Abstract
The cardiac NK-2 transcription factors are the vertebrate relatives of the Drosophila tinman gene. Without the Drosophila tinman gene, fruit flies fail to form their heart (“dorsal vessel”), and mutations or altered expression of cardiac NK-2 genes may lead to abnormal heart formation in vertebrates. Although the cardiac NK-2 gene NKX2–5 is recognized as an important factor in cases of human congenital heart disease and heart development in vertebrates, the roles of the other cardiac NK-2 genes are less clear. This report reviews what is known about the cardiac NK-2 genes in cardiac development, comparing studies in several different model systems.
Keywords: Abnormal heart formation, Cardiac NK-2 genes, Cardiac NK-2 transcription factors, Congenital heart disease, Early heart development
One of the molecules critical to normal cardiac development across vertebrate species is the transcription factor Nkx2–5. Nkx2–5 is a member of a family of transcription factors that uses the Drosophila tinman protein, essential for dorsal vessel (the heart of the fly) formation in the fly, as a prototype. Mutations in NKX2–5 are among the most common genetic causes of nonsyndromic congenital heart disease in humans, and this homeodomain-containing transcription factor has been central to models of cardiac development.
This review aims to introduce Nkx2–5 and closely related proteins expressed in cardiac tissue. Although studies investigating a variety of organisms are discussed, we highlight studies examining this family of proteins that have used embryos of the South African clawed frog, Xenopus laevis.
What Characterizes the Nkx2–5 Family of Proteins?
Several features of Nkx2–5 make it recognizable across species (Fig. 1). The official name of Nkx2–5 is NK-2 transcription factor related, locus 5. This name refers to the grouping of homeodomain-containing proteins into 20 different classes, one of which is the NK-2 class. The abbreviation NK refers to Nirenberg and Kim [26], who carried out a search for Drosophila genes containing homeodomain-encoding genes. The “x” in Nkx was added to designate vertebrate members of the family. Within the NK-2 genes expressed in vertebrates, a subset, referred to as the “cardiac group” [20], includes NKX2–3, NKX2–5, NKX2–6, nkx2–7, nkx-8, and nkx2–10.
Fig. 1.
General domain structure of the cardiac NK-2 family of proteins. Four conserved motifs are found consistently in the NK-2 family of proteins. The domains and the amino acid sequence from human NKX2–5 corresponding to each domain starting from the amino terminus of the protein are the TN domain, the homeodomain, the NK-2 domain, and the sequence GIRAW. One of the distinguishing features of the homeodomain is the presence of a tyrosine (Y) at amino acid position 54
At the outset, it is worth noting that NKX2–3, although found in zebra fish, frog, chicken, mouse, rat, and human, is only expressed in the cardiac region in frog and chicken [30]. The results from analysis of multiple species encourage a model in which NKX2–5 and at least one other family member is involved directly in early cardiac formation.
The protein domains that define the NK-2 family are shown in Fig. 1. The DNA-binding homeodomain is 60 amino acids long with a tyrosine at position 54. In addition to the homeodomain, these proteins have an amino terminal 10-amino-acid-long TN domain. In vertebrates, but not in Drosophila tinman, a 16-amino-acid NK-2 domain lies just beyond the C-terminal residue of the homeodomain. Typically, cardiac NK-2 proteins also have a C-terminal amino acid sequence of GIRAW. The sequence conservation in each of these domains is very high. Using the human NKX2–5 protein sequence as a comparative standard, the vertebrate members of the cardiac NK-2 protein family have an 80% or greater identity in the TN domain and the homeodomain, and more than a 64% identity in the NK2 domain.
How Do These Domains Relate to Protein Function?
Full understanding concerning the function of each of the domains remains a work in progress. However, ample evidence shows that the homeodomain itself is critical. The cardiac NK-2 genes encode DNA-binding transcription factors, with the consensus binding site of 5’T(C/T)AAGTG [9, 37] bound by the homeodomain. Sparrow et al. [37] reported subtle differences using in vitro DNA-binding assays for direct comparison of the three cardiac NK-2 proteins found in Xenopus. Although these subtle differences may be selective for one NK-2 protein over another, the authors favor a model in which NK-2 regulation of specific genes depends on other modulators.
Although NK-2 proteins are capable of binding DNA as monomers, the association is much stronger with DNA as a dimer [25]. The NK-2 proteins can form homodimers and heterodimers. Heterodimer-binding partners may be other NK-2 proteins or a variety of other transcription factors including GATA4 and Tbx5 [1, 15, 22]. Once a protein complex containing an NK-2 protein is bound to a DNA target site, transcription may be stimulated, as seen for the ANF gene and alpha cardiac actin, or inhibited, as seen for β-catenin [35].
At least part of the regulatory posture assumed by the NK-2 proteins is due to the choice of the heterodimer binding partner. For example, ANF transcription is stimulated by a Nkx2–5 GATA4 dimer [14] and inhibited by an Nkx2–5-Tbx-2 dimer [19]. Dimer formation is coordinated via the C-terminus of the homeodomain and enhanced by amino acids that extend beyond the C terminus [23, 25]. The NK-2 domain (also called the NK-2-SD domain) is thought to mediate interaction with other proteins, and some assays indicate a role when NK-2 family members repress rather than activate gene expression [36].
Synteny Among Cardiac NK-2 Family Members
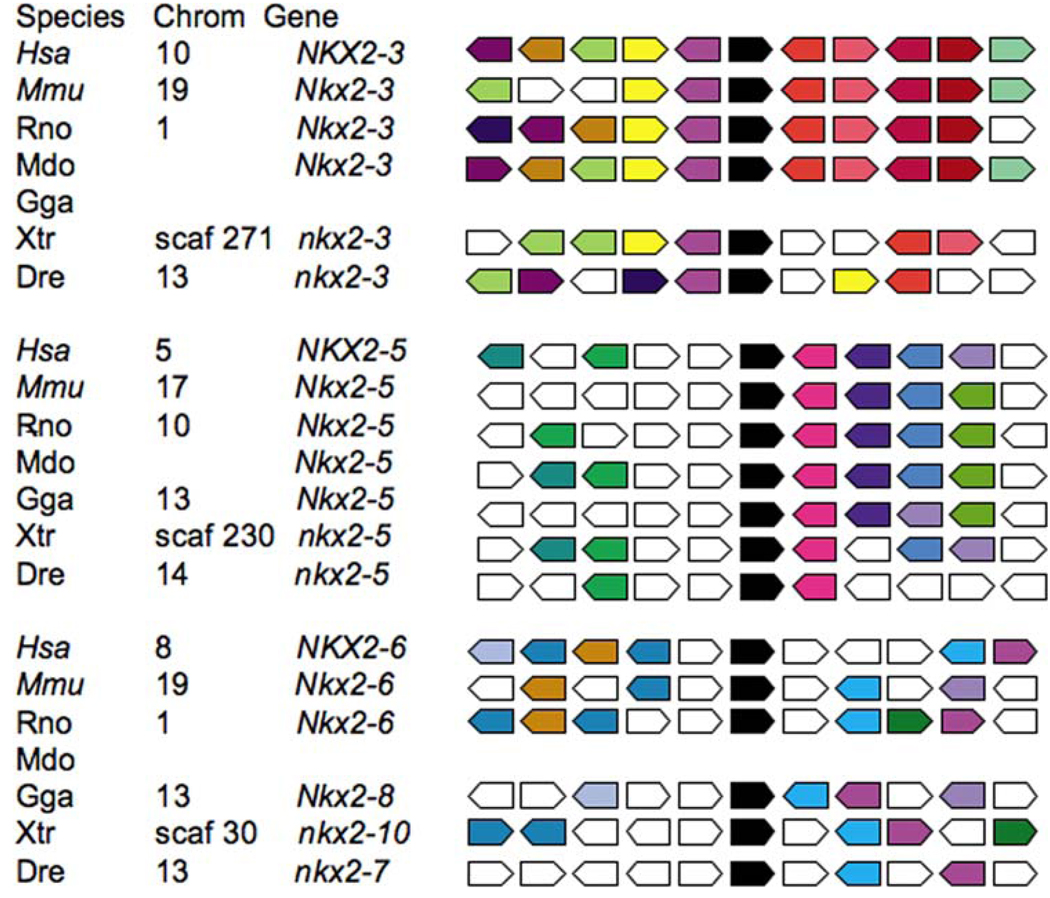
The growing catalog of genomic sequences from a variety of organisms opens the possibility of performing syntenic analysis. In the context of this review, synteny is used to describe the conservation of placement among other genes in the genome. Conservation of protein domains does not guarantee conserved function, and neither does synteny. However, synteny does imply a common genetic past for a group of genes. Using the program Metazome hosted by the Joint Genomic Institute, it is possible to perform such an analysis on the cardiac group of NK-2 genes (Fig. 2). This type of analysis inspects the genes that surround each of the NK-2 genes and color codes gene identity. Among vertebrates, the genes identified as NKX2–5 and NKX2–3 assemble into separate syntenic groups. The analysis suggests that, using the human NKX2–6 gene as the positional reference, the chicken Nkx2–8, Xenopus nkx2–10, and the zebra fish nkx2–7 gene are possible syntenic matches. We note that the search using NK-2 as a keyword did not identify a chicken Nkx2–3 or opossum Nkx2–6 for syntenic analysis. However, with these data in mind, it may be reasonable to think about the cardiac NK-2 genes as defined by three syntenic groupings: NKX2–3, NKX2–5, or NKX2–6.
Fig. 2.
The cardiac NK-2 factors can be sorted into three syntenic groups. Using the program Metazome (http://www.metazome.net/) and the genomic database at the Joint Genome Institute, a search for all chordate NK-2 genes was conducted, and a syntenic cluster analysis was performed. In each case, the cardiac NK-2 gene is indicated in black. Genes with a common identity are color coded, and the non-matching sequence is in white. Clustering based on NKX2–3, NKX2–5, and NKX 2–6 for human (Hsa), mouse (Mmu), rat (Rno), opossum (Mdo), chicken (Gga), frog (Xtr), and zebra fish (Dre) are included in the analysis
Assaying Cardiac NK-2 Genes in Developing Embryos
Functional analysis of cardiac NK-2 proteins supports both unique and redundant roles in cardiac development. Although temporal and spatial expressions are overlapping for different genes, expression patterns are not identical, according to in situ analysis of varied organisms. The answer to the question “How important is each individual gene in heart development?” may depend on the organism and how the question is addressed. For example, if the question is asked in relation to genetic causes of congenital heart defects in humans, NKX2–5 stands out as a key factor for normal morphologic and physiologic development [6], and NKX2–6 is implicated in normal great vessel development [21].
The mouse homolog of NKX2–5 also is critical for heart development [7, 9, 15, 24, 25, 28, 29]. Mice null for Nkx2–5 start to form a heart and express a variety of cardiac specific genes but fail to progress much beyond the linear heart tube stage [29]. Mice carrying Nkx2–5 mutations have phenotypes that include compromised physiology, morphology, and function of the heart. Alternatively, mice null for Nkx2–6 are not reported to have a cardiac phenotype [38].
Tanaka et al. [38] postulated that the loss of Nkx2–6 was compensated by an increase in Nkx2–5 expression. In zebra fish, nkx2–5 and nkx2–7 are expressed in the heart-forming region [10]. Tu et al. [39] recently published studies concluding that loss of nkx2–5 expression alone has little effect on heart development but that loss of nxk2–7 or both nkx2–7 and 2–5 had significant effects. In their studies, the morpholino-induced loss of nkx2.5 and 2.7 protein led to defects that could be at least partially rescued by adding back either nkx2.5 or nkx2.7 mRNA, although nkx2.7 was effective at much lower doses. Reminiscent of Nkx2–6 loss in mouse, nkx2–7 inhibition in zebra fish increases expression of nkx2–5. Studies comparing mouse and zebra fish showed a difference in which cardiac NK-2 protein is most important. However, the loss of both did not lead to a complete loss of cardiac structure in either organism, so neither is both necessary and sufficient for at the least initial steps in cardiac formation.
Assays on the Trio of Cardiac NK-2 Genes in Xenopus
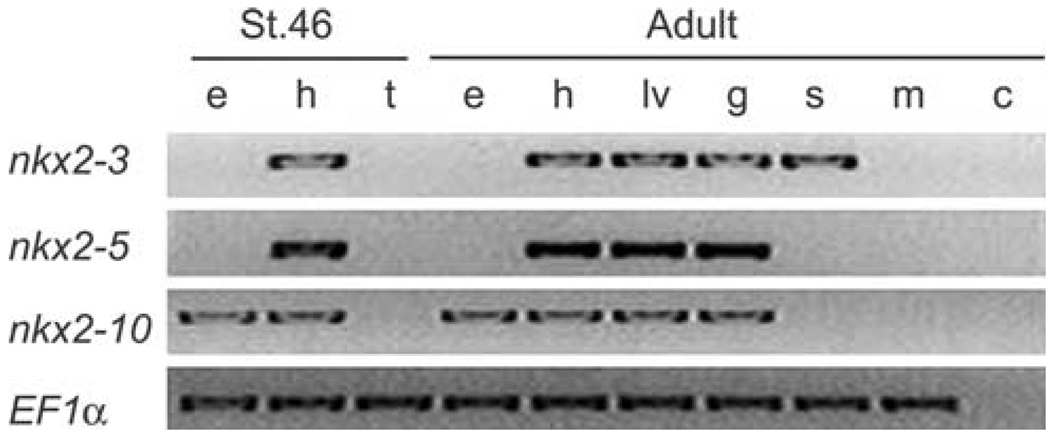
Compromised expression of NKX2–3 in human and mouse gives rise to defects in gut and spleen [32, 40]. However, in frog and chicken, it joins nkx2–5 and nkx2–10 (or Nkx2–8 in chicken) among NK-2 genes expressed in the heart. Although initial studies of nkx2–10 suggested that it was transiently expressed during early cardiac development [31, 37], a more recent analysis suggests that after a transient decrease, expression returns [3]. All the Xenopus cardiac NK-2 genes can be found in RNA isolated from adult heart, liver, and gut. Nkx2–3 also was expressed in skin, whereas nkx2–10 was present in the eye (Fig. 3 [3]).
Fig. 3.
Examination of cardiac Nk-2 gene expression in Xenopus laevis. Findings show mRNA isolated from: Xenopus stage 46 embryo eyes (e), heart (h), and embryo tail (t) or Xenopus adult eye (e), heart (h), liver (lv), gut (g), skin (s), skeletal muscle (m); and negative control (c). This figure is modified from Allen, Allen-Brady, and Weeks [3]
Overexpression of nkx2–3 or nkx2–5 via injection of mRNA in early embryos gives a transient increase in both cell number and size of the heart with little known consequence to noncardiac tissue [12]. A similar nkx2–5 dose-dependent increase in heart size was reported using zebra fish [10], although reduction in heart size and morphologic defects with further increases in nkx2–5 levels also has been reported [39]. Overexpression of Xenopus nkx2–10 has no obvious cardiac effect [3, 31].
In frog, the loss of function of each cardiac NK-2 gene has been tested [3, 17, 18]. Grow and Krieg [18] generated dominant negative nkx2–3 and nkx2–5 through a single mutation in the homeodomain that disrupted DNA binding. Their dominant effects would be through binding to and potentially titrating away proteins that normally would be interacting with nkx2–5 (or nkx2–3) bound to DNA. They judged the consequences for heart development by monitoring the reduction of several cardiac-specific mRNAs with in situ hybridization (cardiac troponin 1, myosin light chain 2, and alpha cardiac actin). Dominant negative nkx2– 3 or nkx2–5 reduced expression of the tested cardiac genes and reduced heart size (to no heart at all in some cases) in 40% to 60% of the embryos scored. The effect of the dominant negative was partially mitigated by co-injection with wild type nkx2–5 mRNA. Importantly, these authors reported few consequences of the dominant negative mutations unrelated to cardiac development.
A slightly different approach was taken by Fu et al. [17]. Their dominant negative construct comprised the isolated homeodomain of nkx2–3 or nkx2–5 fused to the engrailed repressor. They allowed a repressor to occupy promoter sites that normally would have bound to any of the cardiac NK-2 proteins. The expression of either nkx2–5 or nkx2–3 repressor fusion reduced cardiac differentiation as assayed by decreased expression of cardiac marker genes. When both repressor fusion proteins were added together, cardiac development was even worse.
Studies examining nkx2–10 loss of function used oligonucleotide-mediated inhibition of protein expression [3]. In these studies, the loss of nkx2–10 led to both general anterior and cardiac defects. The defective hearts were smaller than control hearts and had decreased trabeculation as well as abnormal anterior repositioning of the atria. Leftward bending of the outflow tract occurred, although the outflow tract was clearly underdeveloped. The loss of nkx2–10 did not seem to change the quantitative embryonic expression of either nkx2–3 or nkx2–5 when these genes were assayed after lateral field fusion. This is a different result than reported for the loss of either Nkx2–6 in mouse or nkx2–7 in zebra fish, in which a loss led to an increase in expression of Nkx2–5.
These studies indicate that in Xenopus, all three cardiac NK-2 genes are playing an active role in cardiac development.
Regulation of the Cardiac NK-2 Genes in the Heart
Most of the studies on the promoters of cardiac NK-2 genes have focused on regulation of homologs of NKX2–5. Transcription of this gene demands both temporal and spatial control during embryogenesis. Transacting regulation is mediated through a variety of transcription factors including BMP/Smad, GATA family members [8, 34, 37] FGF [4], and NFAT [11]. Other reviews (e.g., [1, 13, 16, 27]) and their references can be consulted for a more complete picture of NKX2–5 regulation. However, the coordinated regulation of cardiac NK-2 genes in a single background is relatively unstudied beyond those using in situ hybridization. In Xenopus, relatively detailed in situ hybridization of all three cardiac NK-2 genes has been presented [33, 37], indicating both overlapping and unique spatial and temporal regulation of each gene.
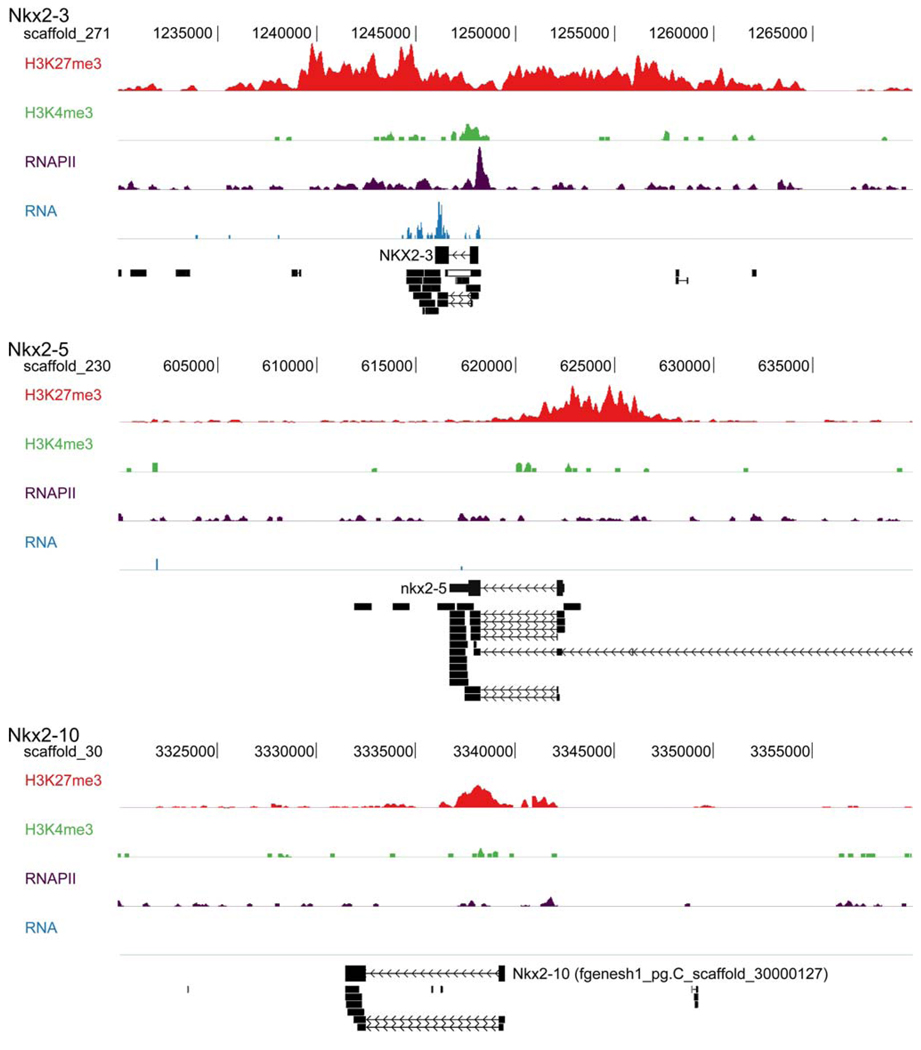
A relatively new technique, chromatin immunoprecipitation (ChIP) followed by deep sequencing (ChIP-seq), can be used to generate genome-wide maps of chromatin modifications and transcriptional activity. The analysis of such an experiment with Xenopus tropicalis embryos [2] provides an interesting insight into the regulatory state of the cardiac NK-2 genes of Xenopus just before gene activation can be detected by in situ hybridization. Antibodies directed against trimethylated histone H3 lysine 27 (H3K27me3) were used to identify regions repressed by Polycomb repressor proteins. Antibodies against trimethylated lysine 4 of the same protein (H3K4me3) were used to identify active promoters, whereas antibodies directed against RNA polymerase II (RNAPII) identified transcribed regions.
In addition, cDNA obtained from embryonic polyadenylated mRNA was sequenced to identify mature transcripts (RNA). The profiles of the sequences captured are displayed over the genomic regions of the cardiac NK-2 genes in Fig. 4. All three cardiac NK-2 genes are decorated with H3K27me3, indicating Polycomb-mediated repression. Concordantly, nkx2–5 and nkx2–10 are not transcribed at this early stage of embryogenesis. However, the promoter of nkx2–3 is relatively depleted for the H3K27me3 marker, is enriched for the active histone modification (H3K4me3), and recruits RNA polymerase 2. In addition, some nkx2–3 RNA was detected in these early embryos (Fig. 4). Moreover, the nkx2–3 promoter and the promoter proximal region respectively recruit TATA binding protein, a key transcription initiation factor, and monomethylated histone H3 lysine 4, a marker for putative enhancers (data not shown).
Fig. 4.
Analysis of proteins bound to genomic regions of the cardiac NK-2 genes of Xenopus tropicalis. A ChIP-seq analysis of gastrulastage embryonic chromatin was performed using antibodies to H3K27me3, H3K4me3, and RNApolII. Analysis of each antibody is presented for nkx2–3, nkx2–5, and nkx2–10. Deep sequencing results of cDNA (RNA-seq) are indicated as well as known gene annotation and sequenced ESTs (bottom of each panel). Scaffold positions are across the top of the analysis for each gene. Direction or transcription is always from right to left in this figure
Together, these data indicate that the first of the three cardiac NK-2 genes activated in the frog embryo is nkx2– 3. The other two NK-2 genes are repressed. The promoter regions of nkx2–5 and nkx2–10 are covered by H3K27me3, consistent with the need for chromatin remodeling as a necessary step before their activation. Ample precedent has been found for the regulation of cardiac genes by dynamic chromatin modification [5]. The ChIP-seq profiles suggest that neither nkx2–5 nor nkx2–10 contain the stalled or paused RNA polymerase reported for many developmentally important genes in Drosophila [41], whereas nkx2–3 shows strong enrichment for RNA polymerase II directly at the transcription start site but much less enrichment further into the gene body, suggestive of pausing. It will be interesting to determine whether the genes regulated by the cardiac NK-2 genes are associated with repressive chromatin or whether they have paused polymerase in anticipation of activation. Another important direction for further research is to determine whether the sequential activation of nkx2–3 and nkx2–5/nkx2–10 represents a cross-regulatory hierarchy in which the cardiac gene regulatory network is progressively established or whether the early activation of nkx2–3 is reflective of a parallel regulatory pathway that, independently of the other NK-2 genes, contributes to the cardiac program.
Conclusion
Although a great deal has been learned about the role of NKX2–5 in heart development, less is known about the other cardiac NK-2 proteins. Unification of their roles in cardiac development may not be possible, but we suggest that it is worth examining these genes as members of three syntenic groups. It is unclear which, if any, combination of cardiac NK-2 genes will be responsible for full activation of cells that can contribute to heart development. However, a great deal still remains to be uncovered before we understand all the roles cardiac NK-2 genes assume in normal heart development.
Acknowledgments
The authors thank the members of their laboratories and the American Heart Association, the National Institutes of Health, and the Netherlands Organization for Scientific Research (NWO-ALW VIDI) for support.
Contributor Information
Heather Bartlett, Department of Pediatrics, University of Iowa, Bowen Science Building, 51 Newton Road, Iowa City, IA 52242, USA.
Gert Jan C. Veenstra, Department of Molecular Biology, Faculty of Science, Nijmegen Centre for Molecular Life Sciences, Radboud University Nijmegen, Nijmegen, The Netherlands
Daniel L. Weeks, Department of Biochemistry, University of Iowa, Bowen Science Building, 51 Newton Road, Iowa City, IA 52242, USA, daniel-weeks@uiowa.edu
References
- 1.Akazawa H, Komuro I. Cardiac transcription factor Csx/Nkx2–5: its role in cardiac development and diseases. Pharmacol Ther. 2005;107:252–268. doi: 10.1016/j.pharmthera.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Veenstra GJ. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell. 2009;17:425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen BG, Allen-Brady K, Weeks DL. Reduction of XNkx2–10 expression leads to anterior defects and malformation of the embryonic heart. Mech Dev. 2006;123:719–729. doi: 10.1016/j.mod.2006.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alsan BH, Schultheiss TM. Regulation of avian cardiogenesis by Fgf8 signaling. Development. 2002;129:1935–1943. doi: 10.1242/dev.129.8.1935. [DOI] [PubMed] [Google Scholar]
- 5.Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res. 2006;98:15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- 6.Benson DW, Silberbach GM, Kavanaugh-McHugh A, Cottrill C, Zhang Y, Riggs S, Smalls O, Johnson MC, Watson MS, Seidman JG, Seidman CE, Plowden J, Kugler JD. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J Clin Invest. 1999;104:1567–1573. doi: 10.1172/JCI8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biben C, Harvey RP. Homeodomain factor Nkx-2.5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes Dev. 1997;11:1357–1369. doi: 10.1101/gad.11.11.1357. [DOI] [PubMed] [Google Scholar]
- 8.Brown CO, III, Chi X, Garcia-Gras E, Shirai M, Feng XH, Schwartz RJ. The cardiac determination factor, Nkx2–5, is activated by mutual cofactors GATA-4 and Smad1/4 via a novel upstream enhancer. J Biol Chem. 2004;279:10659–10669. doi: 10.1074/jbc.M301648200. [DOI] [PubMed] [Google Scholar]
- 9.Chen CY, Schwartz RJ. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J Biol Chem. 1995;270:15628–15633. doi: 10.1074/jbc.270.26.15628. [DOI] [PubMed] [Google Scholar]
- 10.Chen JN, Fishman MC. Zebra fish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 1996;122:3809–3816. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Yuen WH, Fu J, Huang G, Melendez AJ, Ibrahim FB, Lu H, Cao X. The mitochondrial respiratory chain controls intracellular calcium signaling and NFAT activity essential for heart formation in Xenopus laevis. Mol Cell Biol. 2007;27:6420–6432. doi: 10.1128/MCB.01946-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleaver OB, Patterson KD, Krieg PA. Overexpression of the tinman-related genes XNkx-2.5 and XNkx-2.3 in Xenopus embryos results in myocardial hyperplasia. Development. 1996;122:3549–3556. doi: 10.1242/dev.122.11.3549. [DOI] [PubMed] [Google Scholar]
- 13.Cripps RM, Olson EN. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- 14.Durocher D, Chen CY, Ardati A, Schwartz RJ, Nemer M. The atrial natriuretic factor promoter is a downstream target for Nkx-2.5 in the myocardium. Mol Cell Biol. 1996;16:4648–4655. doi: 10.1128/mcb.16.9.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. The cardiac transcription factors Nkx2–5 and GATA-4 are mutual cofactors. Embo J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foley A, Mercola M. Heart induction: embryology to cardiomyocyte regeneration. Trends Cardiovasc Med. 2004;14:121–125. doi: 10.1016/j.tcm.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Fu Y, Yan W, Mohun TJ, Evans SM. Vertebrate tinman homologues XNkx2–3 and XNkx2–5 are required for heart formation in a functionally redundant manner. Development. 1998;125:4439–4449. doi: 10.1242/dev.125.22.4439. [DOI] [PubMed] [Google Scholar]
- 18.Grow MW, Krieg PA. Tinman function is essential for vertebrate heart development: elimination of cardiac differentiation by dominant inhibitory mutants of the tinman-related genes, XNkx2–3 and XNkx2–5. Dev Biol. 1998;204:187–196. doi: 10.1006/dbio.1998.9080. [DOI] [PubMed] [Google Scholar]
- 19.Habets PE, Moorman AF, Clout DE, van Roon MA, Lingbeek M, van Lohuizen M, Campione M, Christoffels VM. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16:1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harvey RP. NK-2 homeobox genes and heart development. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- 21.Heathcote K, Braybrook C, Abushaban L, Guy M, Khetyar ME, Patton MA, Carter ND, Scambler PJ, Syrris P. Common arterial trunk associated with a homeodomain mutation of NKX2.6. Hum Mol Genet. 2005;14:585–593. doi: 10.1093/hmg/ddi055. [DOI] [PubMed] [Google Scholar]
- 22.Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, Komuro I. Tbx5 associates with Nkx2–5 and synergistically promotes cardiomyocyte differentiation. Nat Genet. 2001;28:276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- 23.Inga A, Reamon-Buettner SM, Borlak J, Resnick MA. Functional dissection of sequence-specific NKX2–5 DNA binding domain mutations associated with human heart septation defects using a yeast-based system. Hum Mol Genet. 2005;14:1965–1975. doi: 10.1093/hmg/ddi202. [DOI] [PubMed] [Google Scholar]
- 24.Kasahara H, Lee B, Schott JJ, Benson DW, Seidman JG, Seidman CE, Izumo S. Loss of function and inhibitory effects of human CSX/NKX2.5 homeoprotein mutations associated with congenital heart disease. J Clin Invest. 2000;106:299–308. doi: 10.1172/JCI9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasahara H, Usheva A, Ueyama T, Aoki H, Horikoshi N, Izumo S. Characterization of homo- and heterodimerization of cardiac Csx/Nkx2.5 homeoprotein. J Biol Chem. 2001;276:4570–4580. doi: 10.1074/jbc.M004995200. [DOI] [PubMed] [Google Scholar]
- 26.Kim Y, Nirenberg M. Drosophila NK-homeobox genes. Proc Natl Acad Sci U S A. 1989;86:7716–7720. doi: 10.1073/pnas.86.20.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lien CL, McAnally J, Richardson JA, Olson EN. Cardiac-specific activity of an Nkx2–5 enhancer requires an evolutionarily conserved Smad binding site. Dev Biol. 2002;244:257–266. doi: 10.1006/dbio.2002.0603. [DOI] [PubMed] [Google Scholar]
- 28.Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:969. doi: 10.1242/dev.119.3.969. [DOI] [PubMed] [Google Scholar]
- 29.Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeobox gene Nkx2–5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 30.Newman CS, Krieg PA. Tinman-related genes expressed during heart development in Xenopus. Dev Genet. 1998;22:230–238. doi: 10.1002/(SICI)1520-6408(1998)22:3<230::AID-DVG5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 31.Newman CS, Reecy J, Grow MW, Ni K, Boettger T, Kessel M, Schwartz RJ, Krieg PA. Transient cardiac expression of the tinman-family homeobox gene, XNkx2–10. Mech Dev. 2000;91:369–373. doi: 10.1016/s0925-4773(99)00291-9. [DOI] [PubMed] [Google Scholar]
- 32.Pabst O, Zweigerdt R, Arnold HH. Targeted disruption of the homeobox transcription factor Nkx2–3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development. 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- 33.Patterson KD, Cleaver O, Gerber WV, Grow MW, Newman CS, Krieg PA. Homeobox genes in cardiovascular development. Curr Top Dev Biol. 1998;40:1–44. doi: 10.1016/s0070-2153(08)60363-4. [DOI] [PubMed] [Google Scholar]
- 34.Peterkin T, Gibson A, Patient R. GATA-6 maintains BMP-4 and Nkx2 expression during cardiomyocyte precursor maturation. Embo J. 2003;22:4260–4273. doi: 10.1093/emboj/cdg400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riazi AM, Takeuchi JK, Hornberger LK, Zaidi SH, Amini F, Coles J, Bruneau BG, Van Arsdell GS. NKX2–5 regulates the expression of beta-catenin and GATA4 in ventricular myocytes. PLoS One. 2009;4:e5698. doi: 10.1371/journal.pone.0005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sepulveda JL, Belaguli N, Nigam V, Chen CY, Nemer M, Schwartz RJ. GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: role for regulating early cardiac gene expression. Mol Cell Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sparrow DB, Cai C, Kotecha S, Latinkic B, Cooper B, Towers N, Evans SM, Mohun TJ. Regulation of the tinman homologues in Xenopus embryos. Dev Biol. 2000;227:65–79. doi: 10.1006/dbio.2000.9891. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka M, Schinke M, Liao HS, Yamasaki N, Izumo S. Nkx2.5 and Nkx2.6, homologs of Drosophila tinman, are required for development of the pharynx. Mol Cell Biol. 2001;21:4391–4398. doi: 10.1128/MCB.21.13.4391-4398.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tu CT, Yang TC, Tsai HJ. Nkx2.7 and Nkx2.5 function redundantly and are required for cardiac morphogenesis of zebra fish embryos. PLoS One. 2009;4:e4249. doi: 10.1371/journal.pone.0004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamazaki K, Takahashi A, Takazoe M, Kubo M, Onouchi Y, Fujino A, Kamatani N, Nakamura Y, Hata A. Positive association of genetic variants in the upstream region of NKX2–3 with Crohn’s disease in Japanese patients. Gut. 2009;58:228–232. doi: 10.1136/gut.2007.140764. [DOI] [PubMed] [Google Scholar]
- 41.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]