Singleton 2008].
The competing hypothesis to explain the genetic basis of complex diseases is the “Common Disease-Rare Variant” (CD-RV) theory, which suggests that multiple rare variants underlie susceptibility to such diseases [Fearnhead, et al. 2005]. Although the CD-CV versus CD-RV is a false dichotomy, it has major implications for the future of the research in this field, since different techniques are used to find common versus rare alleles. By definition, rare variants have low frequencies (MAF > 0.1% to 2–3%) and individually small contributions to the overall inherited susceptibility of a disease. In this way, rare variants will not be detectable by GWAS. Instead, the choice of candidate genes and appropriate case groups has been essential to uncover this type of variants[Bodmer and Bonilla 2008]. The new sequencing technologies will facilitate this approach by allowing the assessment of all the genes in the genome. Nonetheless, the extensive DNA resequencing of the whole genome in large numbers of individuals, and the assessment of the functional consequences of the variants found, poses difficult bioinformatic and data management challenges.
There is no doubt that GWAS have uncovered previously unknown polymorphic variants and genes with significant effects on AD risk. However, considering the studies so far carried out in AD (see next section for details), the associations still to be found will have ORs lower than 1.2. This has raised the question as to whether it is worthwhile to pursue even larger GWAS in order to identify variants with small effects disease risk. Certainly, in the future, sequencing and association strategies will be employed together to fully dissect the genetic architecture of the risk of AD and other complex disorders [Bodmer and Bonilla 2008].
Since APOE has been the only risk factor consistently associated with LOAD and some reports have estimated a heritability of AD between 60 and 80% [Gatz, et al. 2006, Pedersen, et al. 2001] it is believed that genetic variability plays a critical role in LOAD and that several risk factors are still to be uncovered. Daw and colleagues had estimated the existence of four to seven additional genes contributing to this genetic variability [Daw, et al. 2000].
Several GWAS in AD have now been published. The first study of this type was performed by Grupe et al., in which the approach used was to first generate a short list of candidate SNPs by analyzing >17,000 SNPs in DNA pools from one set of cases and controls. In truth, this study only covers a small proportion of the genome, probably around 5%, and is not strictly “genome-wide”. In this study, markers meeting a previously defined significance criteria were then typed in DNA pools from a second set of samples and, again the SNPs satisfying the criteria were then individually genotyped in four sets of cases and controls and finally in a fifth sample. In total, nearly 4000 samples (~2000 AD cases, ~2000 controls) from the United States and the United Kingdom were studied. Three SNPs on chromosome 19 in LD to APOE represented the most significant associations, of which two (rs157581 in TOMM40 and rs405509 in APOE) achieved genome-wide significance. SNPs in other genes reached nominal significance but did not achieve genome wide significance [Grupe, et al. 2007].
The first studies covering the majority of the genome were published by Coon et al. [Coon, et al. 2007] and Reiman et al. [Reiman, et al. 2007] (~1000 cases and controls). In these studies on largely the same dataset, only a single SNP in LD with APOE (rs4420638) reached genome wide significance although SNPs in GAB2 (rs2373115) reached significance when the AD cases were stratified by APOE genotype [Reiman, et al. 2007]. An analysis of the top hits from AlzGene in the same dataset found that only SNPs is ACE reached nominal significance [Webster J, et al. 2009].
Liu et al. reported on a genome screen of 402 microsatellite markers in 103 LOAD patients and 170 first-degree relatives from a pedigree with 4645 members from The Netherlands. Multipoint analysis revealed four significant (1q21, 1q25, 10q22-24 and 3q22-24) and one suggestive (11q24-25) linkage peaks. Several of these regions coincide with previously reported loci (Table 1) with the strongest linkage association found for chromosome 1q21. Following these results, the authors tested for association between cognitive function and 4,173 SNPs in the linked regions in an independent sample consisting of 197 individuals from the same Genetic Research in Isolated Populations program. After adjusting for multiple testing, significant associations were identified in four (1q25, 3q22-24, 10q22-24 and 11q25) of the previous five regions (Table 2) [Liu, et al. 2007].
Table 2.
Main features and results of genome wide association studies performed in AD.
| Study | Platform | #SNPs | Population | Genotype data publicly available |
# Subjects | Replication stage(s) | Significant results | |
|---|---|---|---|---|---|---|---|---|
| Cases | Ctrls | |||||||
|
Grupe, 2007 [Grupe, et al. 2007] |
Celera (cSNPs) | 17343 | UK and USA | No | 380 | 396 | 4 replication tiers (UK2, 309 cases+349 controls; UK3, 503 cases+643 controls; WU, 376 cases+344 controls; SD, 240 cases+330 controls) Total number of samples (4 tiers) = 1428 cases + 1666 controls |
rs157581 (TOMM40, chr 19q13.32) rs405509 (APOE, chr 19q13.32) rs3745833 (GALP, chr 19q13.42) rs1554948 (TNK1, chr 17p13.1) rs1132899 (APOC2, chr 19q13.32) rs11622883 (14q32.13) rs8192708 (PCK1, chr 20q13.31) rs505058 (LMNA, chr 1q22) rs3800324 (PGBD1, chr 6p22.1) rs6907175 (LOC651924, chr 6q24.1) rs1859849 (7p15.2) rs41310885 (hCV22274641, FAM63A, chr 1q21.2) rs2074877 (MYH13, chr 17p13.1) rs41271951 (hCV15746640, CTSS, chr 1q21.2) rs444013 (UBD, chr 6p22.1) rs9608099 (BCR, chr 22q11.23) rs2882676 (ACAN, chr 15q26.1) rs13022344 (TRAK2, chr 2q33.1) rs11016976 (EBF3, chr 10q26.3) |
|
Coon, 2007 [Coon, et al. 2007] |
Affymetrix (500K) |
502627 | USA, Netherlands; overlaps with Reiman, 2007 |
No | 664 | 422 | No | rs4420638 (APOC1, chr 19q13.32) |
|
Reiman, 2007 [Reiman, et al. 2007] |
Affymetrix (500K) |
312316 | USA, Netherlands; overlaps with Coon, 2007 |
Yes | 446 | 290 | Neuropathological replication cohort (197 cases + 114 controls); Clinical replication cohort (218 cases + 146 controls) Total number of samples (2 cohorts) = 415 cases + 260 controls |
rs901104; rs1385600; rs1007837; rs2510038; rs4945261; rs7101429; rs10793294; rs4291702; rs7115850; rs2373115 (GAB2, chr 11q14.1) |
|
Liu, 2007 [Liu, et al. 2007] |
Affymetrix (500K) |
262000 | Netherlands | No | 103 LOAD patients and 170 first- degree relatives from a pedigree with 4645 members |
- | 4173 SNPs in 197 unrelated subjects from the GRIP population |
Chr 1q25 (RGSL2, RALGPS2, C1orf49); Chr 3q22-24 (NMNAT3, CLSTN2); Chr 10q22-24 (HTR7, MPHOSPH1, CYP2C); Chr 11q25 (OPCML, HNT) |
|
Li, 2008 [Li, et al. 2008] |
Affymetrix (500K) |
469438 | Canada and UK |
Yes | 753 | 736 | 120 SNPs in 418 cases + 249 controls |
rs10868366; rs7019241 (GOLM1, chr 9q21.33) rs4420638 (APOC1, chr 19q13.32) also significant in Coon, 2007; rs10519262 (chr 15q21.2); rs9886784 (chr 9p24.3) |
|
Abraham , 2008 [Abraham, et al. 2008] |
Illumina HumanHap300 + Illumina Sentrix HumanHap240 S |
561494 | UK | No | 1082 LOAD pooled |
1239 contro ls poole d |
114 SNPs were individually genotyped in the cases and controls used in the pools and 1400 controls were added |
ApoE; rs727153 (~13 kb from the start of transcription of LRAT, chr 4q32.1), also showed increased significance (10 fold) when additional controls were combined |
|
Bertram, 2008 [Bertram, et al. 2008] |
Affymetrix 500K |
484522 | Self-reported European descent from NIMH Genetics Initiative Study sample |
No | 1376 AD from 410 families |
- | 4 SNPs in 3 independent AD family samples: NIA- 1040 samples from 329 families; NCRAD- 1108 samples from 331 families; CAG- 483 samples from 215 sibships (Total: 2689 samples 1816 affected and 845 unaffected) |
Rs4420638 (located 340 bp 3’ of APC1 and probably reflects the effects of apoE E4 allele); rs 11159647 (in predicted gene NT_026437.1360, chr14q31.2), also significant in replication samples and in TGEN’s data; rs179943 (in ATXN1, chr 6p22.3), trend for significance in replication samples; rs3826656 (in predicted gene NT_011109.848, chr 19q13.33), also significant in replication samples; rs2049161 (in cDNA BC040718, chr 18p11.31) |
|
Beechma m, 2009 [Beecham, et al. 2009] |
Illumina HumanHap550 |
532000 | USA | No | 492 LOAD | 496 | 1 SNP in 238 cases + 220 controls |
ApoE; rs11610206 (downstream of FAM113B, chr 12q13), also significant in replication sample 1q42; 4q28; 6q14; 19q13 |
|
Carrasqui llo, 2009 [Carrasquillo, et al. 2009] |
Illumina HumanHap300 |
313504 | USA | No | 844 LOAD | 1255 | 25 SNPs in 1547 cases + 1209 controls |
ApoE; rs5984894, rs2573905 (in PCDH11X, chr Xq21.3), also significant in replication sample |
|
Harold, 2009 [Harold, et al. 2009] |
Illumina 610 quadchip (Illumina HumanHap550 and HumanHap300 in some samples) |
529205 | Europe USA |
Yes | 3941 | 7848 | 2 significant SNPs in 2023 cases + 2340 controls |
ApoE; rs11136000 (in CLU, chr 8p21- p12), also significant in replication sample rs3851179 (5’ of PICALM, chr 11q14, also significant in replication sample) |
|
Lambert, 2009 [Lambert, et al. 2009a] |
Illumina Human 610-Quad BeadChip |
537029 | France Europe |
No | 2032 | 5328 | Significant SNPs in s3978 probable AD cases + 3297 controls from Belgium, Finland, Italy and Spain |
rs11136000, rs2279590, rs9331888 (in CLU, chr 8p21- p12), also significant in replication sample rs6656401 (in CR1, chr 1q32), also significant in replication sample |
Abraham and colleagues performed a genome-wide association study in pooled DNA samples of ~2000 LOAD cases and >1000 controls. They identified a set of 109 SNPs with a significant association with AD and genotyped them individually. In addition to APOE, one SNP (rs727153), located approximately 13 kb apart from the start of LRAT transcription site, was suggested as associated with disease [Abraham, et al. 2008].
Li et al., 2008 studied a hypothesis-generating cohort from Canada and identified rs4420638 within APOC1 to be strongly associated with AD, due to LD with APOE [Coon, et al. 2007] but nothing else reached genome wide significance [Li, et al. 2008].
Bertram et al., 2008 used samples from 410 AD families to identify five SNPs significantly or marginally associated with a multivariate phenotype combining age at onset of the disease and affection status. The marker presenting the most significant association was again rs4420638 (located 340 bp 3’ of APOC1) and almost certainly reflecting the effects of APOE ε4 allele. Once more, no other SNP reached genome wide significance [Bertram, et al. 2008].
Beecham and colleagues analyzed ~500 LOAD cases and ~500 cognitive controls followed by a further >200 cases and >200 controls used as a validation data set for SNPs that reached nominal, but not genome-wide significance. They too, were able to confirm association with the APOE locus and they suggest an association with the 12q13 locus, which they replicated in the validation data set. The associated SNP (rs11610206) is close to the hypothetical gene FAM113B. Additionally, there are a number of nearby candidate genes, such as VDR and AMIGO2. In order to validate associated SNPs with p values <0.0001 and nominally associated candidate genes, they imputed SNPs from a previously published GWAS [Reiman, et al. 2007]. Four additional highly associated signals were replicated with the use of the imputed data set: 1q42-within DISC1; 4q28-200 kb proximal to PCDH18; 6q14 – nearest gene BCKDHB; and 19q13 – within ZNF224 [Beecham, et al. 2009].
By analyzing ~1000 LOAD cases and ~1000 controls and evaluating the 25 SNPs with the most significant allelic association in four additional series, Carrasquillo and colleagues suggested an association with the X chromosome SNP rs5984894 in PCDH11X (Xq21.3) [Carrasquillo, et al. 2009] but again, this association did not reach genome wide significance.
Feulner et al. generated genome-wide data in a German cohort of ~500 AD patients and ~500 controls. The results obtained were analyzed only for the genes included in the top results list on the AlzGene database. Additionally to APOE, nominally significant associations were found for six of the ten studied genes (CH25H, PGBD1, LMNA, PCK1, MAPT and SORL1) [Feulner, et al. 2009] but none of these putative signals reached genome wide significance.
All the above studies, which each used <2000 Alzheimer cases in their analysis, were able to pick up the signal at the APOE locus, but despite tantalizing results, none were able to identify other loci at a level which passes the threshold for genome wide significance, and no two of them identified the same locus. The data from the Reiman et al. and by Li et al. studies, were made publicly available (http://www.tgen.org/research/neuro_gab2.cfm and http://www.GSK.com, respectively) to enable their additive use in other studies. Overall, these data pointed to the need for larger studies to identify risk loci with smaller effect sizes. Two such studies have now been reported.
One reported the analysis of ~4000 cases and ~8000 controls in stage 1 and ~2000 cases and ~2000 controls in stage 2. Two SNPs significantly associated with AD, outside the APOE locus, were identified: rs11136000 located in an intron of CLU on chromosome 8 and rs3851179 located close to the gene PICALM on chromosome 11. Although the risks associated with CLU and PICALM genes are relatively small (APOE odds ratio~4; CLU/PICALM~1.1) these associations reached genome wide significance [Harold, et al. 2009]. The other large AD GWAS reported on 2000 cases and ~5000 controls in the first stage, and ~3000 cases and ~3000 controls in the replication stage. An association between AD and ApoE, CLU and CR1 locus on chromosome 1q32 (OR=1.2) was found [Lambert, et al. 2009b].
These aforementioned three genes (CLU, CR1 and PICALM) will clearly be the subject of intense research. Here we will discuss the main features of each of these genes, as well as the potential pathobiological pathways in which they may be involved in the context of AD.
Clusterin or apolipoprotein J is, like APOE, a lipoprotein expressed in most mammalian tissues with higher levels present in brain, ovary, testis and liver [de Silva, et al. 1990]. CLU interacts with different molecules, including lipids, amyloid proteins, components of the complement membrane attack complex (MAC) and immunoglobulins [Jones and Jomary 2002]. Accordingly, it has been proposed to be involved in a number of physiological processes such as ongoing synapse turnover [Danik, et al. 1993], apoptosis [Jenne and Tschopp 1992, Wong, et al. 1993], cytoprotection at fluid-tissue boundaries, membrane recycling during development and in response to injury and regulation of complement-mediated MAC [Jones and Jomary 2002, Oda, et al. 1994]. Clusterin has also been proposed to be a form of secreted heat-shock protein or chaperone molecule [Michel, et al. 1997, Wilson and Easterbrook-Smith 2000].
Several lines of evidence suggest that CLU has a central (either protective or pathogenic) role in the pathway leading to Alzheimer’s disease. First, CLU mRNA has been reported to be elevated in AD affected brain areas such as hippocampus, either when brains from AD patients were compared to one Huntington patient [Duguid, et al. 1989], or to controls [May, et al. 1990]. Likewise, Oda et al. reported a statistically significant difference in the Clusterin protein content of extracts from cortex and hippocampus when comparing 10 non-AD individuals and 25 AD patients [Oda, et al. 1994]. Second, Clusterin is one of the components of amyloid plaques [Kida, et al. 1995, McGeer, et al. 1992, McGeer, et al. 1994, Takamaru 1994]. Third, is able to bind soluble Aβ through a specific, reversible and high-affinity interaction in cerebrospinal fluid [Ghiso, et al. 1993, Golabek, et al. 1995] to form complexes able to cross the blood-brain barrier by a high affinity receptor mediated process involving transcytosis [Zlokovic 1996]. Fourth, reduced levels of APOE and increased levels of CLU have been correlated with the number of E4 alleles, suggesting a compensatory induction of CLU in the brain of AD individuals with the ε4 allele of APOE presenting low brain levels of APOE [Bertrand, et al. 1995]. Moreover, CLU was shown to prevent aggregation and polymerization of synthetic Aβ and to enhance the oxidative stress caused by A in vitro [Matsubara, et al. 1996, Oda, et al. 1995], and to facilitate Aβ uptake in cell culture experiments [Hammad, et al. 1997]. Clusterin appears to regulate the toxicity and conversion of Aβ into soluble forms [Boggs, et al. 1996, DeMattos, et al. 2002, Matsubara, et al. 1996, Oda, et al. 1995]. Together with APOE, suppresses Aβ deposition [DeMattos, et al. 2004] and may modify Aβ clearance at the blood brain barrier [Bell, et al. 2007].
PICALM, encodes the phosphatidylinositol-binding clathrin assembly protein, also known as CALM: clathrin assembly lymphoid-myeloid leukemia gene. It is ubiquitously expressed with particularly high levels in neurons. This gene has been associated with leukemia, thus its relation to AD may appear not as direct as the one observed for CLU (Figure 1). Nonetheless, its involvement in clathrin-mediated endocytosis (essential to the intracellular trafficking of proteins and lipids) [Kim and Kim 2001] and in the fusion of synaptic vesicles to the presynaptic membrane by directing the trafficking of VAMP2 [Harel, et al. 2008] have lead Harold et al. to propose two interesting hypotheses for the role of Picalm in AD. In this way, genetic variability in PICALM may result on synapse perturbations, possibly through synaptic vesicle cycling, or on alterations of APP processing through endocytic pathways, culminating in changes in Aβ levels [Harold, et al. 2009].
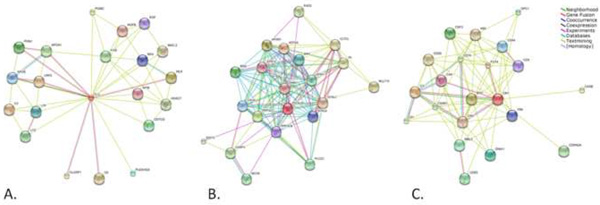
Figure 1. Predicted interactions for Clu (A), Picalm (B) and CR1 (C).
The software STRING 8.0 (available at http://string.embl.de/newstring_cgi/show_input_page.pl?UserId=75H_lKjgP5Xi&sessionId=9vx3bNEK6tmB) was used to establish a network of predicted interactions for Clu (A), Picalm (B) and CR1 (C) proteins. Accessed on August 2009. STRING is a database of known and predicted protein interactions. The interactions include direct (physical) and indirect (functional) associations; they are derived from four sources: genomic context, high-throughput experiments, (conserved) coexpression and previous knowledge. STRING quantitatively integrates interaction data from these sources for a large number of organisms, and transfers information between these organisms where applicable. The database currently covers 2,483,276 proteins from 630 organisms.
From the three represented proteins, CLU is the one with a more direct relation with AD: APOE is directly connected to CLU. Interestingly, CLU also interacts with proteins present in CR1 network (like C3). No other genes consistently associated with AD are present in PICALM or CR1 networks, indicating that these proteins may be involved in new pathobiological pathways.
CR1, the complement component (3b/4b) receptor 1 (Knops blood group) is a member of the receptors of complement activation family. The gene encodes a monomeric single-pass type I membrane glycoprotein found on erythrocytes, leukocytes, glomerular podocytes, and splenic follicular dendritic cells that mediates cellular binding to particles and immune complexes that have activated complement [Ahearn and Fearon 1989].
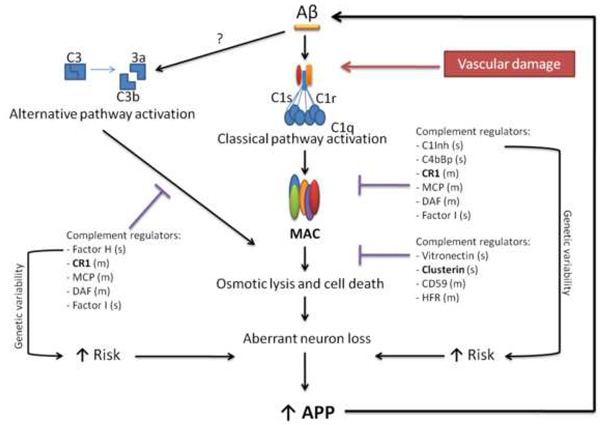
Three complement pathways are known: the classical, alternative and lectin-mediated cascades, which have different activation triggers, but all terminate with the production of the membrane attack complex. High enough concentrations of MAC result in cell lysis. This may lead to tissue damaging when the complement activation tight regulation, (that occurs through the action of several different endogenous complement inhibitor proteins), is deficient [Sjoberg, et al. 2009]. Typically, the complement classical pathway is the one associated with AD [Akiyama, et al. 2000], mainly due to three facts: 1) C1q, the first protein in this pathway, efficiently binds to aggregated Aβ, activating the pathway and further enhancing Aβ aggregation and fibril formation [Rogers, et al. 1992, Webster, et al. 1995]; 2) early complement activation proteins (C1q, C4 and C3) and the MAC have been found to co-localize with senile plaques, NFTs and dystrophic neurites in AD brains [McGeer, et al. 1989, Veerhuis, et al. 1996, Webster, et al. 1997]; and 3) increased mRNA levels of complement proteins are present in AD brains when compared to controls [Walker and McGeer 1992]. More recently, activation of the alternative pathway in AD brains has also been demonstrated: Aβ activates the alternative pathway in vitro [Bradt, et al. 1998]; factor B mRNA is present in AD frontal cortex, and factor D cleaved split products of factor B (Bb and Ba) are significantly increased in AD brains [Strohmeyer, et al. 2000]. After the discovery that the complement system can be activated in the brain by several senile plaques and neurofibrillary tangle related components, in the absence of antibodies, and that neurons are a source of complement proteins in the brain, the involvement of the complement system in AD has been widely accepted [McGeer and McGeer 2001]. However, whether this involvement has a protective or deleterious effect has been extensively debated. In fact, it has been proposed that binding of C1q to misfolded proteins in early AD, together with C4BP that decreases MAC activation, are favorable and enable clearance of the misfolded material. But, when the system is overwhelmed by amyloid, this protein binds extensively to C1q leading to the full activation of the complement, ultimately leading to detrimental inflammation and neurodegeneration [Sjoberg, et al. 2009].
In summary, as in other aging degenerative diseases, the complement system has an important role in AD, and one may expect that the activation of the classical or the alternative pathways (or both) by Aβ will lead to neurodegeneration in individuals with a genetic predisposition [Zipfel 2009]. This will possibly result from an unbalance between the expression of regulator proteins, and one or more cascade proteins (Figure 2). This model is able to explain, at least partially, the presence of neuropathological changes in the brains of non-demented individuals [Hof, et al. 1996] since the genetic variability in the complement genes may be responsible for different complement reactions to the presence of NFT and senile plaques. Interestingly, as mentioned above, one function attributed to clusterin is in the regulation of complement-mediated membrane attack complex. Together with vitronectin, clusterin binds to the nascent amphiphilic C5b-9 complex, rendering it water soluble and lytically inactive, raising the possibility that the genetic risk conferred by clusterin for the development of AD, may arise from its regulation role in the complement system.
Figure 2. Genetic variability and the role of the complement system in AD.
Aβ and probably NFTs are able to activate the classical and alternative complement pathways. These are regulated by several membrane (m) and soluble (s) proteins at different stages. The genetic variability in CR1 is now known to be associated with the risk of developing AD. Increased numbers of samples are needed to know if the same is true for any other component of these pathways. Drawn from the work of Tenner [Tenner 2001] and McGeer and McGeer [McGeer and McGeer 2002]
VASCULAR RISK FACTORS AND AD
These genetic findings point to the role of tissue and vascular damage in AD pathogenesis and thus to vascular risk factors in its aetiology. It is noteworthy that many of the genes now implicated potentially have a direct role at the blood brain interface: this includes APOE, ACE and the complement cascade components including CR1 and CLU. As such, these findings are consistent with the epidemiologic literature, which has consistently reported an association between vascular risk factors and AD. The evidence base for the prevention of AD and related brain pathologies is strongest for control of vascular risk factors [Gustafson D and I 2009]. Overweight and obesity is a cornerstone of vascular risk, which predisposes to hypertension, hypercholesterolemia, diabetes, and cardiovascular disease. Indeed, the low risk ratios observed in genetic studies attempting to identify new susceptibility genes for AD may be due to lack of information on presence and/or severity of this vascular involvement, as well as to the differential expression and clustering of vascular and metabolic traits. AD brain pathologies exist against a background continuum of vascular pathologies, which may modulate risk for clinically manifest disease. Vascular factors most directly related to newly identified susceptibility genes are hypertension, hypercholesterolemia, and overweight and obesity.
Hypertension is a risk factor for stroke, ischemic white matter lesions, silent infarcts, general atherosclerosis, myocardial infarction and cardiovascular morbidity and mortality. This risk increases with increasing blood pressure also at apparently healthy blood pressure ranges [Kannel 2000]. Several longitudinal studies have suggested an association between AD and previous hypertension [Kivipelto, et al. 2002, Launer, et al. 2000, Qiu, et al. 2005, Skoog, et al. 1996, Stewart, et al. 2009]. ACE, which is currently (accessed on November 16th) the fifth top hit on Alzgene database [Bertram, et al. 2007], plays a classical role in blood pressure regulation as part of the renin-angiotensin system [Goossens, et al. 2003]. This system may play a role in dementia pathogenesis because of its effects on vascular and metabolic homeostasis, as well as amyloid metabolism. The gene encoding for ACE is an AD susceptibility gene whereby effect modification is observed by vascular phenotype, particularly with population stratification by vascular factors such as APOE ε4 allele, systolic blood pressure, body mass index, and obesity indices [Gustafson, et al. 2008, Katzov, et al. 2004]. Thus, the renin angiotensin system may also provide a link between obesity, hypertension, and vascular syndromes, such as type 2 diabetes, and health of the brain [Goossens, et al. 2003, Katzov, et al. 2004] because human brain and adipose tissue express the renin-angiotensin system [Strazzullo, et al. 2003].
Cholesterol is important in AD, not only because of its relationship with cardiovascular disease, but due to its role in amyloid metabolism [Sparks, et al. 1994]. APOE and CLU are two proteins involved in lipid transport in the peripheral and central nervous systems [Nuutinen, et al. 2009]. APOE regulates cholesterol homeostasis in astrocytes and microglia, and is related to blood cholesterol levels [Hoglund and Blennow 2007]. In addition, mutations in the APP gene upregulate Aβ40 and Aβ42 production; [Pangalos, et al. 2005] and Aβ processing is sensitive to cholesterol levels and lipid trafficking. Brain cholesterol levels increase during AD progression [Hoglund and Blennow 2007]. The epidemiologic evidence associating high blood cholesterol levels with AD and other forms of dementia is mixed. High cholesterol levels in mid-life may increase risk for subsequent dementia and AD [Kivipelto, et al. 2002, Notkola, et al. 1998, Whitmer, et al. 2005], however, in late-life, low cholesterol levels have been predictive of subsequent dementia [Mielke, et al. 2005, Reitz, et al. 2004] or no association has been observed [Li, et al. 2005, Yoshitake, et al. 1995]. Nevertheless, even within the mid-life cholesterol literature, results are conflicting, as some studies have not found high cholesterol to predict later dementia [Kalmijn, et al. 2000, Stewart, et al. 2007, Tan, et al. 2003]. While there remain a number of questions regarding the amyloid hypothesis in relationship to AD, the potential link to cholesterol metabolism and vascular damage is noteworthy [Hardy 2009].
Finally, while overweight and obesity appear to increase risk for dementia independently of other vascular factors, there is limited evidence related to adipose-specific mechanisms of action in AD, particularly in relationship to these newly identified susceptibility genes. Mid-life total or central obesity measured decades before dementia onset has been linked to higher risk of dementia in late life [Fitzpatrick, et al. 2009, Gustafson, et al. 2009, Kivipelto, et al. 2005, Whitmer, et al. 2007, Whitmer, et al. 2008]. Risky effects of high BMI as late as in the eighth decade of life have also been observed [Gustafson, et al. 2003, Hayden, et al. 2006]. During the prodromal phase of dementia, higher rates of body weight or BMI decline occur among those developing dementia [Barrett-Connor, et al. 1996, Buchman, et al. 2005, Stewart, et al. 2005]. Thus, one cannot deny the role of excess adiposity as enhanced substrate for CR1-related inflammatory events, nor its potential role in hypertension and hyperlipidemia. However, also of importance are the implications of declining metabolic parameters, such as BMI [Barrett-Connor, et al. 1996, Buchman, et al. 2005, Stewart, et al. 2005], blood pressure [Stewart, et al. 2009] and cholesterol [Stewart, et al. 2007] in AD, for which these newly identified susceptibility genes may enhance our precision in identifying subgroups of AD for whom interventions are more advantageous.
POSSIBLE ROLE OF HOMOZYGOSITY AND RECESSIVE CASES
AD, as many other diseases occurring sporadically, recurs within families more often than expected by chance alone. However, in the majority of the cases, the pattern of familial recurrence is not compatible with simple Mendelian transmission and this model is typically presumed to reflect a multifactorial determination with contributions from multiple genes and/or environmental factors [Altshuler, et al. 2008]. However, the observed familial recurrence could also be attributed to genetic loci with large phenotypic effects and reduced penetrance (possibly recessive loci). In this case, one would not necessarily expect to see recurrence of the disease in multiple generations, nor a high recurrence rate among siblings, and the disease would be sporadic in the population. Although without a definite confirmation of pathogenicity, two rare potentially disease-associated mutations (Q170H and R181G) in ADAM10 (an alpha-secretase capable of anti-amyloidogenic proteolysis of the amyloid precursor protein) were recently reported to be associated with LOAD [Kim, et al. 2009]. Other mutations in PSEN1 (A79V) [Kauwe, et al. 2007] and PSEN2 (N141I) [Levy-Lahad, et al. 1995] have also been reported to be present in families with non-carriers affected individuals.
Recessive contributions can be inferred when populations with high degrees of consanguinity present higher prevalence of the disease than the general population [Mani, et al. 2002]. The Wadi Ara population is one example of this premise: an unusually high prevalence of AD (20% of those over 65 years and 60% of those over 85 years) in a population where the 4 allele of APOE is relatively uncommon [Bowirrat, et al. 2000, Bowirrat, et al. 2001]. In this regard, the study of families with a recessive mode of inheritance may not only identify the cause of disease in the respective family, but also be of utility in the identification of risk factors contributing to the sporadic form of disease.
Populations that have been largely isolated and subjected to extensive inbreeding during considerable periods in their recent history represent a powerful resource for the study of new genetic variants for common diseases. These populations provide several advantages for genetic research, such as longer stretches of linkage between neighboring markers, high levels of genetic and environmental homogeneity and a simpler genetic architecture for complex traits. Although these long homozygous tracts of uninterrupted sequences may represent deletion polymorphisms, loss of heterozygosity or segmental uniparental disomy, recent data suggests that these, most likely, represent autozygosity (homozygosity by virtue of parental descent from a common ancestor) [Devilee, et al. 2001, Li, et al. 2006, Raghavan, et al. 2005, Woods, et al. 2004]. In this way, an obvious application of whole genome platforms in relation to autozygosity is in the genetic analysis of consanguineous families. This can be considered as an analogous approach to linkage analysis, in which researchers aim to define shared regions of autozygosity and/or overlapping structural variants in order to determine the role of autozygosity in a particular disease. Autozygosity mapping has long been recognized as a rapid and cost effective way to identify loci underlying recessive disease [Lander and Botstein 1987] and several genes underlying different disorders have been identified using this methodology [Camargos, et al. 2008, Paisan-Ruiz, et al. 2009]. Specifically in AD, we have generated the first catalog of autozygosity in EOAD by studying a consanguineous Israeli family. Although we were unable to pinpoint a specific gene due to the small number of samples we had available for analysis, we were able to generate a catalog that may be used in future studies of other families [Clarimon, et al. 2008].
Furthermore, even in outbred populations, more individuals have a high frequency of these autozygous tracts than previously expected [Gibson, et al. 2006, Li, et al. 2006]. Our group and others have recently reported the unexpected high degree of apparent parental consanguinity in control individuals from North America (~10% of studied individuals harboring homozygous tracts larger than 5Mb) [Simon-Sanchez, et al. 2007]. Similar numbers (~6%) were presented by Li and collaborators, when studying an outbred population of unrelated Han Chinese [Li, et al. 2006] and by Gibson et al. who reported that >1000 tracts exceeding 1Mb in length were observed in the ~200 unrelated HapMap individuals studied [Gibson, et al. 2006]. These observations prompted us to study autozygosity in LOAD in an outbred population. By comparing measures of extended homozygosity (greater than 1 Mb in length) in a population of >800 LOAD cases and >550 controls we were able to identify one homozygous region on chromosome 8 (8p12, not including the CLU locus), significantly associated with LOAD. Additionally, the comparison of the total numbers of homozygous runs and the total length of these runs between cases and controls, revealed a suggestive difference in these measures (p-values 0.052–0.062), most likely symptomatic of a recessive component in the etiology of LOAD [Nalls, et al. 2009]. The role of recessive mutations in AD has been considerably overlooked: in addition to the recent works describing recessive APP mutations (discussed above in this review) [Di Fede, et al. 2009, Tomiyama, et al. 2008], only two other studies of isolated populations with a high incidence of the disease have been reported where the primary analyses were performed using dominant or additive modes of inheritance [Farrer, et al. 2003, Liu, et al. 2007]. The new technologies now available will allow us to overcome this gap in the near future.
THE NEXT STEPS
Two major and clearly non-exclusive pathways have been recently discussed as the future guidelines in the genome wide analysis of complex disorders [Goldstein 2009, Hirschhorn 2009, Kraft and Hunter 2009]. The first is the extension of the assembly of studies containing larger (tens or even hundreds of thousands) and representative samples in order to identify variants with lower frequencies that may have been missed until now and that could explain the so elusive fractions of “missing heritability” in AD. The increase in the number of studied samples will inevitably result in the discovery of new variants and probably new genes associated with AD, but the real net value of these variants is highly disputable. Therefore, the actual dilemma now is to know how far one should take these studies in order to keep a positive balance between the resources applied and the gathered genetic returns [Goldstein 2009, Hardy and Singleton 2009]. Clearly, still to discover rare variants and variants with small effect sizes will be difficult to replicate, due to reduced power and restriction to specific populations, respectively. Nonetheless, in order to identify these rare variants one may speculate the need to use new chips with a better coverage of rare variants and the resequencing of previously identified regions. One may also predict that, being AD a complex disorder, more emphasis will be put in the study of endophenotypes, as happened in the study by Liu et al. that used cognitive function as an endophenotype of AD and identified the RGSL2, RALGPS2 and C1orf49 genes as potential causative genes located in one of the associated genomic regions [Liu, et al. 2007]. This approach will obviously require precise and clearly defined clinical assessments.
The second line of investigation will rely in the sequencing of whole exomes and whole genomes, expectantly unveiling several new rarer risk variants. These higher risk variants may be related to the commoner lower risk variants found with GWAS. In fact, a classical example comes from the study of Parkinson’s disease genetics: rare high-risk variants in the α-synuclein gene are the cause of monogenic PD [Polymeropoulos, et al. 1997], while a common haplotype of this same gene has been established as a moderate risk cause of sporadic disease [Simon-Sanchez, et al. 2009].
Both lines of research will be followed by large resequencing efforts, in order to identify the real risk variants. Many GWAS have identified regions of the genome associated with AD, but in many cases the real risk/causal changes are not known. Additionally, in the cases where SNPs have been associated with a disease, these SNPs identified may in fact be in LD with the real variants causing the association. Further functional data, (for instance, the effects of a determined variant in gene expression), although not always possible to obtain, will be essential not only to validate the previously identified variants, but also because many variants map to non coding protein sequences, gene deserts, or genomic regions without any functional elements. The integration of these functional studies with information on multiple variants from the same gene, in different populations and the effects of epigenetics and epistasis will be vital. The interpretation of these results, however, will be complex and will most likely require the combination of disciplines as integrative genomics and systems biology. Although difficult, this approach will ultimately allow a more profound understanding of the molecular pathways underlying AD and AD risk, as well as the subsequent identification of effective biomarkers and drugs.
ACKNOWLEDGEMENTS
This work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services; project number Z01 AG000950-06 and Fundacao para a Ciencia e Tecnologia, Portugal grant SFRH/BD/27442/2006. DRG was supported by the Swedish MFR and JH by an MRC Returning Scientist Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE STATEMENT
None of the authors has any actual or potential conflicts of interest including any financial, personal or other relationships with other people or organizations within three years of beginning the work submitted that could inappropriately bias their work.
REFERENCES
- Abraham R, Moskvina V, Sims R, Hollingworth P, Morgan A, Georgieva L, Dowzell K, Cichon S, Hillmer AM, O'Donovan MC, Williams J, Owen MJ, Kirov G. A genome-wide association study for late-onset Alzheimer's disease using DNA pooling. BMC Med Genomics. 2008;144 doi: 10.1186/1755-8794-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahearn JM, Fearon DT. Structure and function of the complement receptors, CR1 (CD35) and CR2 (CD21) Adv Immunol. 1989:46183–46219. doi: 10.1016/s0065-2776(08)60654-9. [DOI] [PubMed] [Google Scholar]
- Akiyama H, Barger S, Barnum S, Bradt B, Bauer J, Cole GM, Cooper NR, Eikelenboom P, Emmerling M, Fiebich BL, Finch CE, Frautschy S, Griffin WS, Hampel H, Hull M, Landreth G, Lue L, Mrak R, Mackenzie IR, McGeer PL, O'Banion MK, Pachter J, Pasinetti G, Plata-Salaman C, Rogers J, Rydel R, Shen Y, Streit W, Strohmeyer R, Tooyoma I, Van Muiswinkel FL, Veerhuis R, Walker D, Webster S, Wegrzyniak B, Wenk G, Wyss-Coray T. Inflammation and Alzheimer's disease. Neurobiol Aging. 2000;21(3):383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322(5903):881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashford JW. APOE genotype effects on Alzheimer's disease onset and epidemiology. J Mol Neurosci. 2004;23(3):157–165. doi: 10.1385/JMN:23:3:157. [DOI] [PubMed] [Google Scholar]
- Ashley-Koch AE, Shao Y, Rimmler JB, Gaskell PC, Welsh-Bohmer KA, Jackson CE, Scott WK, Haines JL, Pericak-Vance MA. An autosomal genomic screen for dementia in an extended Amish family. Neurosci Lett. 2005;379(3):199–204. doi: 10.1016/j.neulet.2004.12.065. [DOI] [PubMed] [Google Scholar]
- Avramopoulos D, Fallin MD, Bassett SS. Linkage to chromosome 14q in Alzheimer's disease (AD) patients without psychotic symptoms. Am J Med Genet B Neuropsychiatr Genet. 2005;132B(1):9–13. doi: 10.1002/ajmg.b.30074. [DOI] [PubMed] [Google Scholar]
- Bacanu SA, Devlin B, Chowdari KV, DeKosky ST, Nimgaonkar VL, Sweet RA. Linkage analysis of Alzheimer disease with psychosis. Neurology. 2002;59(1):118–120. doi: 10.1212/wnl.59.1.118. [DOI] [PubMed] [Google Scholar]
- Barrett-Connor E, Edelstein SL, Corey-Bloom J, Wiederholt WC. Weight loss precedes dementia in community-dwelling older adults. J Am Geriatr Soc. 1996;44(10):1147–1152. doi: 10.1111/j.1532-5415.1996.tb01362.x. [DOI] [PubMed] [Google Scholar]
- Bassett SS, Avramopoulos D, Fallin D. Evidence for parent of origin effect in late-onset Alzheimer disease. Am J Med Genet. 2002;114(6):679–686. doi: 10.1002/ajmg.10648. [DOI] [PubMed] [Google Scholar]
- Beecham GW, Martin ER, Li YJ, Slifer MA, Gilbert JR, Haines JL, Pericak-Vance MA. Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease. Am J Hum Genet. 2009;84(1):35–43. doi: 10.1016/j.ajhg.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekris LM, Galloway NM, Montine TJ, Schellenberg GD, Yu CE. APOE mRNA and protein expression in postmortem brain are modulated by an extended haplotype structure. Am J Med Genet B Neuropsychiatr Genet. 2009 doi: 10.1002/ajmg.b.30993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RD, Sagare AP, Friedman AE, Bedi GS, Holtzman DM, Deane R, Zlokovic BV. Transport pathways for clearance of human Alzheimer's amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system. J Cereb Blood Flow Metab. 2007;27(5):909–918. doi: 10.1038/sj.jcbfm.9600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BM, Hooli B, Divito J, Ionita I, Jiang H, Laird N, Moscarillo T, Ohlsen KL, Elliott K, Wang X, Hu-Lince D, Ryder M, Murphy A, Wagner SL, Blacker D, Becker KD, Tanzi RE. Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83(5):623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. Thirty years of Alzheimer's disease genetics: the implications of systematic meta-analyses. Nat Rev Neurosci. 2008;9(10):768–778. doi: 10.1038/nrn2494. [DOI] [PubMed] [Google Scholar]
- Bertrand P, Poirier J, Oda T, Finch CE, Pasinetti GM. Association of apolipoprotein E genotype with brain levels of apolipoprotein E and apolipoprotein J (clusterin) in Alzheimer disease. Brain Res Mol Brain Res. 1995;33(1):174–178. doi: 10.1016/0169-328x(95)00097-c. [DOI] [PubMed] [Google Scholar]
- Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RC, Mahoney A, Beaty T, Fallin MD, Avramopoulos D, Chase GA, Folstein MF, McInnis MG, Bassett SS, Doheny KJ, Pugh EW, Tanzi RE. Results of a high-resolution genome screen of 437 Alzheimer's disease families. Hum Mol Genet. 2003;12(1):23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- Blom ES, Viswanathan J, Kilander L, Helisalmi S, Soininen H, Lannfelt L, Ingelsson M, Glaser A, Hiltunen M. Low prevalence of APP duplications in Swedish and Finnish patients with early-onset Alzheimer's disease. Eur J Hum Genet. 2008;16(2):171–175. doi: 10.1038/sj.ejhg.5201966. [DOI] [PubMed] [Google Scholar]
- Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40(6):695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs LN, Fuson KS, Baez M, Churgay L, McClure D, Becker G, May PC. Clusterin (Apo J) protects against in vitro amyloid-beta (1–40) neurotoxicity. J Neurochem. 1996;67(3):1324–1327. doi: 10.1046/j.1471-4159.1996.67031324.x. [DOI] [PubMed] [Google Scholar]
- Bowirrat A, Friedland RP, Chapman J, Korczyn AD. The very high prevalence of AD in an Arab population is not explained by APOE epsilon4 allele frequency. Neurology. 2000;55(5):731. doi: 10.1212/wnl.55.5.731. [DOI] [PubMed] [Google Scholar]
- Bowirrat A, Treves TA, Friedland RP, Korczyn AD. Prevalence of Alzheimer's type dementia in an elderly Arab population. Eur J Neurol. 2001;8(2):119–123. doi: 10.1046/j.1468-1331.2001.00183.x. [DOI] [PubMed] [Google Scholar]
- Bradt BM, Kolb WP, Cooper NR. Complement-dependent proinflammatory properties of the Alzheimer's disease beta-peptide. J Exp Med. 1998;188(3):431–438. doi: 10.1084/jem.188.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman AS, Wilson RS, Bienias JL, Shah RC, Evans DA, Bennett DA. Change in body mass index and risk of incident Alzheimer disease. Neurology. 2005;65(6):892–897. doi: 10.1212/01.wnl.0000176061.33817.90. [DOI] [PubMed] [Google Scholar]
- Butler AW, Ng MY, Hamshere ML, Forabosco P, Wroe R, Al-Chalabi A, Lewis CM, Powell JF. Meta-analysis of linkage studies for Alzheimer's disease--a web resource. Neurobiol Aging. 2009;30(7):1037–1047. doi: 10.1016/j.neurobiolaging.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Cabrejo L, Guyant-Marechal L, Laquerriere A, Vercelletto M, De la Fourniere F, Thomas-Anterion C, Verny C, Letournel F, Pasquier F, Vital A, Checler F, Frebourg T, Campion D, Hannequin D. Phenotype associated with APP duplication in five families. Brain. 2006;129(Pt 11):2966–2976. doi: 10.1093/brain/awl237. [DOI] [PubMed] [Google Scholar]
- Camargos S, Scholz S, Simon-Sanchez J, Paisan-Ruiz C, Lewis P, Hernandez D, Ding J, Gibbs JR, Cookson MR, Bras J, Guerreiro R, Oliveira CR, Lees A, Hardy J, Cardoso F, Singleton AB. DYT16, a novel young-onset dystonia-parkinsonism disorder: identification of a segregating mutation in the stress-response protein PRKRA. Lancet Neurol. 2008;7(3):207–215. doi: 10.1016/S1474-4422(08)70022-X. [DOI] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22(3):231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, Younkin SG, Younkin CS, Younkin LH, Bisceglio GD, Ertekin-Taner N, Crook JE, Dickson DW, Petersen RC, Graff-Radford NR, Younkin SG. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer's disease. Nat Genet. 2009;41(2):192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A. Population genetics--making sense out of sequence. Nat Genet. 1999;21(1) Suppl:56–60. doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Parfitt M, Legrain S, Perez-Tur J, Brousseau T, Evans A, Berr C, Vidal O, Roques P, Gourlet V, et al. Apolipoprotein E, epsilon 4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer's disease: analysis of the 19q13.2 chromosomal region. Hum Mol Genet. 1994;3(4):569–574. doi: 10.1093/hmg/3.4.569. [DOI] [PubMed] [Google Scholar]
- Choudhry S, Coyle NE, Tang H, Salari K, Lind D, Clark SL, Tsai HJ, Naqvi M, Phong A, Ung N, Matallana H, Avila PC, Casal J, Torres A, Nazario S, Castro R, Battle NC, Perez-Stable EJ, Kwok PY, Sheppard D, Shriver MD, Rodriguez-Cintron W, Risch N, Ziv E, Burchard EG. Population stratification confounds genetic association studies among Latinos. Hum Genet. 2006;118(5):652–664. doi: 10.1007/s00439-005-0071-3. [DOI] [PubMed] [Google Scholar]
- Clarimon J, Djaldetti R, Lleo A, Guerreiro RJ, Molinuevo JL, Paisan-Ruiz C, Gomez-Isla T, Blesa R, Singleton A, Hardy J. Whole genome analysis in a consanguineous family with early onset Alzheimer's disease. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium IH. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, Zismann VL, Beach TG, Leung D, Bryden L, Halperin RF, Marlowe L, Kaleem M, Walker DG, Ravid R, Heward CB, Rogers J, Papassotiropoulos A, Reiman EM, Hardy J, Stephan DA. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J Clin Psychiatry. 2007;68(4):613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- Corbo RM, Scacchi R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE*4 a 'thrifty' allele? Ann Hum Genet. 1999;63(Pt 4):301–310. doi: 10.1046/j.1469-1809.1999.6340301.x. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, Jr, Rimmler JB, Locke PA, Conneally PM, Schmader KE, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7(2):180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Curtis D, North BV, Sham PC. A novel method of two-locus linkage analysis applied to a genome scan for late onset Alzheimer's disease. Ann Hum Genet. 2001;65(Pt 5):473–481. doi: 10.1017/S0003480001008776. [DOI] [PubMed] [Google Scholar]
- Danik M, Chabot JG, Hassan-Gonzalez D, Suh M, Quirion R. Localization of sulfated glycoprotein-2/clusterin mRNA in the rat brain by in situ hybridization. J Comp Neurol. 1993;334(2):209–227. doi: 10.1002/cne.903340205. [DOI] [PubMed] [Google Scholar]
- Daw EW, Payami H, Nemens EJ, Nochlin D, Bird TD, Schellenberg GD, Wijsman EM. The number of trait loci in late-onset Alzheimer disease. Am J Hum Genet. 2000;66(1):196–204. doi: 10.1086/302710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawn Teare M, Barrett JH. Genetic linkage studies. Lancet. 2005;366(9490):1036–1044. doi: 10.1016/S0140-6736(05)67382-5. [DOI] [PubMed] [Google Scholar]
- de Silva HV, Harmony JA, Stuart WD, Gil CM, Robbins J. Apolipoprotein J: structure and tissue distribution. Biochemistry. 1990;29(22):5380–5389. doi: 10.1021/bi00474a025. [DOI] [PubMed] [Google Scholar]
- De Strooper B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active gamma-Secretase complex. Neuron. 2003;38(1):9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- DeMattos RB, Cirrito JR, Parsadanian M, May PC, O'Dell MA, Taylor JW, Harmony JA, Aronow BJ, Bales KR, Paul SM, Holtzman DM. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 2004;41(2):193–202. doi: 10.1016/s0896-6273(03)00850-x. [DOI] [PubMed] [Google Scholar]
- DeMattos RB, O'Dell MA, Parsadanian M, Taylor JW, Harmony JA, Bales KR, Paul SM, Aronow BJ, Holtzman DM. Clusterin promotes amyloid plaque formation and is critical for neuritic toxicity in a mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2002;99(16):10843–10848. doi: 10.1073/pnas.162228299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devilee P, Cleton-Jansen AM, Cornelisse CJ. Ever since Knudson. Trends Genet. 2001;17(10):569–573. doi: 10.1016/s0168-9525(01)02416-7. [DOI] [PubMed] [Google Scholar]
- Di Fede G, Catania M, Morbin M, Rossi G, Suardi S, Mazzoleni G, Merlin M, Giovagnoli AR, Prioni S, Erbetta A, Falcone C, Gobbi M, Colombo L, Bastone A, Beeg M, Manzoni C, Francescucci B, Spagnoli A, Cantu L, Del Favero E, Levy E, Salmona M, Tagliavini F. A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science. 2009;323(5920):1473–1477. doi: 10.1126/science.1168979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguid JR, Bohmont CW, Liu NG, Tourtellotte WW. Changes in brain gene expression shared by scrapie and Alzheimer disease. Proc Natl Acad Sci U S A. 1989;86(18):7260–7264. doi: 10.1073/pnas.86.18.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch FS, Keim PS, Beattie EC, Blacher RW, Culwell AR, Oltersdorf T, McClure D, Ward PJ. Cleavage of amyloid beta peptide during constitutive processing of its precursor. Science. 1990;248(4959):1122–1124. doi: 10.1126/science.2111583. [DOI] [PubMed] [Google Scholar]
- Farrer LA, Bowirrat A, Friedland RP, Waraska K, Korczyn AD, Baldwin CT. Identification of multiple loci for Alzheimer disease in a consanguineous Israeli-Arab community. Hum Mol Genet. 2003;12(4):415–422. doi: 10.1093/hmg/ddg037. [DOI] [PubMed] [Google Scholar]
- Fearnhead NS, Winney B, Bodmer WF. Rare variant hypothesis for multifactorial inheritance: susceptibility to colorectal adenomas as a model. Cell Cycle. 2005;4(4):521–525. doi: 10.4161/cc.4.4.1591. [DOI] [PubMed] [Google Scholar]
- Feulner TM, Laws SM, Friedrich P, Wagenpfeil S, Wurst SH, Riehle C, Kuhn KA, Krawczak M, Schreiber S, Nikolaus S, Forstl H, Kurz A, Riemenschneider M. Examination of the current top candidate genes for AD in a genome-wide association study. Mol Psychiatry. 2009 doi: 10.1038/mp.2008.141. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick AL, Kuller LH, Lopez OL, Diehr P, O'Meara ES, Longstreth WT, Jr, Luchsinger JA. Midlife and late-life obesity and the risk of dementia: cardiovascular health study. Arch Neurol. 2009;66(3):336–342. doi: 10.1001/archneurol.2008.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63(2):168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- Ghiso J, Matsubara E, Koudinov A, Choi-Miura NH, Tomita M, Wisniewski T, Frangione B. The cerebrospinal-fluid soluble form of Alzheimer's amyloid beta is complexed to SP-40,40 (apolipoprotein J), an inhibitor of the complement membrane-attack complex. Biochem J. 1993;293(Pt 1):27–30. doi: 10.1042/bj2930027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J, Morton NE, Collins A. Extended tracts of homozygosity in outbred human populations. Hum Mol Genet. 2006;15(5):789–795. doi: 10.1093/hmg/ddi493. [DOI] [PubMed] [Google Scholar]
- Golabek A, Marques MA, Lalowski M, Wisniewski T. Amyloid beta binding proteins in vitro and in normal human cerebrospinal fluid. Neurosci Lett. 1995;191(1–2):79–82. doi: 10.1016/0304-3940(95)11565-7. [DOI] [PubMed] [Google Scholar]
- Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360(17):1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- Goossens GH, Blaak EE, van Baak MA. Possible involvement of the adipose tissue renin-angiotensin system in the pathophysiology of obesity and obesity-related disorders. Obes Rev. 2003;4(1):43–55. doi: 10.1046/j.1467-789x.2003.00091.x. [DOI] [PubMed] [Google Scholar]
- Grupe A, Abraham R, Li Y, Rowland C, Hollingworth P, Morgan A, Jehu L, Segurado R, Stone D, Schadt E, Karnoub M, Nowotny P, Tacey K, Catanese J, Sninsky J, Brayne C, Rubinsztein D, Gill M, Lawlor B, Lovestone S, Holmans P, O'Donovan M, Morris JC, Thal L, Goate A, Owen MJ, Williams J. Evidence for novel susceptibility genes for late-onset Alzheimer's disease from a genome-wide association study of putative functional variants. Hum Mol Genet. 2007;16(8):865–873. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- Guerreiro RJ, Baquero M, Blesa R, Boada M, Bras JM, Bullido MJ, Calado A, Crook R, Ferreira C, Frank A, Gomez-Isla T, Hernandez I, Lleo A, Machado A, Martinez-Lage P, Masdeu J, Molina-Porcel L, Molinuevo JL, Pastor P, Perez-Tur J, Relvas R, Oliveira CR, Ribeiro MH, Rogaeva E, Sa A, Samaranch L, Sanchez-Valle R, Santana I, Tarraga L, Valdivieso F, Singleton A, Hardy J, Clarimon J. Genetic screening of Alzheimer's disease genes in Iberian and African samples yields novel mutations in presenilins and APP. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.06.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson DIS. Control of vascular risk factors. In: L-O Wahlund TE, Gauthier S, editors. Vascular dementia in clinical practice. Cambridge University Press; 2009. [Google Scholar]
- Gustafson D, Rothenberg E, Blennow K, Steen B, Skoog I. An 18-year follow-up of overweight and risk of Alzheimer disease. Arch Intern Med. 2003;163(13):1524–1528. doi: 10.1001/archinte.163.13.1524. [DOI] [PubMed] [Google Scholar]
- Gustafson DR, Backman K, Waern M, Ostling S, Guo X, Zandi P, Mielke MM, Bengtsson C, Skoog I. Adiposity indicators and dementia over 32 years in Sweden. Neurology. 2009;73(19):1559–1566. doi: 10.1212/WNL.0b013e3181c0d4b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson DR, Melchior L, Eriksson E, Sundh V, Blennow K, Skoog I. The ACE Insertion Deletion polymorphism relates to dementia by metabolic phenotype, APOEvarepsilon4, and age of dementia onset. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Haass C, Schlossmacher MG, Hung AY, Vigo-Pelfrey C, Mellon A, Ostaszewski BL, Lieberburg I, Koo EH, Schenk D, Teplow DB, et al. Amyloid beta-peptide is produced by cultured cells during normal metabolism. Nature. 1992;359(6393):322–325. doi: 10.1038/359322a0. [DOI] [PubMed] [Google Scholar]
- Hahs DW, McCauley JL, Crunk AE, McFarland LL, Gaskell PC, Jiang L, Slifer SH, Vance JM, Scott WK, Welsh-Bohmer KA, Johnson SR, Jackson CE, Pericak-Vance MA, Haines JL. A genome-wide linkage analysis of dementia in the Amish. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(2):160–166. doi: 10.1002/ajmg.b.30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad SM, Ranganathan S, Loukinova E, Twal WO, Argraves WS. Interaction of apolipoprotein J-amyloid beta-peptide complex with low density lipoprotein receptor-related protein-2/megalin. A mechanism to prevent pathological accumulation of amyloid beta-peptide. J Biol Chem. 1997;272(30):18644–18649. doi: 10.1074/jbc.272.30.18644. [DOI] [PubMed] [Google Scholar]
- Hamshere ML, Holmans PA, Avramopoulos D, Bassett SS, Blacker D, Bertram L, Wiener H, Rochberg N, Tanzi RE, Myers A, Wavrant-De Vrieze F, Go R, Fallin D, Lovestone S, Hardy J, Goate A, O'Donovan M, Williams J, Owen MJ. Genome-wide linkage analysis of 723 affected relative pairs with late-onset Alzheimer's disease. Hum Mol Genet. 2007;16(22):2703–2712. doi: 10.1093/hmg/ddm224. [DOI] [PubMed] [Google Scholar]
- Hardy J. Amyloid, the presenilins and Alzheimer's disease. Trends Neurosci. 1997;20(4):154–159. doi: 10.1016/s0166-2236(96)01030-2. [DOI] [PubMed] [Google Scholar]
- Hardy J. The amyloid hypothesis for Alzheimer's disease: a critical reappraisal. J Neurochem. 2009;110(4):1129–1134. doi: 10.1111/j.1471-4159.2009.06181.x. [DOI] [PubMed] [Google Scholar]
- Hardy J, Crook R. Presenilin mutations line up along transmembrane alpha-helices. Neurosci Lett. 2001;306(3):203–205. doi: 10.1016/s0304-3940(01)01910-3. [DOI] [PubMed] [Google Scholar]
- Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360(17):1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Wu F, Mattson MP, Morris CM, Yao PJ. Evidence for CALM in directing VAMP2 trafficking. Traffic. 2008;9(3):417–429. doi: 10.1111/j.1600-0854.2007.00694.x. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O'Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley AT, McCarthy MI. What makes a good genetic association study? Lancet. 2005;366(9493):1315–1323. doi: 10.1016/S0140-6736(05)67531-9. [DOI] [PubMed] [Google Scholar]
- Hayden KM, Zandi PP, Lyketsos CG, Khachaturian AS, Bastian LA, Charoonruk G, Tschanz JT, Norton MC, Pieper CF, Munger RG, Breitner JC, Welsh-Bohmer KA. Vascular risk factors for incident Alzheimer disease and vascular dementia: the Cache County study. Alzheimer Dis Assoc Disord. 2006;20(2):93–100. doi: 10.1097/01.wad.0000213814.43047.86. [DOI] [PubMed] [Google Scholar]
- Henricson A, Kall L, Sonnhammer EL. A novel transmembrane topology of presenilin based on reconciling experimental and computational evidence. Febs J. 2005;272(11):2727–2733. doi: 10.1111/j.1742-4658.2005.04691.x. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN. Genomewide association studies--illuminating biologic pathways. N Engl J Med. 2009;360(17):1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- Hof PR, Glannakopoulos P, Bouras C. The neuropathological changes associated with normal brain aging. Histol Histopathol. 1996;11(4):1075–1088. [PubMed] [Google Scholar]
- Hoglund K, Blennow K. Effect of HMG-CoA reductase inhibitors on beta-amyloid peptide levels: implications for Alzheimer's disease. CNS Drugs. 2007;21(6):449–462. doi: 10.2165/00023210-200721060-00002. [DOI] [PubMed] [Google Scholar]
- Hollingworth P, Hamshere ML, Holmans PA, O'Donovan MC, Sims R, Powell J, Lovestone S, Myers A, DeVrieze FW, Hardy J, Goate A, Owen M, Williams J. Increased familial risk and genomewide significant linkage for Alzheimer's disease with psychosis. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(7):841–848. doi: 10.1002/ajmg.b.30515. [DOI] [PubMed] [Google Scholar]
- Holmans P, Hamshere M, Hollingworth P, Rice F, Tunstall N, Jones S, Moore P, Wavrant DeVrieze F, Myers A, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, O'Donovan M, Jones L, Hardy J, Goate A, Lovestone S, Owen M, Williams J. Genome screen for loci influencing age at onset and rate of decline in late onset Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet. 2005;135B(1):24–32. doi: 10.1002/ajmg.b.30114. [DOI] [PubMed] [Google Scholar]
- Hyman BT, Hedley-Whyte ET, Rebeck GW, Vonsattel JP, West HL, Growdon JH. Apolipoprotein E epsilon4/4 in a neuropathologically normal very elderly individual. Arch Neurol. 1996;53(3):215. doi: 10.1001/archneur.1996.00550030017010. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29(3):306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- Jenne DE, Tschopp J. Clusterin: the intriguing guises of a widely expressed glycoprotein. Trends Biochem Sci. 1992;17(4):154–159. doi: 10.1016/0968-0004(92)90325-4. [DOI] [PubMed] [Google Scholar]
- Jones SE, Jomary C. Clusterin. Int J Biochem Cell Biol. 2002;34(5):427–431. doi: 10.1016/s1357-2725(01)00155-8. [DOI] [PubMed] [Google Scholar]
- Kalmijn S, Foley D, White L, Burchfiel CM, Curb JD, Petrovitch H, Ross GW, Havlik RJ, Launer LJ. Metabolic cardiovascular syndrome and risk of dementia in Japanese-American elderly men. The Honolulu-Asia aging study. Arterioscler Thromb Vasc Biol. 2000;20(10):2255–2260. doi: 10.1161/01.atv.20.10.2255. [DOI] [PubMed] [Google Scholar]
- Kannel WB. Risk stratification in hypertension: new insights from the Framingham Study. Am J Hypertens. 2000;13(1 Pt 2):3S–10S. doi: 10.1016/s0895-7061(99)00252-6. [DOI] [PubMed] [Google Scholar]
- Kasuga K, Shimohata T, Nishimura A, Shiga A, Mizuguchi T, Tokunaga J, Ohno T, Miyashita A, Kuwano R, Matsumoto N, Onodera O, Nishizawa M, Ikeuchi T. Identification of independent APP locus duplication in Japanese patients with early-onset Alzheimer disease. J Neurol Neurosurg Psychiatry. 2009;80(9):1050–1052. doi: 10.1136/jnnp.2008.161703. [DOI] [PubMed] [Google Scholar]
- Katzov H, Bennet AM, Kehoe P, Wiman B, Gatz M, Blennow K, Lenhard B, Pedersen NL, de Faire U, Prince JA. A cladistic model of ACE sequence variation with implications for myocardial infarction, Alzheimer disease and obesity. Hum Mol Genet. 2004;13(21):2647–2657. doi: 10.1093/hmg/ddh286. [DOI] [PubMed] [Google Scholar]
- Kauwe JS, Jacquart S, Chakraverty S, Wang J, Mayo K, Fagan AM, Holtzman DM, Morris JC, Goate AM. Extreme cerebrospinal fluid amyloid beta levels identify family with late-onset Alzheimer's disease presenilin 1 mutation. Ann Neurol. 2007;61(5):446–453. doi: 10.1002/ana.21099. [DOI] [PubMed] [Google Scholar]
- Kehoe P, Wavrant-De Vrieze F, Crook R, Wu WS, Holmans P, Fenton I, Spurlock G, Norton N, Williams H, Williams N, Lovestone S, Perez-Tur J, Hutton M, Chartier-Harlin MC, Shears S, Roehl K, Booth J, Van Voorst W, Ramic D, Williams J, Goate A, Hardy J, Owen MJ. A full genome scan for late onset Alzheimer's disease. Hum Mol Genet. 1999;8(2):237–245. doi: 10.1093/hmg/8.2.237. [DOI] [PubMed] [Google Scholar]
- Kida E, Choi-Miura NH, Wisniewski KE. Deposition of apolipoproteins E and J in senile plaques is topographically determined in both Alzheimer's disease and Down's syndrome brain. Brain Res. 1995;685(1–2):211–216. doi: 10.1016/0006-8993(95)00482-6. [DOI] [PubMed] [Google Scholar]
- Kim JA, Kim HL. Cell-free expression and functional reconstitution of CALM in clathrin assembly. Exp Mol Med. 2001;33(2):89–94. doi: 10.1038/emm.2001.16. [DOI] [PubMed] [Google Scholar]
- Kim M, Suh J, Romano D, Truong MH, Mullin K, Hooli B, Norton D, Tesco G, Elliott K, Wagner SL, Moir RD, Becker KD, Tanzi RE. Potential late-onset Alzheimer's disease-associated mutations in the ADAM10 gene attenuate {alpha}-secretase activity. Hum Mol Genet. 2009;18(20):3987–3996. doi: 10.1093/hmg/ddp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivipelto M, Laakso MP, Tuomilehto J, Nissinen A, Soininen H. Hypertension and hypercholesterolaemia as risk factors for Alzheimer's disease: potential for pharmacological intervention. CNS Drugs. 2002;16(7):435–444. doi: 10.2165/00023210-200216070-00001. [DOI] [PubMed] [Google Scholar]
- Kivipelto M, Ngandu T, Fratiglioni L, Viitanen M, Kareholt I, Winblad B, Helkala EL, Tuomilehto J, Soininen H, Nissinen A. Obesity and vascular risk factors at midlife and the risk of dementia and Alzheimer disease. Arch Neurol. 2005;62(10):1556–1560. doi: 10.1001/archneur.62.10.1556. [DOI] [PubMed] [Google Scholar]
- Kovacs DM, Fausett HJ, Page KJ, Kim TW, Moir RD, Merriam DE, Hollister RD, Hallmark OG, Mancini R, Felsenstein KM, Hyman BT, Tanzi RE, Wasco W. Alzheimer-associated presenilins 1 and 2: neuronal expression in brain and localization to intracellular membranes in mammalian cells. Nat Med. 1996;2(2):224–229. doi: 10.1038/nm0296-224. [DOI] [PubMed] [Google Scholar]
- Kraft P, Hunter DJ. Genetic risk prediction--are we there yet? N Engl J Med. 2009;360(17):1701–1703. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Araria-Goumidi L, Myllykangas L, Ellis C, Wang JC, Bullido MJ, Harris JM, Artiga MJ, Hernandez D, Kwon JM, Frigard B, Petersen RC, Cumming AM, Pasquier F, Sastre I, Tienari VB, Frank A, Sulkava R, Morris JC, St Clair D, Mann DM, Wavrant-DeVrieze F, Ezquerra-Trabalon M, Amouyel P, Hardy J, Haltia M, Valdivieso F, Goate AM, Perez-Tur J, Lendon CL, Chartier-Harlin MC. Contribution of APOE promoter polymorphisms to Alzheimer's disease risk. Neurology. 2002;59(1):59–66. doi: 10.1212/wnl.59.1.59. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009a;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009b doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Perez-Tur J, Dupire MJ, Galasko D, Mann D, Amouyel P, Hardy J, Delacourte A, Chartier-Harlin MC. Distortion of allelic expression of apolipoprotein E in Alzheimer's disease. Hum Mol Genet. 1997;6(12):2151–2154. doi: 10.1093/hmg/6.12.2151. [DOI] [PubMed] [Google Scholar]
- Lander ES. The new genomics: global views of biology. Science. 1996;274(5287):536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science. 1987;236(4808):1567–1570. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- Laudon H, Hansson EM, Melen K, Bergman A, Farmery MR, Winblad B, Lendahl U, von Heijne G, Naslund J. A nine-transmembrane domain topology for presenilin 1. J Biol Chem. 2005;280(42):35352–35360. doi: 10.1074/jbc.M507217200. [DOI] [PubMed] [Google Scholar]
- Launer LJ, Ross GW, Petrovitch H, Masaki K, Foley D, White LR, Havlik RJ. Midlife blood pressure and dementia: the Honolulu-Asia aging study. Neurobiol Aging. 2000;21(1):49–55. doi: 10.1016/s0197-4580(00)00096-8. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Graff-Radford N, Foroud T, Mayeux R. Analyses of the National Institute on Aging Late-Onset Alzheimer's Disease Family Study: implication of additional loci. Arch Neurol. 2008;65(11):1518–1526. doi: 10.1001/archneur.65.11.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Santana V, Williamson J, Lantigua R, Medrano M, Arriaga A, Stern Y, Tycko B, Rogaeva E, Wakutani Y, Kawarai T, St George-Hyslop P, Mayeux R. Expanded genomewide scan implicates a novel locus at 3q28 among Caribbean hispanics with familial Alzheimer disease. Arch Neurol. 2006;63(11):1591–1598. doi: 10.1001/archneur.63.11.1591. [DOI] [PubMed] [Google Scholar]
- Lee JH, Mayeux R, Mayo D, Mo J, Santana V, Williamson J, Flaquer A, Ciappa A, Rondon H, Estevez P, Lantigua R, Kawarai T, Toulina A, Medrano M, Torres M, Stern Y, Tycko B, Rogaeva E, St George-Hyslop P, Knowles JA. Fine mapping of 10q and 18q for familial Alzheimer's disease in Caribbean Hispanics. Mol Psychiatry. 2004;9(11):1042–1051. doi: 10.1038/sj.mp.4001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Lahad E, Poorkaj P, Wang K, Fu YH, Oshima J, Mulligan J, Schellenberg GD. Genomic structure and expression of STM2, the chromosome 1 familial Alzheimer disease gene. Genomics. 1996;34(2):198–204. doi: 10.1006/geno.1996.0266. [DOI] [PubMed] [Google Scholar]
- Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K, et al. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science. 1995;269(5226):973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- Li G, Shofer JB, Kukull WA, Peskind ER, Tsuang DW, Breitner JC, McCormick W, Bowen JD, Teri L, Schellenberg GD, Larson EB. Serum cholesterol and risk of Alzheimer disease: a community-based cohort study. Neurology. 2005;65(7):1045–1050. doi: 10.1212/01.wnl.0000178989.87072.11. [DOI] [PubMed] [Google Scholar]
- Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M, Coletta N, Delisle R, Dhalla D, Ehm MG, Feldman HH, Fornazzari L, Gauthier S, Goodgame N, Guzman D, Hammond S, Hollingworth P, Hsiung GY, Johnson J, Kelly DD, Keren R, Kertesz A, King KS, Lovestone S, Loy-English I, Matthews PM, Owen MJ, Plumpton M, Pryse-Phillips W, Prinjha RK, Richardson JC, Saunders A, Slater AJ, St George-Hyslop PH, Stinnett SW, Swartz JE, Taylor RL, Wherrett J, Williams J, Yarnall DP, Gibson RA, Irizarry MC, Middleton LT, Roses AD. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65(1):45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- Li LH, Ho SF, Chen CH, Wei CY, Wong WC, Li LY, Hung SI, Chung WH, Pan WH, Lee MT, Tsai FJ, Chang CF, Wu JY, Chen YT. Long contiguous stretches of homozygosity in the human genome. Hum Mutat. Y.T;27(11):1115–1121. doi: 10.1002/humu.20399. [DOI] [PubMed] [Google Scholar]
- Li YJ, Scott WK, Hedges DJ, Zhang F, Gaskell PC, Nance MA, Watts RL, Hubble JP, Koller WC, Pahwa R, Stern MB, Hiner BC, Jankovic J, Allen FA, Jr, Goetz CG, Mastaglia F, Stajich JM, Gibson RA, Middleton LT, Saunders AM, Scott BL, Small GW, Nicodemus KK, Reed AD, Schmechel DE, Welsh-Bohmer KA, Conneally PM, Roses AD, Gilbert JR, Vance JM, Haines JL, Pericak-Vance MA. Age at onset in two common neurodegenerative diseases is genetically controlled. Am J Hum Genet. 2002;70(4):985–993. doi: 10.1086/339815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Arias-Vasquez A, Sleegers K, Aulchenko YS, Kayser M, Sanchez-Juan P, Feng BJ, Bertoli-Avella AM, van Swieten J, Axenovich TI, Heutink P, van Broeckhoven C, Oostra BA, van Duijn CM. A genomewide screen for late-onset Alzheimer disease in a genetically isolated Dutch population. Am J Hum Genet. 2007;81(1):17–31. doi: 10.1086/518720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240(4852):622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- Mahley RW, Rall SC., Jr Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000:1507–1537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- Mani A, Meraji SM, Houshyar R, Radhakrishnan J, Mani A, Ahangar M, Rezaie TM, Taghavinejad MA, Broumand B, Zhao H, Nelson-Williams C, Lifton RP. Finding genetic contributions to sporadic disease: a recessive locus at 12q24 commonly contributes to patent ductus arteriosus. Proc Natl Acad Sci U S A. 2002;99(23):15054–15059. doi: 10.1073/pnas.192582999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara E, Soto C, Governale S, Frangione B, Ghiso J. Apolipoprotein J and Alzheimer's amyloid beta solubility. Biochem J. 1996;316(Pt 2):671–679. doi: 10.1042/bj3160671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May PC, Lampert-Etchells M, Johnson SA, Poirier J, Masters JN, Finch CE. Dynamics of gene expression for a hippocampal glycoprotein elevated in Alzheimer's disease and in response to experimental lesions in rat. Neuron. 1990;5(6):831–839. doi: 10.1016/0896-6273(90)90342-d. [DOI] [PubMed] [Google Scholar]
- McGeer EG, McGeer PL. Innate immunity in Alzheimer's disease: a model for local inflammatory reactions. Mol Interv. 2001;1(1):22–29. [PubMed] [Google Scholar]
- McGeer PL, Akiyama H, Itagaki S, McGeer EG. Activation of the classical complement pathway in brain tissue of Alzheimer patients. Neurosci Lett. 1989;107(1–3):341–346. doi: 10.1016/0304-3940(89)90843-4. [DOI] [PubMed] [Google Scholar]
- McGeer PL, Kawamata T, Walker DG. Distribution of clusterin in Alzheimer brain tissue. Brain Res. 1992;579(2):337–341. doi: 10.1016/0006-8993(92)90071-g. [DOI] [PubMed] [Google Scholar]
- McGeer PL, Klegeris A, Walker DG, Yasuhara O, McGeer EG. Pathological proteins in senile plaques. Tohoku J Exp Med. 1994;174(3):269–277. doi: 10.1620/tjem.174.269. [DOI] [PubMed] [Google Scholar]
- McGeer PL, McGeer EG. The possible role of complement activation in Alzheimer disease. Trends Mol Med. 2002;8(11):519–523. doi: 10.1016/s1471-4914(02)02422-x. [DOI] [PubMed] [Google Scholar]
- Michel D, Chatelain G, North S, Brun G. Stress-induced transcription of the clusterin/apoJ gene. Biochem J. 1997;328(Pt 1):45–50. doi: 10.1042/bj3280045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke MM, Zandi PP, Sjogren M, Gustafson D, Ostling S, Steen B, Skoog I. High total cholesterol levels in late life associated with a reduced risk of dementia. Neurology. 2005;64(10):1689–1695. doi: 10.1212/01.WNL.0000161870.78572.A5. [DOI] [PubMed] [Google Scholar]
- Myers A, Wavrant De-Vrieze F, Holmans P, Hamshere M, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Knowles H, Morris JC, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, Rice F, Giles P, Tunstall N, Jones L, Lovestone S, Williams J, Owen MJ, Hardy J, Goate A. Full genome screen for Alzheimer disease: stage II analysis. Am J Med Genet. 2002;114(2):235–244. doi: 10.1002/ajmg.10183. [DOI] [PubMed] [Google Scholar]
- Nalls MA, Guerreiro RJ, Simon-Sanchez J, Bras JT, Traynor BJ, Gibbs JR, Launer L, Hardy J, Singleton AB. Extended tracts of homozygosity identify novel candidate genes associated with late-onset Alzheimer's disease. Neurogenetics. 2009 doi: 10.1007/s10048-009-0182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notkola IL, Sulkava R, Pekkanen J, Erkinjuntti T, Ehnholm C, Kivinen P, Tuomilehto J, Nissinen A. Serum total cholesterol, apolipoprotein E epsilon 4 allele, and Alzheimer's disease. Neuroepidemiology. 1998;17(1):14–20. doi: 10.1159/000026149. [DOI] [PubMed] [Google Scholar]
- Nuutinen T, Suuronen T, Kauppinen A, Salminen A. Clusterin: A forgotten player in Alzheimer's disease. Brain Res Rev. 2009 doi: 10.1016/j.brainresrev.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Oda T, Pasinetti GM, Osterburg HH, Anderson C, Johnson SA, Finch CE. Purification and characterization of brain clusterin. Biochem Biophys Res Commun. 1994;204(3):1131–1136. doi: 10.1006/bbrc.1994.2580. [DOI] [PubMed] [Google Scholar]
- Oda T, Wals P, Osterburg HH, Johnson SA, Pasinetti GM, Morgan TE, Rozovsky I, Stine WB, Snyder SW, Holzman TF, et al. Clusterin (apoJ) alters the aggregation of amyloid beta-peptide (A beta 1–42) and forms slowly sedimenting A beta complexes that cause oxidative stress. Exp Neurol. 1995;136(1):22–31. doi: 10.1006/exnr.1995.1080. [DOI] [PubMed] [Google Scholar]
- Olson JM, Goddard KA, Dudek DM. A second locus for very-late-onset Alzheimer disease: a genome scan reveals linkage to 20p and epistasis between 20p and the amyloid precursor protein region. Am J Hum Genet. 2002;71(1):154–161. doi: 10.1086/341034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Bhatia KP, Li A, Hernandez D, Davis M, Wood NW, Hardy J, Houlden H, Singleton A, Schneider SA. Characterization of PLA2G6 as a locus for dystonia-parkinsonism. Ann Neurol. 2009;65(1):19–23. doi: 10.1002/ana.21415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangalos MN, Jacobsen SJ, Reinhart PH. Disease modifying strategies for the treatment of Alzheimer's disease targeted at modulating levels of the beta-amyloid peptide. Biochem Soc Trans. 2005;33(Pt 4):553–558. doi: 10.1042/BST0330553. [DOI] [PubMed] [Google Scholar]
- Pedersen NL, Posner SF, Gatz M. Multiple-threshold models for genetic influences on age of onset for Alzheimer disease: findings in Swedish twins. Am J Med Genet. 2001;105(8):724–728. doi: 10.1002/ajmg.1608. [DOI] [PubMed] [Google Scholar]
- Pericak-Vance MA, Bass MP, Yamaoka LH, Gaskell PC, Scott WK, Terwedow HA, Menold MM, Conneally PM, Small GW, Vance JM, Saunders AM, Roses AD, Haines JL. Complete genomic screen in late-onset familial Alzheimer disease. Evidence for a new locus on chromosome 12. Jama. 1997;278(15):1237–1241. [PubMed] [Google Scholar]
- Pericak-Vance MA, Grubber J, Bailey LR, Hedges D, West S, Santoro L, Kemmerer B, Hall JL, Saunders AM, Roses AD, Small GW, Scott WK, Conneally PM, Vance JM, Haines JL. Identification of novel genes in late-onset Alzheimer's disease. Exp Gerontol. 2000;35(9–10):1343–1352. doi: 10.1016/s0531-5565(00)00196-0. [DOI] [PubMed] [Google Scholar]
- Podewils LJ, Guallar E, Kuller LH, Fried LP, Lopez OL, Carlson M, Lyketsos CG. Physical activity, APOE genotype, and dementia risk: findings from the Cardiovascular Health Cognition Study. Am J Epidemiol. 2005;161(7):639–651. doi: 10.1093/aje/kwi092. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276(5321):2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Qiu C, Winblad B, Fratiglioni L. The age-dependent relation of blood pressure to cognitive function and dementia. Lancet Neurol. 2005;4(8):487–499. doi: 10.1016/S1474-4422(05)70141-1. [DOI] [PubMed] [Google Scholar]
- Rademakers R, Cruts M, Van Broeckhoven C. Genetics of early-onset Alzheimer dementia. ScientificWorldJournal. 2003:3497–3519. doi: 10.1100/tsw.2003.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan M, Lillington DM, Skoulakis S, Debernardi S, Chaplin T, Foot NJ, Lister TA, Young BD. Genome-wide single nucleotide polymorphism analysis reveals frequent partial uniparental disomy due to somatic recombination in acute myeloid leukemias. Cancer Res. 2005;65(2):375–378. [PubMed] [Google Scholar]
- Raux G, Guyant-Marechal L, Martin C, Bou J, Penet C, Brice A, Hannequin D, Frebourg T, Campion D. Molecular diagnosis of autosomal dominant early onset Alzheimer's disease: an update. J Med Genet. 2005;42(10):793–795. doi: 10.1136/jmg.2005.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Huentelman MJ, Craig DW, Coon KD, Liang WS, Herbert RH, Beach T, Rohrer KC, Zhao AS, Leung D, Bryden L, Marlowe L, Kaleem M, Mastroeni D, Grover A, Heward CB, Ravid R, Rogers J, Hutton ML, Melquist S, Petersen RC, Alexander GE, Caselli RJ, Kukull W, Papassotiropoulos A, Stephan DA. GAB2 alleles modify Alzheimer's risk in APOE epsilon4 carriers. Neuron. 2007;54(5):713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz C, Tang MX, Luchsinger J, Mayeux R. Relation of plasma lipids to Alzheimer disease and vascular dementia. Arch Neurol. 2004;61(5):705–714. doi: 10.1001/archneur.61.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remes AM, Finnila S, Mononen H, Tuominen H, Takalo R, Herva R, Majamaa K. Hereditary dementia with intracerebral hemorrhages and cerebral amyloid angiopathy. Neurology. 2004;63(2):234–240. doi: 10.1212/01.wnl.0000129988.68657.fa. [DOI] [PubMed] [Google Scholar]
- Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T, et al. Familial Alzheimer's disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature. 1995;376(6543):775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- Rogaeva E. The solved and unsolved mysteries of the genetics of early-onset Alzheimer's disease. Neuromolecular Med. 2002;2(1):1–10. doi: 10.1385/NMM:2:1:01. [DOI] [PubMed] [Google Scholar]
- Rogers J, Cooper NR, Webster S, Schultz J, McGeer PL, Styren SD, Civin WH, Brachova L, Bradt B, Ward P, et al. Complement activation by beta-amyloid in Alzheimer disease. Proc Natl Acad Sci U S A. 1992;89(21):10016–10020. doi: 10.1073/pnas.89.21.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovelet-Lecrux A, Frebourg T, Tuominen H, Majamaa K, Campion D, Remes AM. APP locus duplication in a Finnish family with dementia and intracerebral haemorrhage. J Neurol Neurosurg Psychiatry. 2007;78(10):1158–1159. doi: 10.1136/jnnp.2006.113514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerriere A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, Dubas F, Frebourg T, Campion D. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38(1):24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- Sanan DA, Weisgraber KH, Russell SJ, Mahley RW, Huang D, Saunders A, Schmechel D, Wisniewski T, Frangione B, Roses AD, et al. Apolipoprotein E associates with beta amyloid peptide of Alzheimer's disease to form novel monofibrils. Isoform apoE4 associates more efficiently than apoE3. J Clin Invest. 1994;94(2):860–869. doi: 10.1172/JCI117407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed-Tabatabaei FA, Houwing-Duistermaat JJ, van Duijn CM, Witteman JC. Angiotensin-converting enzyme gene polymorphism and carotid artery wall thickness: a meta-analysis. Stroke. 2003;34(7):1634–1639. doi: 10.1161/01.STR.0000077926.49330.64. [DOI] [PubMed] [Google Scholar]
- Scott WK, Hauser ER, Schmechel DE, Welsh-Bohmer KA, Small GW, Roses AD, Saunders AM, Gilbert JR, Vance JM, Haines JL, Pericak-Vance MA. Ordered-subsets linkage analysis detects novel Alzheimer disease loci on chromosomes 2q34 and 15q22. Am J Hum Genet. 2003;73(5):1041–1051. doi: 10.1086/379083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375(6534):754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Shoji M, Golde TE, Ghiso J, Cheung TT, Estus S, Shaffer LM, Cai XD, McKay DM, Tintner R, Frangione B, et al. Production of the Alzheimer amyloid beta protein by normal proteolytic processing. Science. 1992;258(5079):126–129. doi: 10.1126/science.1439760. [DOI] [PubMed] [Google Scholar]
- Siest G, Pillot T, Regis-Bailly A, Leininger-Muller B, Steinmetz J, Galteau MM, Visvikis S. Apolipoprotein E: an important gene and protein to follow in laboratory medicine. Clin Chem. 1995;41(8 Pt 1):1068–1086. [PubMed] [Google Scholar]
- Sillen A, Andrade J, Lilius L, Forsell C, Axelman K, Odeberg J, Winblad B, Graff C. Expanded high-resolution genetic study of 109 Swedish families with Alzheimer's disease. Eur J Hum Genet. 2008;16(2):202–208. doi: 10.1038/sj.ejhg.5201946. [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J, Scholz S, Fung HC, Matarin M, Hernandez D, Gibbs JR, Britton A, de Vrieze FW, Peckham E, Gwinn-Hardy K, Crawley A, Keen JC, Nash J, Borgaonkar D, Hardy J, Singleton A. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16(1):1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, Kruger R, Federoff M, Klein C, Goate A, Perlmutter J, Bonin M, Nalls MA, Illig T, Gieger C, Houlden H, Steffens M, Okun MS, Racette BA, Cookson MR, Foote KD, Fernandez HH, Traynor BJ, Schreiber S, Arepalli S, Zonozi R, Gwinn K, van der Brug M, Lopez G, Chanock SJ, Schatzkin A, Park Y, Hollenbeck A, Gao J, Huang X, Wood NW, Lorenz D, Deuschl G, Chen H, Riess O, Hardy JA, Singleton AB, Gasser T. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet. 2009;41(12):1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Sanchez J, Singleton A. Genome-wide association studies in neurological disorders. Lancet Neurol. 2008;7(11):1067–1072. doi: 10.1016/S1474-4422(08)70241-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoberg AP, Trouw LA, Blom AM. Complement activation and inhibition: a delicate balance. Trends Immunol. 2009;30(2):83–90. doi: 10.1016/j.it.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Skoog I, Lernfelt B, Landahl S, Palmertz B, Andreasson LA, Nilsson L, Persson G, Oden A, Svanborg A. 15-year longitudinal study of blood pressure and dementia. Lancet. 1996;347(9009):1141–1145. doi: 10.1016/s0140-6736(96)90608-x. [DOI] [PubMed] [Google Scholar]
- Sleegers K, Brouwers N, Gijselinck I, Theuns J, Goossens D, Wauters J, Del-Favero J, Cruts M, van Duijn CM, Van Broeckhoven C. APP duplication is sufficient to cause early onset Alzheimer's dementia with cerebral amyloid angiopathy. Brain. 2006;129(Pt 11):2977–2983. doi: 10.1093/brain/awl203. [DOI] [PubMed] [Google Scholar]
- Sparks DL, Scheff SW, Hunsaker JC, 3rd, Liu H, Landers T, Gross DR. Induction of Alzheimer-like beta-amyloid immunoreactivity in the brains of rabbits with dietary cholesterol. Exp Neurol. 1994;126(1):88–94. doi: 10.1006/exnr.1994.1044. [DOI] [PubMed] [Google Scholar]
- Staessen JA, Wang JG, Ginocchio G, Petrov V, Saavedra AP, Soubrier F, Vlietinck R, Fagard R. The deletion/insertion polymorphism of the angiotensin converting enzyme gene and cardiovascular-renal risk. J Hypertens. 1997;15(12 Pt 2):1579–1592. doi: 10.1097/00004872-199715120-00059. [DOI] [PubMed] [Google Scholar]
- Stewart R, Masaki K, Xue QL, Peila R, Petrovitch H, White LR, Launer LJ. A 32-year prospective study of change in body weight and incident dementia: the Honolulu-Asia Aging Study. Arch Neurol. 2005;62(1):55–60. doi: 10.1001/archneur.62.1.55. [DOI] [PubMed] [Google Scholar]
- Stewart R, White LR, Xue QL, Launer LJ. Twenty-six-year change in total cholesterol levels and incident dementia: the Honolulu-Asia Aging Study. Arch Neurol. 2007;64(1):103–107. doi: 10.1001/archneur.64.1.103. [DOI] [PubMed] [Google Scholar]
- Stewart R, Xue QL, Masaki K, Petrovitch H, Ross GW, White LR, Launer LJ. Change in blood pressure and incident dementia: a 32-year prospective study. Hypertension. 2009;54(2):233–240. doi: 10.1161/HYPERTENSIONAHA.109.128744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazzullo P, Iacone R, Iacoviello L, Russo O, Barba G, Russo P, D'Orazio A, Barbato A, Cappuccio FP, Farinaro E, Siani A. Genetic variation in the renin-angiotensin system and abdominal adiposity in men: the Olivetti Prospective Heart Study. Ann Intern Med. 2003;138(1):17–23. doi: 10.7326/0003-4819-138-1-200301070-00007. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Weisgraber KH, Goedert M, Saunders AM, Huang D, Corder EH, Dong LM, Jakes R, Alberts MJ, Gilbert JR, et al. Hypothesis: microtubule instability and paired helical filament formation in the Alzheimer disease brain are related to apolipoprotein E genotype. Exp Neurol. 1994;125(2):163–171. doi: 10.1006/exnr.1994.1019. discussion 172-164. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Weisgraber KH, Huang DY, Dong LM, Salvesen GS, Pericak-Vance M, Schmechel D, Saunders AM, Goldgaber D, Roses AD. Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90(17):8098–8102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmeyer R, Shen Y, Rogers J. Detection of complement alternative pathway mRNA and proteins in the Alzheimer's disease brain. Brain Res Mol Brain Res. 2000;81(1–2):7–18. doi: 10.1016/s0169-328x(00)00149-2. [DOI] [PubMed] [Google Scholar]
- Takamaru Y. [Analysis of the 80 kDa antigen in senile plaque amyloid] Hokkaido Igaku Zasshi. 1994;69(2):191–201. [PubMed] [Google Scholar]
- Tan ZS, Seshadri S, Beiser A, Wilson PW, Kiel DP, Tocco M, D'Agostino RB, Wolf PA. Plasma total cholesterol level as a risk factor for Alzheimer disease: the Framingham Study. Arch Intern Med. 2003;163(9):1053–1057. doi: 10.1001/archinte.163.9.1053. [DOI] [PubMed] [Google Scholar]
- Tenner AJ. Complement in Alzheimer's disease: opportunities for modulating protective and pathogenic events. Neurobiol Aging. 2001;22(6):849–861. doi: 10.1016/s0197-4580(01)00301-3. [DOI] [PubMed] [Google Scholar]
- Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. 2008;17(R2):R143–R150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomiyama T, Nagata T, Shimada H, Teraoka R, Fukushima A, Kanemitsu H, Takuma H, Kuwano R, Imagawa M, Ataka S, Wada Y, Yoshioka E, Nishizaki T, Watanabe Y, Mori H. A new amyloid beta variant favoring oligomerization in Alzheimer's-type dementia. Ann Neurol. 2008;63(3):377–387. doi: 10.1002/ana.21321. [DOI] [PubMed] [Google Scholar]
- Veerhuis R, Janssen I, Hack CE, Eikelenboom P. Early complement components in Alzheimer's disease brains. Acta Neuropathol. 1996;91(1):53–60. doi: 10.1007/s004019570001. [DOI] [PubMed] [Google Scholar]
- Walker DG, McGeer PL. Complement gene expression in human brain: comparison between normal and Alzheimer disease cases. Brain Res Mol Brain Res. 1992;14(1–2):109–116. doi: 10.1016/0169-328x(92)90017-6. [DOI] [PubMed] [Google Scholar]
- Webster J, Myers A, Reiman EM, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Huentelman MJ, Craig DW, Coon KD, Beach T, Roher KC, Zhao AS, Leung D, Bryden L, Marlowe L, Kaleem M, Mastroeni D, Grover A, Rogers J, Heun R, Jessen F, Kölsch H, Heward CB, Ravid R, Hutton ML, Melquist S, Petersen RC, Caselli RJ, Papassotiropoulos A, Stephan DA, J, H Whole genome association analysis shows that ACE is a risk factor for Alzheimer’s disease and fails to replicate most candidates from Meta-analysis. Int J Mol Epidemiol Genet. 2009 (in press) [PMC free article] [PubMed] [Google Scholar]
- Webster S, Glabe C, Rogers J. Multivalent binding of complement protein C1Q to the amyloid beta-peptide (A beta) promotes the nucleation phase of A beta aggregation. Biochem Biophys Res Commun. 1995;217(3):869–875. doi: 10.1006/bbrc.1995.2852. [DOI] [PubMed] [Google Scholar]
- Webster S, Lue LF, Brachova L, Tenner AJ, McGeer PL, Terai K, Walker DG, Bradt B, Cooper NR, Rogers J. Molecular and cellular characterization of the membrane attack complex, C5b-9, in Alzheimer's disease. Neurobiol Aging. 1997;18(4):415–421. doi: 10.1016/s0197-4580(97)00042-0. [DOI] [PubMed] [Google Scholar]
- Weisgraber KH. Apolipoprotein E: structure-function relationships. Adv Protein Chem. 1994:45249–45302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- Whitmer RA, Gunderson EP, Quesenberry CP, Jr, Zhou J, Yaffe K. Body mass index in midlife and risk of Alzheimer disease and vascular dementia. Curr Alzheimer Res. 2007;4(2):103–109. doi: 10.2174/156720507780362047. [DOI] [PubMed] [Google Scholar]
- Whitmer RA, Gustafson DR, Barrett-Connor E, Haan MN, Gunderson EP, Yaffe K. Central obesity and increased risk of dementia more than three decades later. Neurology. 2008;71(14):1057–1064. doi: 10.1212/01.wnl.0000306313.89165.ef. [DOI] [PubMed] [Google Scholar]
- Whitmer RA, Sidney S, Selby J, Johnston SC, Yaffe K. Midlife cardiovascular risk factors and risk of dementia in late life. Neurology. 2005;64(2):277–281. doi: 10.1212/01.WNL.0000149519.47454.F2. [DOI] [PubMed] [Google Scholar]
- Wilson MR, Easterbrook-Smith SB. Clusterin is a secreted mammalian chaperone. Trends Biochem Sci. 2000;25(3):95–98. doi: 10.1016/s0968-0004(99)01534-0. [DOI] [PubMed] [Google Scholar]
- Wong P, Pineault J, Lakins J, Taillefer D, Leger J, Wang C, Tenniswood M. Genomic organization and expression of the rat TRPM-2 (clusterin) gene, a gene implicated in apoptosis. J Biol Chem. 1993;268(7):5021–5031. [PubMed] [Google Scholar]
- Woods CG, Valente EM, Bond J, Roberts E. A new method for autozygosity mapping using single nucleotide polymorphisms (SNPs) and EXCLUDEAR. J Med Genet. 2004;41(8):e101. doi: 10.1136/jmg.2003.016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikai S, Sasaki H, Doh-ura K, Furuya H, Sakaki Y. Genomic organization of the human amyloid beta-protein precursor gene. Gene. 1990;87(2):257–263. doi: 10.1016/0378-1119(90)90310-n. [DOI] [PubMed] [Google Scholar]
- Yoshitake T, Kiyohara Y, Kato I, Ohmura T, Iwamoto H, Nakayama K, Ohmori S, Nomiyama K, Kawano H, Ueda K, et al. Incidence and risk factors of vascular dementia and Alzheimer's disease in a defined elderly Japanese population: the Hisayama Study. Neurology. 1995;45(6):1161–1168. doi: 10.1212/wnl.45.6.1161. [DOI] [PubMed] [Google Scholar]
- Zannis VI, Just PW, Breslow JL. Human apolipoprotein E isoprotein subclasses are genetically determined. Am J Hum Genet. 1981;33(1):11–24. [PMC free article] [PubMed] [Google Scholar]
- Zipfel PF. Complement and Immune Defense: From Innate Immunity to Human Diseases. Immunol Lett. 2009 doi: 10.1016/j.imlet.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Zlokovic BV. Cerebrovascular transport of Alzheimer's amyloid beta and apolipoproteins J and E: possible anti-amyloidogenic role of the blood-brain barrier. Life Sci. 1996;59(18):1483–1497. doi: 10.1016/0024-3205(96)00310-4. [DOI] [PubMed] [Google Scholar]