Abstract
Abba is a member of the I-BAR-domain protein family that is characterized by a convex-shaped membrane-binding motif. Overexpression of GFP-tagged Abba in murine fibroblasts potentiated PDGF-mediated formation of membrane ruffles and lamellipodia. Immunofluorescent microscopy and pull-down analysis revealed that GFP-Abba colocalized with an active form of Rac1 in the membrane ruffles and enhanced the Rac GTPase activity in response to PDGF stimulation. Further immunoprecpitation assays demonstrated that GFP-Abba bound to both wild-type and constitutively active Rac1 and that the binding to either of the Rac1 forms was significantly enhanced upon PDGF stimulation. On the other hand, an Abba mutant deficient in Rac1 binding failed to promote membrane ruffling and Rac1 activation in response to PDGF. However, the cells overexpressing a truncated mutant carrying the I-BAR domain alone displayed numerous filopodia-like microspikes in a manner independent of growth factors. Also, the Rac binding activity of the mutant was not affected by PDGF treatment. Our data indicates that the interaction between full-length Abba and Rac1 is implicated in membrane deformation and subjected to a growth factor-mediated regulation through the C-terminal sequence.
Introduction
In response to extracellular signals the plasma membrane undergoes dynamic changes such as protrusion, invagination and tubulation during cell migration, endocytosis and organelle trafficking. While the detailed mechanism for the membrane curvature upon extracellular stimuli remains elusive, it is often sensed or initiated by BAR-domain proteins, which bend the plasma membrane and interact either directly or indirectly with the actin cytoskeleton [1]. All the members of the BAR-domain family contain a helix-rich folder that dimerizes into a rigid module, or BAR-domain, for lipid binding through a positively charged surface, and thereby drives the membrane into a specific curvature. This module was initially recognized as a conserved motif shared by the metazoan Bin/amphiphysin and Rvs161/Rvs167 proteins [1;2]. Crystal structural and sequence analyses have revealed that the BAR-domain superfamily can be divided into several subfamilies: classical BARs, F-BARs and inverse BARs (I-BARs) [3–5]. While most BAR domains form a concave-shaped lipid-binding surface, the I-BAR domain, which is also called IRSp53-MIM domain (IMD), displays a shallow convex geometry and binds to the inner leaflet of phosphatidylinositol 4,5 biphosphate-2-enriched membranes. Consequently, I-BARs drive the formation of dynamic membrane tubules in the opposite orientation to that of conventional BARs and promote membrane protrusions such as lamellipodia, ruffles and filopodia [6]. In mammalian cells, the I-BAR subfamily can be further divided into two subsets: the MIM-like subset, which includes Abba and missing-in-metastasis (MIM); and the IRSp53 subset, which includes IRSp53, IRTKS and FLJ22582. IRSp53 differs from MIM by having an SH3 domain, a partial Cdc42/Rac interactive binding (CRIB) site, and by the lack of a C-terminal Wiscott–Aldrich-syndrome-protein homology 2 (WH2) domain.
In addition to the membrane-binding property, I-BAR proteins associate with and activate small GTPase Rac through the I-BAR domain [7;8]. The ability of IRSp53 to interact with Rac has been thought to provide a link between WAVE/SCAR-2 and the Arp2/3 complex [9], the primary machinery responsible for polymerization of branched actin filaments in the cell cortex. Unlike IRSp53, which binds to both Rac and Cdc42 through the I-BAR domain and the CRIB site respectively [10;11], MIM and Abba proteins do not contain any apparent CRIB-like domain and show no Cdc42-binding activity in vitro [12;13]. Nevertheless, overexpression of MIM or its I-BAR domain peptide triggers the formation of prominent lamellipodia and filopodia [7;14;15], the structures that are known to be regulated by Rac and Cdc42, respectively [16]. However, it remains uncertain whether the Rac-binding activity of I-BARs would be subjected to a regulation in vivo and plays a role in the membrane deformation upon stimulation by extracellular signals. In this study, we examined the effect of Abba on the morphological change induced by PDGF. We found that overexpression of Abba in NIH3T3 cells increases the binding of Abba to Rac1 upon platelet derived growth factor (PDGF) stimulation and promotes membrane ruffling in response to PDGF- and integrin-mediated signaling. Furthermore, we show that the ruffle-promoting activity is dependent upon Rac1 binding. Interestingly, while PDGF regulates the binding of intact Abba protein to Rac1, it has little effect on a mutant carrying the Abba's BAR domain alone. Our data provides evidence for the regulation of I-BAR's Rac-binding activity and indicates a mechanism for the membrane remodeling mediated by the I-BAR proteins.
Materials and Methods
Reagents and antibodies
All the chemicals unless otherwise indicated were obtained from Sigma Aldrich. Monoclonal antibody against Rac1 and monoclonal antibody against α-tubulin were purchased from Millipore; monoclonal antibody against Myc epitope was a gift of Dudley Strickland; and polyclonal antibody against green fluorescent protein (GFP) was raised as described previously [17].
Plasmid construction
Plasmids pGFP-Abba and pFlag-Abba, both of which carry a human Abba cDNA, were obtained as a gift from Akiko Yamagishi [14]. pGFP-Abba-IMD and pGFP-AbbaK/D constructs were made by polymerase chain reaction using pGFP-Abba as the template and primers corresponding to the sequence flanking to the mutation. All the constructs were confirmed by direct DNA sequencing. Plasmids pMyc-Rac1 and pMyc-RacG12V were obtained from Alan Hall.
Cell culture
NIH3T3 cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum, streptomycin and penicillin. To establish cell populations stably expressing GFP-Abba, 293GPG packaging cells were transfected with pMGIN-GFP-Abba. The resulting viruses were collected and used to infect NIH3T3 cells as described previously [18]. The infected cells were maintained in medium supplemented with 0.4 mg/ml G418. For transient transfection, cells growing at the log phase were transfected with 2 μg of plasmid DNA using Superfect (Qiagen) according to the manufacturer's instructions. For PDGF treatment, cells were plated on a fibronectin-coated glass coverslip or on an 8-chamber slide in medium supplemented with 10% calf serum. The medium was changed to DMEM containing 0.2% calf serum when cells attached. After incubation for 24 h, the medium was added with 30 ng/ml PDGF (R&D Systems) and the cells were incubated for indicated times.
Analysis of Rac activation
GTP-bound Rac (GTP-Rac) in cells was measured by pull-down assay using recombinant protein GST-PAK-CRIB, which was purified from bacteria as described previously [19]. Purified GST-PAK-CRIB proteins were immobilized on glutathione beads at approximately 3 mg/ml. To isolate GTP-Rac proteins in cells, 5× 105 cells were lysed in 0.5 ml of pre-cold lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) containing freshly added protease-inhibitor- cocktail tablets (Roche) and 1 mM phenylmethanesulfonyl fluoride. The lysates were clarified by centrifugation at 14,000 rpm for 5 min. The clarified lysates were incubated with 30 μl of 50% (v/v) GST-PAK-CRIB beads at 4°C for 10 min on a rotation wheel and followed by brief centrifugation for 10 sec. The pellet was washed twice with the lysis buffer and dissolved in 30 μl of 2 × sample buffer [20]. The proteins in the precipitates were separated by 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by electroblotting to a cellulose membrane. The transferred membrane was blotted with monoclonal Rac antibody followed by incubation for 30 min with horseradish phosphatase-conjugated secondary antibody. The membrane was developed with SuperSignal-West-Dura-chemiluminesecent substrate (Thermo Scientific) and digitally scanned with ImageStation 2000 (Kodak).
Results
Abba induces membrane ruffling in response to PDGF and integrin signaling
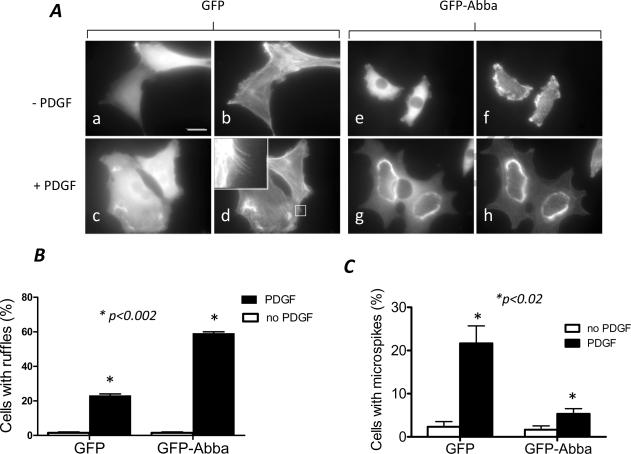
The cellular function of Abba was investigated with NIH3T3 cells infected by retrovirus carrying a human Abba cDNA tagged with GFP at the N-terminus. In addition, cells infected with the viral vector encoding GFP alone were also analyzed in parallel. When cells were arrested in 0.2% serum-containing medium and stained with GFP antibody and phalloidin for filamentous (F)-actin, no dramatic difference in morphology were recognized except GFP-Abba cells displayed retracted and more folded actin-rich structures or ruffles at the periphery (Fig. 1A). To study the effect of Abba on growth factor-mediated cell shape changes, we examined the morphological response of those cells to PDGF. The PDGF treatment rapidly triggered characteristic ruffling on the dorsal surface in about 58% GFP-Abba cells (Fig. 1B). In contrast, only 24% GFP cells developed prominent dorsal ruffles under the same condition. We also examined the formation of filopodia, the structure that is known to be promoted by overexpression of IRSp53 [11;14] and by MIM to a less degree [21;22]. We found that 60% less GFP-Abba cells showing prominent microspikes than GFP cells after PDGF stimulation (Fig. 1C).
Figure 1. Overexpression of Abba promotes the PDGF-mediated membrane ruffling.
(A) NIH3T3 cells infected with retroviruses carrying GFP (a–d) and GFP-Abba (e–h) were starved in 0.2% serum-containing medium. After 24 h, cells were exposed to PDGF at 30 ng/ml for 10 min (c, d, g and h) followed by staining with GFP antibody (a, c, e and g) and phalloidin (b, d, f and h). The scale bar: 20 μm. The inset of d is an enlarged box as indicated to show typical filopodia-like microspikes. (B) Quantification of PDGF-treated cells with prominent dorsal membrane ruffles. (C) Quantification of the formation of microspikes. The data presented were mean ± SEM of three independent experiments, and p values (student's t-test) refer to the difference between GFP and GFP-Abba cells as indicated by *.
Abba binds to and activates Rac protein
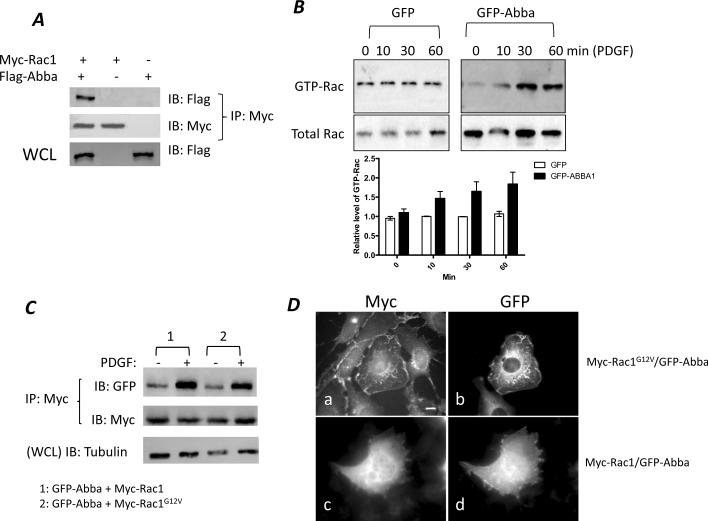
To analyze whether the ruffle-promoting activity of Abba involves Rac activation, we first analyzed the interaction of Rac1 with Abba. Because of extremely low level of endogenous Abba in 3T3 cells (data not shown), we analyzed 3T3 cells transiently transfected with plasmids encoding Flag epitope tagged Abba (Flag-Abba) and Myc epitope tagged Rac1 (Myc-Rac1). The transfected cells were then subjected to immunoprecipitation using Myc antibody. Co-precipitation of Flag-Abba with Myc-Rac1 was detected by Western blot of the precipitates with Flag antibody. Flag-Abba protein was readily detected in the immunoprecipitates derived from cells expressing both Flag-Abba and Myc-Rac1 but not from cells expressing Flag-Abba or Myc-Rac1 alone (Fig. 2A).
Figure 2. Abba interacts with and activates Rac1.
(A) Abba interacts with Rac1. 3T3 cells were co-transfected with Myc-Rac1 and Flag-Abba (lane 1), Myc-Rac1 alone (lane 2) and Flag-Abba alone (lane 3). After transfection, the cell lysates were immunoprecipitated with Myc antibody, and Rac1-associated Flag-Abba proteins in the pellets were measured by Western blot using Flag antibody. The same blot was re-probed with Myc antibody to show equal immunoprecipitation. The expression level of Flag-Abba in the transfected cells was determined by immunoblotting of whole cell lysates (WCL) with Flag antibody as shown in the bottom panel. (B–C) Analysis of the effect of overexpression of GFP-Abba on Rac activation in response to PDGF. Cells were starved at 0.2% serum-containing medium for 24 h and stimulated with PDGF for 10 to 60 min. GTP-bound Rac (GTP-Rac) in cell lysates was determined by pull-down with GST-PAK-CRIB followed by Western blot with Rac1 antibody. Aliquots of the cell lysates were also immunoblotted with Rac antibody as the input control. The chart represents the normalized levels of GTP-Rac to the input Rac1, and error bars were standard derivations of three independent experiments. (C) Cells co-expressing GFP-Abba with Myc-Rac1 or with Myc-Rac1G12Vcells were incubated in 0.2% serum-containing medium for 24 h and stimulated with PDGF for 30 min. The lysates derived from treated cells were immunoprecipitated with Myc antibody, and the pellets were immunoblotted with GFP and Myc antibody, respectively. Aliquots of cell lysates were also blotted with α-tubulin antibody for the loading control. (D) Cells stably expressing Myc-Rac1G12V (a and b) or Myc-Rac1 (c and d) were transiently transfected with GFP-Abba, and stained with phalloidin (a and c) and GFP antibody (b and d). A cell co-expressing GFP-Abba and Myc-Rac1G12V developed prominent ruffles. Scale bar: 10 μm.
To analyze the potential of Abba to activate Rac in response to growth factors, we utilized GST-PAK-CRIB, a peptide that preferentially recognizes GTP-bound Rac (GTP-Rac), the activated form of Rac protein [23]. GST-PAK-CRIB immobilized on beads was used to pull down GTP-Rac in cell lysates after PDGF stimulation for various times. As shown in Fig. 2B, the level of GTP-Rac in GFP-Abba cells at 30 and 60 min was significantly higher than that in GFP cells.
Binding to Rac is required for and correlated with Abba-mediated membrane remodeling
To determine whether the Abba-mediated Rac activation was dependent upon Rac binding, we measured the Rac1-binding activity in response to PDGF in the cells co-expressing GFP-Abba with either Myc-Rac1 or Myc-Rac1G12V, a constitutively-active form of Rac1 [24]. As shown in Fig. 2C, in a quiescent status GFP-Abba was poorly co-precipitated with Myc-Rac1 or Myc-Rac1G12V. Upon PDGF treatment, the interaction of GFP-Abba with either form of Rac1 was markedly enhanced.
To reinforce the functional relationship between Abba and Rac activation in cells, we examined the cellular distribution of GFP-Abba and Myc-Rac1G12V proteins. As shown in Fig. 2D, a cell co-expressing GFP-Abba and Myc-RacG12V exhibited more dramatic increase in the formation of membrane ruffles and lamellipodia than those expressing Myc-RacG12V alone. Within the ruffles colocalization of GFP-Abba with Myc-RacG12V was evident. However, a cell co-expressing GFP-Abba and Myc-Rac1 failed to display significant membrane ruffles. Thus, Abba promotes the membrane ruffling mediated by activated Rac1.
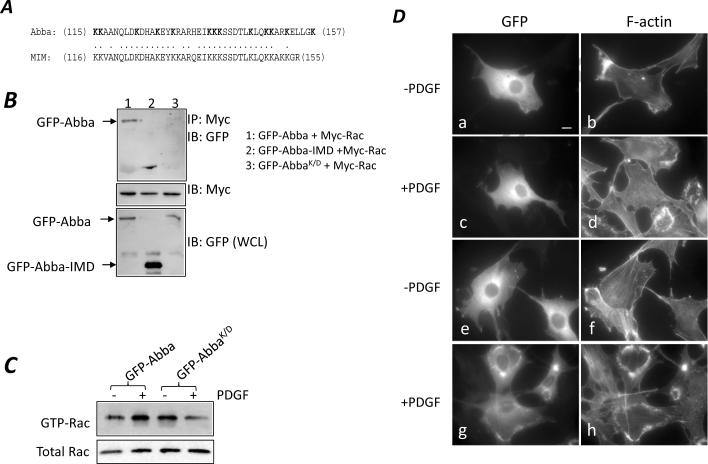
We have also attempted to determine whether binding to Rac1 would be required for Abba to promote the formation of ruffles. Previous studies have established that binding of IRSp53 or MIM to Rac is the property of the I-BAR domain and involves a basic patch that is comprised of multiple lysine residues [25;26]. This basic patch is conserved in the Abba amino acid sequence (Fig. 3A). To confirm the role of the basic patch in the binding to Rac, we prepared an Abba mutant (AbbaK/D) in which all 13 lysine residues within the patch were converted to glutamic acids. We then analyzed the interaction of Myc-Rac1 with GFP-AbbaK/D by co-immunoprecipitation of cells transiently transfected with their corresponding plasmids. The mutant GFP-AbbaK/D reduced markedly the binding activity for Myc-Rac1 as compared to GFP-Abba (Fig. 3B). To verify whether mutation of the basic patch has any effect on Rac activation, 3T3 cells transiently transfected with GFP-AbbaK/D were analyzed for Rac activation in response to PDGF. While the basal level of GTP-Rac in GFP-AbbaK/D cells was similar to that of GFP-Abba cells when starved in 0.2% serum-containing medium, PDGF treatment failed to increase the Rac activity in GFP-AbbaK/D cells (Fig. 3C). This result indicates that the basic patch is necessary for Abba to interact with Rac and potentiate the PDGF-mediated Rac activation.
Figure 3. Mutation at the basic batch abolished the interaction of Abba with Rac1 and Rac1 activation.
(A) Comparison of the basic patch of human Abba with that of MIM. The Lys residues in Abba that had been mutated to Glu are bolded. (B) Cells were transiently co-transfected with constructs as indicated. After 24 h of transfection, cells were analyzed for Rac1 binding as described above. The same membrane was also re-blotted with Myc antibody for equal immunoprecipitation. Also, aliquots of the lysates were immunoblotted with GFP antibody to determine expression of GFP-tagged proteins. (C) Cells were transiently transfected with GFP-Abba and GFP-AbbaK/D, respectively. The level of GTP-Rac in the transfected cells was analyzed as described in the legend of Fig. 3. (D) GFP-AbbaK/D cells (a–d) and GFP-Abba cells (e–h) were plated on fibronectin-coated coverslips and arrested in 0.2% serum-containing medium for 24 h. The arrested cells were treated without (a, b, e and f) or with PDGF (c, d, g and h) for 10 min, and co-stained with GFP antibody (a, c, e and g) and phalloidin (b, d, f and h). Bar: 10 μm.
We also examined Abba mutant cells under the conditions with and without PDGF. When grown in 0.2% serum-containing medium, GFP-AbbaK/D cells showed little difference in morphology as compared to GFP-Abba cells (Fig. 3D, compare a–b with e–f). Upon PDGF stimulation for 10 min GFP-Abba cells formed prominent dorsal ruffles while GFP-AbbaK/D cells failed to do so (Fig. 3D, compare c–d with g–h), indicating that the mutation at the basic patch impaired the morphological response to PDGF.
PDGF does not enhance the interaction between Abba-I-BAR domain and Rac1
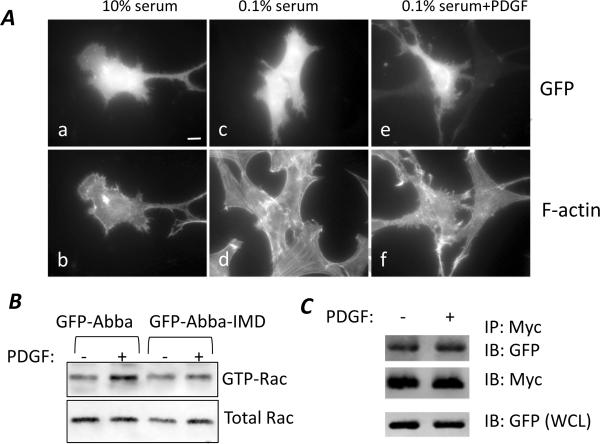
Previous studies have indicated that the I-BAR domain is the sole motif responsible for Rac1 binding as analyzed in vitro [25;26], raising an issue whether the I-BAR domain itself contains a regulatory element in response to PDGF. Thus, we examined the cells overexpressing GFP-Abba-IMD, a mutant that contains only the N-terminal 250 amino acids corresponding to the entire I-BAR domain. Interestingly, cells overexpressing GFP-Abba-IMD displayed a morphology distinguished from that of GFP-Abba expressing cells (Fig. 4A). Instead of forming membrane ruffles, those cells developed numerous filopodia-like microspikes on the periphery when grown in either 10% (a and b) or 0.1% serum (c and d). The similar morphological change remained apparent even after 10 min of PDGF stimulation (e and f), suggesting that the change was not subjected to regulation by growth factors. To verify this, we examined the Rac activation in GFP-Abba-IMD expressing cells with and without PDGF treatment. As shown in Fig. 4B, while the Rac activity in GFP-Abba expressing cells was enhanced upon PDGF stimulation, the increased Rac activation was not evident in PDGF-treated GFP-Abba-IMD cells. To analyze whether PDGF regulates the Rac1 binding activity of the mutant, stable Myc-Rac1G12V expressing cells were transfected with GFP-Abba-IMD and subsequently subjected to Rac1 binding analysis under the condition with and without PDGF treatment. As shown in Fig. 4C, no dramatic difference in the binding of GFP-Abba-IMD to Myc-Rac1G12V was observed before and after PDGF treatment.
Figure 4. Interaction between Abba-IMD alone and Rac1 was not regulated by PDGF.
Cells transfected with GFP-Abba-IMD were grown in 10% serum-containing medium (a and b), 0.1% serum-containing medium (c and d), and 0.1% serum medium plus PDGF for 10 min (e and f). The morphology of the transfected cells was examined by immunofluorescent microscopy after staining with GFP antibody (a, c and e) and phalloidin (b, d and f). Bar: 10 μm. (B) Serum-starved 3T3 cells transiently transfected with GFP-Abba and GFP-Abba-IMD were stimulated with PDGF for 30 min. Rac activation was measured by the pull-down assay. Aliquots of lysates of the samples were also analyzed for Rac input. (C) 3T3 cells stably expressing Myc-RacG12V were transiently transfected with GFP-Abba-IMD and treated with PDGF for 30 min. The interaction between GFP-Abba-IMD and Myc-RacG12V in the treated cells was analyzed as described in the legend of Fig. 4.
Discussion
It has been recently speculated that association of I-BARs with Rac would provide an important link between the dynamic actin cytoskeleton and membrane protrusions [27]. However, it is unclear whether the Rac binding is essential for membrane protrusions and subjected to regulation by the extracellular stimuli that trigger Rac activation. The study presented here was aimed to address this issue by investigating NIH3T3, a cellular model that has been widely used to study I-BARs and growth factor signal transduction. The prominent morphological change manifested by Abba overexpressors is the enhanced formation of ruffles and lamellipodia upon stimuli by PDGF, suggesting that Abba promotes preferentially membrane ruffles. This finding agrees with a previous report that depletion of Abba impairs the lamellipodial dynamics and membrane extension of radial glial cells where Abba is abundantly expressed [13]. Our further study indicates that binding to Rac is required for the function of Abba because the ruffle-promoting activity was diminished with a mutant deficient in the Rac binding. The Rac binding site of Abba is apparently located within the basic patch of the I-BAR domain and conserved in MIM. Mutation at four lysine residues within the same region of MIM was known to abolish its Rac binding activity [12]. Depletion of the similar region of MIM also impaired PDGF-mediated membrane ruffle formation as we described previously [28], confirming the necessity of this domain and the Rac binding in the function of I-BARs.
We demonstrated that PDGF regulates the interaction of GFP-Abba with either wild type or constitutively activated Myc-Rac1 proteins. Unexpectedly, we did not observe the similar regulation with GFP-Abba-IMD, a truncated mutant that carries the I-BAR domain only. In addition, the activity of this mutant is significantly different from that of the full-length GFP-Abba. While GFP-Abba promotes membrane ruffles and lamellipodia formation, overexpression of GFP-Abba-IMD promotes formation of membrane tubulation and filopodia-like microspikes. The similar difference in inducing different types of membrane curvatures between the I-BAR domain and full-length of MIM has also been previously reported [12]. Thus, the mechanism to regulate Abba likely lies in its C-terminal region. Inspection of the amino acid sequence reveals that Abba protein is nearly 55% identical to that of MIM in the overall sequence, in particular within the I-BAR and the C-terminal WASP homology 2 domains. However, the sequence between I-BAR and WH2 domains varies significantly, suggesting that this region likely plays a regulatory role. Indeed, the sites for several MIM binding proteins, including cortactin, transcriptional factor Gli, the tumor suppressor Sufu, and protein receptor phosphatase δ, are all located within the region [29–32]. In addition, two tyrosine phosphorylated residues in MIM expressing cells are also found downstream of the I-BAR domain [28]. Characterization of the nature of the regulation would ultimately shine light on the dynamic membrane remodeling mediated by extracellular stimuli.
Acknowledgements
We greatly appreciate the gift of human Abba cDNA from Dr. Akiko Yamagishi. We would also like to thank Dr. Jeff Winkles for the critical reading. The work was supported by CA113809, CA CA091984 and MSCF (2008) to XZ, and 07-B-023 and H200824 from Health Department of Jiangsu province to DT.
Abbreviations
- BAR
Bin-Amphiphysin-Rvs
- CRIB
Cdc42/Rac interactive binding
- F-actin
filamentous actin
- GFP
green fluorescent protein
- GST
glutathione s transferase
- IMD
IRSp53-MIM domain
- MIM
missing in metastasis
- PDGF
platelet derived growth factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- [1].Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- [2].Farsad K, Ringstad N, Takei K, Floyd SR, Rose K, De CP. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J.Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gallop JL, McMahon HT. BAR domains and membrane curvature: bringing your curves to the BAR. Biochem.Soc.Symp. 2005:223–231. doi: 10.1042/bss0720223. [DOI] [PubMed] [Google Scholar]
- [4].Saarikangas J, Zhao H, Pykalainen A, Laurinmaki P, Mattila PK, Kinnunen PK, Butcher SJ, Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr.Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- [5].Frost A, Unger VM, De CP. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mattila PK, Pykalainen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J.Cell Biol. 2007;176:953–964. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Suetsugu S, Murayama K, Sakamoto A, Hanawa-Suetsugu K, Seto A, Oikawa T, Mishima C, Shirouzu M, Takenawa T, Yokoyama S. The RAC binding domain/IRSp53-MIM homology domain of IRSp53 induces RAC-dependent membrane deformation. J.Biol.Chem. 2006;281:35347–35358. doi: 10.1074/jbc.M606814200. [DOI] [PubMed] [Google Scholar]
- [8].Suetsugu S. The direction of actin polymerization for vesicle fission suggested from membranes tubulated by the EFC/F-BAR domain protein FBP17. FEBS Lett. 2009;583:3401–3404. doi: 10.1016/j.febslet.2009.10.019. [DOI] [PubMed] [Google Scholar]
- [9].Miki H, Yamaguchi H, Suetsugu S, Takenawa T. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature. 2000;408:732–735. doi: 10.1038/35047107. [DOI] [PubMed] [Google Scholar]
- [10].Soltau M, Richter D, Kreienkamp HJ. The insulin receptor substrate IRSp53 links postsynaptic shank1 to the small G-protein cdc42. Mol.Cell Neurosci. 2002;21:575–583. doi: 10.1006/mcne.2002.1201. [DOI] [PubMed] [Google Scholar]
- [11].Krugmann S, Jordens I, Gevaert K, Driessens M, Vandekerckhove J, Hall A. Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr.Biol. 2001;11:1645–1655. doi: 10.1016/s0960-9822(01)00506-1. [DOI] [PubMed] [Google Scholar]
- [12].Bompard G, Sharp SJ, Freiss G, Machesky LM. Involvement of Rac in actin cytoskeleton rearrangements induced by MIM-B. J.Cell Sci. 2005;118:5393–5403. doi: 10.1242/jcs.02640. [DOI] [PubMed] [Google Scholar]
- [13].Saarikangas J, Hakanen J, Mattila PK, Grumet M, Salminen M, Lappalainen P. ABBA regulates plasma-membrane and actin dynamics to promote radial glia extension. J.Cell Sci. 2008;121:1444–1454. doi: 10.1242/jcs.027466. [DOI] [PubMed] [Google Scholar]
- [14].Yamagishi A, Masuda M, Ohki T, Onishi H, Mochizuki N. A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J.Biol.Chem. 2004;279:14929–14936. doi: 10.1074/jbc.M309408200. [DOI] [PubMed] [Google Scholar]
- [15].Lin J, Liu J, Wang Y, Zhu J, Zhou K, Smith N, Zhan X. Differential regulation of cortactin and N-WASP-mediated actin polymerization by missing in metastasis (MIM) protein. Oncogene. 2005;24:2059–2066. doi: 10.1038/sj.onc.1208412. [DOI] [PubMed] [Google Scholar]
- [16].Hall A. Ras-related GTPases and the cytoskeleton. Mol.Biol.Cell. 1992;3:475–479. doi: 10.1091/mbc.3.5.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yu D, Zhang H, Blanpied TA, Smith E, Zhan X. Cortactin is implicated in murine zygotic development. Exp.Cell Res. 2009 doi: 10.1016/j.yexcr.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li Y, Tondravi M, Liu J, Smith E, Haudenschild CC, Kaczmarek M, Zhan X. Cortactin potentiates bone metastasis of breast cancer cells. Cancer Res. 2001;61:6906–6911. [PubMed] [Google Scholar]
- [19].Huang C, Ni Y, Wang T, Gao Y, Haudenschild CC, Zhan X. Down-regulation of the filamentous actin cross-linking activity of cortactin by Src-mediated tyrosine phosphorylation. J.Biol.Chem. 1997;272:13911–13915. doi: 10.1074/jbc.272.21.13911. [DOI] [PubMed] [Google Scholar]
- [20].Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- [21].Millard TH, Bompard G, Heung MY, Dafforn TR, Scott DJ, Machesky LM, Futterer K. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 2005;24:240–250. doi: 10.1038/sj.emboj.7600535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mattila PK, Salminen M, Yamashiro T, Lappalainen P. Mouse MIM, a tissue-specific regulator of cytoskeletal dynamics, interacts with ATP-actin monomers through its C-terminal WH2 domain. J.Biol.Chem. 2003;278:8452–8459. doi: 10.1074/jbc.M212113200. [DOI] [PubMed] [Google Scholar]
- [23].Edlund S, Landstrom M, Heldin CH, Aspenstrom P. Transforming growth factor-beta-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol.Biol.Cell. 2002;13:902–914. doi: 10.1091/mbc.01-08-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nobes CD, Hall A. Rho, rac and cdc42 GTPases: regulators of actin structures, cell adhesion and motility. Biochem.Soc.Trans. 1995;23:456–459. doi: 10.1042/bst0230456. [DOI] [PubMed] [Google Scholar]
- [25].Bompard G, Sharp SJ, Freiss G, Machesky LM. Involvement of Rac in actin cytoskeleton rearrangements induced by MIM-B. J.Cell Sci. 2005;118:5393–5403. doi: 10.1242/jcs.02640. [DOI] [PubMed] [Google Scholar]
- [26].Suetsugu S, Murayama K, Sakamoto A, Hanawa-Suetsugu K, Seto A, Oikawa T, Mishima C, Shirouzu M, Takenawa T, Yokoyama S. The RAC binding domain/IRSp53-MIM homology domain of IRSp53 induces RAC-dependent membrane deformation. J.Biol.Chem. 2006;281:35347–35358. doi: 10.1074/jbc.M606814200. [DOI] [PubMed] [Google Scholar]
- [27].Scita G, Confalonieri S, Lappalainen P, Suetsugu S. IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 2008;18:52–60. doi: 10.1016/j.tcb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- [28].Wang Y, Zhou K, Zeng X, Lin J, Zhan X. Tyrosine phosphorylation of missing in metastasis protein is implicated in platelet-derived growth factor-mediated cell shape changes. J.Biol.Chem. 2007;282:7624–7631. doi: 10.1074/jbc.M608448200. [DOI] [PubMed] [Google Scholar]
- [29].Lin J, Liu J, Wang Y, Zhu J, Zhou K, Smith N, Zhan X. Differential regulation of cortactin and N-WASP-mediated actin polymerization by missing in metastasis (MIM) protein. Oncogene. 2005;24:2059–2066. doi: 10.1038/sj.onc.1208412. [DOI] [PubMed] [Google Scholar]
- [30].Gonzalez-Quevedo R, Shoffer M, Horng L, Oro AE. Receptor tyrosine phosphatase-dependent cytoskeletal remodeling by the hedgehog-responsive gene MIM/BEG4. J.Cell Biol. 2005;168:453–463. doi: 10.1083/jcb.200409078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Callahan CA, Ofstad T, Horng L, Wang JK, Zhen HH, Coulombe PA, Oro AE. MIM/BEG4, a Sonic hedgehog-responsive gene that potentiates Gli-dependent transcription. Genes Dev. 2004;18:2724–2729. doi: 10.1101/gad.1221804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Woodings JA, Sharp SJ, Machesky LM. MIM-B, a putative metastasis suppressor protein, binds to actin and to protein tyrosine phosphatase delta. Biochem.J. 2003;371:463–471. doi: 10.1042/BJ20021962. [DOI] [PMC free article] [PubMed] [Google Scholar]