Abstract
GenMapDB (http://genomics.med.upenn.edu/genmapdb) is a repository of human bacterial artificial chromosome (BAC) clones mapped by our laboratory to sequence-tagged site markers. Currently, GenMapDB contains over 3000 mapped clones that span 19 chromosomes, chromosomes 2, 4, 5, 9–22, X and Y. This database provides positional information about human BAC clones from the RPCI-11 human male BAC library. It also contains restriction fragment analysis data and end sequences of the clones. GenMapDB is freely available to the public. The main purpose of GenMapDB is to organize the mapping data and to allow the research community to search for mapped BAC clones that can be used in gene mapping studies and chromosomal mutation analysis projects.
INTRODUCTION
Physical maps are essential in a large number of projects from gene mapping to sequencing. Our laboratory is mapping and characterizing a set of bacterial artificial chromosome (BAC) clones that covers the human genome at about 1-Mb intervals (1). GenMapDB is a relational database designed to manage the data from this mapping project. Two main objectives of GenMapDB are (i) to integrate our experimental data, and (ii) to allow public access to the data we generate. Unlike other resources, our group mapped and characterized all the clones and all the entries are individually inspected.
The data in GenMapDB are organized by chromosome. For each chromosome, we list the sequence-tagged site (STS) markers that are used for mapping and their corresponding BAC clones. There is additional information on the clones such as restriction digest patterns, BAC end sequences (DNA sequences of insert ends) and cytogenetic locations. There are also links to other genome databases such as the radiation hybrid database [RHdb (2); http://www.ebi.ac.uk/RHdb] and GeneMap ’99 [(3); http://www.ncbi.nlm.nih.gov/genemap99] to integrate our data with those in other genome databases.
GenMapDB allows users to obtain information on a set of evenly distributed BAC clones that span the human genome at about 1-Mb intervals. It is also designed for individuals who are interested in BAC clones mapping to specific genomic regions. For example, from GenMapDB, researchers constructing a physical map of a candidate gene region can identify clones that map to their region of interest. Similarly, scientists who are studying tumor cells for genomic rearrangements can obtain clones mapping to regions that correspond to the rearrangements observed in the tumor samples.
DATABASE STRUCTURE
GenMapDB is a relational database implemented using Access (Microsoft Inc., Seattle, WA). The server side of GenMapDB consists of the Apache web server, a collection of Perl CGI scripts and the Perl DBI module for database connectivity.
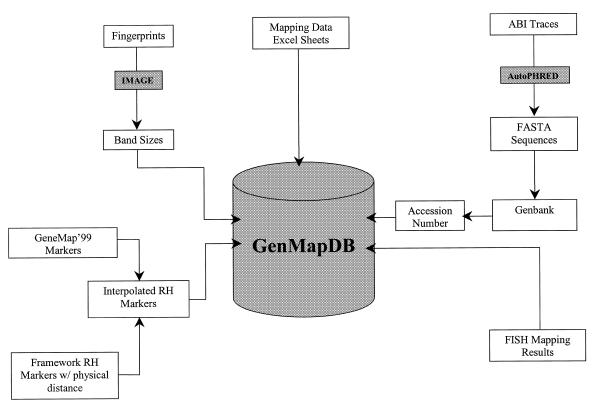
The architecture of the database allows importing of data from different sources using Perl scripts (Fig. 1). Some data such as the mapping information are entered using HTML forms and CGI scripts. Other data, such as fingerprints and BAC end sequences, are generated by image analysis software. In this case, we obtain the data by parsing the text files of the output of the programs.
Figure 1.
A flowchart of GenMapDB structure.
MAPPED HUMAN BAC CLONES
GenMapDB collects and displays human BAC clones mapped by our laboratory. The core of the database is the genomic positions of the BAC clones in relation to STS markers on the GeneBridge4 (GB4) radiation hybrid (RH) map (4). A set of STS markers selected from the GB4 RH map by Greg Schuler at NCBI, is used as anchors for our BAC map. The markers are then used to make radioactively labeled probes for hybridization onto RPCI-11 human male BAC library high-density filters (5). The BAC clones that are identified to contain the STS markers are pulled from the library and grown as single-colony cultures. We then use the single-colony cultures to verify the STS content of the BAC clones by a second hybridization step and by PCR (1). Once verified, the BAC clones corresponding to the STS markers are entered into GenMapDB.
Additional molecular characterizations are performed on the clones. These include HindIII fingerprints and BAC end sequences. To integrate our STS-based map with cytogenetic maps, BAC clones are FISH mapped and the resulting cytogenetic addresses are provided in GenMapDB.
CONTENT
The major classes of data in GenMapDB are mapped clones, HindIII fingerprints, BAC end sequences and cytogenetic locations.
Mapped clones
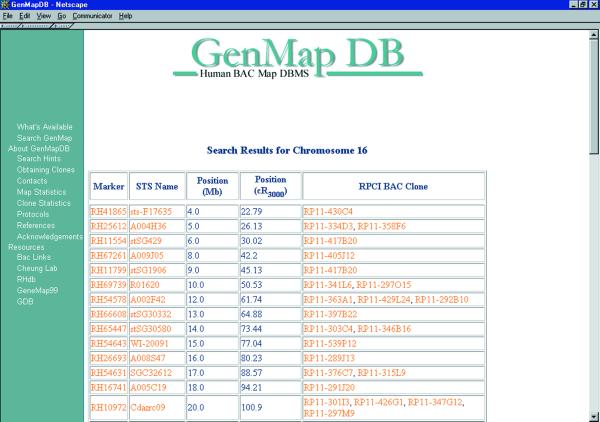
The display of the clones is arranged by chromosome in order of their genomic locations. Each clone entry includes physical location of the clone in cRays (cR3000) and megabases (Mb) from the telomere of the p-arm of the chromosome (pter), and its STS content (Fig. 2). Physical distances in Mb were obtained based on interpolations from distances of framework markers in the RH map. The accuracy of the assignments of map location relies on the accuracy of the RH map and likely varies at different sites along the genome. A preliminary estimate based on results from FISH analyses of 120 mapped clones on chromosomes 14 and 16 show that seven clones (four on chromosome 14, three on chromosome 16) did not map to the chromosomes assigned by RH mapping. For clones that mapped to the same chromosomes on the RH and cytogenetic maps, four clones (all on chromosome 14) mapped to a different location on the chromosome as predicted by the RH map.
Figure 2.
An example of the main output page. Shown here is the output from a search for clones mapping to chromosome 16; the data include STS markers, their physical addresses and BAC clones corresponding to each STS marker.
We provide two names for each STS marker: an RH identification number and a STS name according to GeneMap ’99. These names serve as hyperlinks to RHdb and GeneMap ’99. Each clone name is also internally hyperlinked to a clone page that provides molecular information about the clone (see section below).
HindIII fingerprints
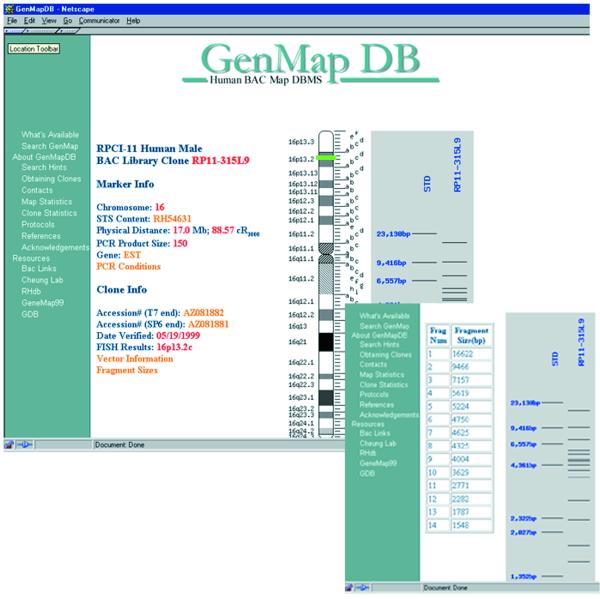
We perform HindIII restriction digests on the clones to provide a unique molecular identification for each clone. The HindIII fingerprints are analyzed using the freeware IMAGE from the Sanger Centre (6). The fragment sizes are presented in tabular form and graphically as reconstructions of their electrophoresis migration patterns (Fig. 3).
Figure 3.
Examples of clone page. Shown here is information on the BAC clone RP11-315L9 from chromosome 16. Included in the page is information on the chromosomal location of the clone, its physical and cytogenetic addresses, its HindIII restriction digest pattern, fragment sizes from the HindIII digest (inset) and the GenBank accession numbers for its end sequences.
BAC end sequences
We sequence the insert ends of the clones to generate information that can be used to identify overlapping clones and to align our map to the human sequence map.
The BAC end sequences are obtained by cycle sequencing using Big-Dye terminator chemistry (PE Biosystems, CA). We pass our sequences through the base-calling program PHRED (7,8) using AutoPHRED, a Perl script. Sequences where every base has a PHRED score > 20, which indicates that there is less than a 1 in 100 chance that the base call is incorrect, are submitted to GenBank. Hyperlinks are available to GenBank to allow users to access the sequence data (Fig. 3).
Cytogenetic results
The clones are FISH mapped in order to validate their ordering and chromosomal assignments as mentioned above. The FISH analyses are performed by the NCI Cancer Chromosome Aberration Project, Cancer Genome Anatomy Project [(9); http://www.ncbi.nlm.nih.gov/CCAP]. The results are available on the CCAP web page and in GenMapDB. We present the FISH data as cytogenetic bands in text and pictorially on idiograms (Fig. 3).
EXTERNAL LINKS
We provide hyperlinks to RHdb, GeneMap ’99 and Unigene to provide additional information about our clones and to integrate our clone map with other genomic maps. This allows users to get additional information on the STS markers that are used as anchors in our map. The links provide primer sequences, PCR conditions for generating the STS amplicons and quality measures of marker positions expressed in LOD scores by the RH consortium. Over 80% of the STS markers are coding sequences for expressed sequence tags or known genes. The link to Unigene is our first step towards annotating our clone map. Integration with databases that contain sequence and mapping information is important since our goal is to provide a set of BAC clones to be used as evenly distributed landmarks along the human genome.
DATABASE ACCESS
GenMapDB is freely available to all users. It can be searched by chromosome or by a specific genomic region. Data in GenMapDB are updated monthly. In order to allow users to search through the database quickly, we have a dedicated 450 MHz Pentium II computer with 128 Mb RAM and 6.4 GB storage space to hold the data.
FUTURE DIRECTIONS
We plan to improve GenMapDB in several directions—integration, annotation and speed. GenMapDB currently contains clones that cover chromosomes 2, 4, 5, 9–22, X and Y. By early 2001, we will have clones covering all the chromosomes in the human genome. With the finishing of the human genome sequence, our main goal is to integrate our map with data in other public resources such as the TIGR BAC end sequence database [(10) www.tigr.org/tdb/humgen/bac_end_search/bac_end_intro.html] and the Washington University BAC fingerprint database [(11) http://genome.wustl.edu/gsc/human/human_database.shtml]. There are also other groups that are establishing resources of mapped BAC clones. We will integrate our map with clones in other mapped clone resources such as the NCI CCAP (http://www.ncbi.nlm.nih.gov/CCAP/bac.cgi), the Roswell Park Cancer Institute BAC resource (http://bacpac.med.buffalo.edu/human/overview.html) and the resource at the University of Washington (http://www.biotech.washington.edu/bacresource/index.shtml). The different resources named here contain large collections of clones that are characterized by HindIII fingerprints, end sequences or cytogenetic addresses. Our collection differs by offering a set of clones that are evenly spaced at 1-Mb intervals. We characterize each of our mapped clones and provide the fingerprint, end sequence and cytogenetic addresses on them. All the clone resources are complementary. GenMapDB will provide links to other resources so that users can obtain different information depending on their research needs.
To bring biological meaning to our clone set, we will annotate our BAC end sequences by defining their sequence features and aligning them against the human genome sequence. We will use gene prediction and alignment software such as GeneScan, BLAST and FASTA.
In addition, we will upgrade our hardware to improve speed and to allow additional storage space. We should be able to move our data to a new database server such as Oracle or to a UNIX platform without significant modifications to the current Perl scripts.
Acknowledgments
ACKNOWLEDGEMENTS
The work was supported by grants from the Merck Genome Research Institute (V.G.C.), National Institutes of Health grants DC00154 and HG01880 (V.G.C.).
References
- 1.Cheung V.G., Dalrymple,H.L., Narasimhan,S.,Watts,J., Schuler,G., Raap,A.K., Morley,M. and Bruzel,A. (1999) A resource of mapped human bacterial artificial chromosome clones. Genome Res., 9, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez-Tomé P. and Lijnzaad,P. (2000) RHdb: the Radiation Hybrid database. Nucleic Acids Res., 28, 146–147. Updated article in this issue: Nucleic Acids Res. (2001), 29, 165–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deloukas P., Schuler,G.D., Gyapay,G., Beasley,E.M., Soderlund,C., Rodriguez-Tomé,P., Hui,L., Matise,T.C., MuKusick,K.B., Beckmann,J.S. et al. (1998) A physical map of 30 000 human genes. Science, 282, 744–746. [DOI] [PubMed] [Google Scholar]
- 4.Gyapay G., Schmitt,K., Fizames,C., Jones,H., Vega-Czarny,N., Spillett,D., Muselet,D., Prud’Homme,J.F., Dib,C., Auffray,C., Morissette,J., Weissenbach,J. and Goodfellow,P.N. (1996) A radiation hybrid map of the human genome. Hum. Mol. Genet., 5, 339–346. [DOI] [PubMed] [Google Scholar]
- 5.Osoegawa K., Woon,P.Y., Zhao,B., Frengen,E., Tateno,M., Catanese,J.J. and de Jong,P.J. (1998) An improved approach for construction of bacterial artificial chromosome libraries. Genomics, 52, 1–8. [DOI] [PubMed] [Google Scholar]
- 6.Sulston J., Mallet,F., Durbin,R. and Horsnell,T. (1989) Image analysis of restriction enzyme fingerprint autoradiograms. Comput. Applic. Biosci., 5, 101–106. [DOI] [PubMed] [Google Scholar]
- 7.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 8.Ewing B., Hillier,L., Wendl,M.C. and Green,P. (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res., 8, 175–185. [DOI] [PubMed] [Google Scholar]
- 9.Kirsch I.R., Green,E.D., Yonescu,R., Strausberg,R., Carter,N., Bentley,D., Leversha,M.A., Dunham,I., Braden,V.V., Hilgenfeld,E. et al. (2000) A systematic, high-resolution linkage of the cytogenetic and physical maps of the human genome. Nature Genet., 24, 339–340. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Z. (2000) Human BAC ends. Nucleic Acids Res., 28, 129–132. Updated article in this issue: Nucleic Acids Res. (2001), 29, 141–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marra M.A., Kucaba,T.A., Dietrich,N.L., Green,E.D., Brownstein,B., Wilson,R.K., McDonald,K.M., Hillier,L.W., McPherson,J.D. and Waterston,R.H. (1997) High throughput fingerprint analysis of large-insert clones. Genome Res., 7, 1072–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]