Abstract
To understand the molecular etiology of osteosarcoma, we isolated and characterized a human osteosarcoma cell line (OS1). OS1 cells have high osteogenic potential in differentiation induction media. Molecular analysis reveals OS1 cells expressing the pocket protein pRB and the runt-related transcription factor Runx2. Strikingly, Runx2 is expressed at higher levels in OS1 cells than in human fetal osteoblasts (HFOB). Both pRB and Runx2 have growth suppressive potential in osteoblasts and are key factors controlling competency for osteoblast differentiation. The high levels of Runx2 clearly suggest osteosarcomas may form from committed osteoblasts that have bypassed growth restrictions normally imposed by Runx2. Interestingly, OS1 cells do not exhibit p53 expression and thus lack a functional p53/p21 DNA damage response pathway as has been observed for other osteosarcoma cell types. Absence of this pathway predicts genomic instability and/or vulnerability to secondary mutations that may counteract the anti-proliferative activity of Runx2 that is normally observed in osteoblasts. We conclude OS1 cells provide a valuable cell culture model to examine molecular events that are responsible for the pathologic conversion of phenotypically normal osteoblast precursors into osteosarcoma cells.
Keywords: human osteosarcoma, osteoblast, p53/p21 pathway, pRB gene, Runx2
Introduction
The generally accepted concept of stepwise mutagenesis leading to tumorigenesis is a model that has best been described in polyposis of the colon leading to colorectal cancer [1]. Similar mechanisms of tumorigenesis have been proposed in the oncogenic conversion of other tissues. It is generally believed osteogenic sarcoma may originate from mesenchymal stem cells [2]. Consistent with this idea, the pluripotent nature of mesenchymal stem cells permits differentiation in vitro into several distinct lineages (e.g., chondroblastic, fibroblastic, and osteoblastic phenotypes) that account for most of the phenotypes exhibited in osteosarcoma.
Osteosarcoma cells develop from genetic events that mediate immortalization and may support metastasis [3–7]. pRB is a key molecule in cell cycle regulation [8–10] and there is a strong relationship between a pRB-null status and the development of osteosarcoma. The presence of pRB normally suppresses growth by attenuating the activity of E2F factors until pRB is phosphorylated by cyclin-dependent kinases (e.g, CDK2/cyclin E) and is released from E2F. Absence of pRB permits proliferation by relieving the suppression of E2F factors and will promote cell cycle progression in osteosarcoma cells. Similarly, null mutations in the human p53 gene are associated with osteosarcoma and p53-null mice develop de novo osteosarcoma [11]. It has been well-established that loss of p53 function compromises DNA damage responses and apoptosis (e.g. due to a failure to induce p21), and thus p53-null osteosarcomas may exhibit genomic instability and may harbor additional mutations that promote immortalization and metastatic potential. Not all osteosarcoma cells have homozygous null mutations in both p53 and pRB mutations. In cells lacking both genes, there may be alternative mutational pathways that could be responsible for the de novo conversion of putative mesenchymal cells into osteosarcoma.
The runt-related transcription factor 2 (Runx2) defines the osteoblastic lineage by mediating the expression of a myriad of osteoblast specific genes, and the regulation of its activity is linked to cell proliferation [12–16]. Runx2-null mutations are known to promote osteoblast proliferation, and forced expression of Runx2 in mesenchymal cells attenuates cell growth. Similar to previous studies [6], we postulate that Runx2 is a critical factor controlling cellular phenotypes, but also that molecular aberrations affecting its function could be involved in the development of osteosarcoma. Based on the anti-proliferative function of Runx2, one could hypothesize that the expression of Runx2 is frequently silenced in osteosarcoma cells, as has been observed for other growth-suppressive proteins, such as pRB and p53. We addressed this hypothesis by examining the expression of Runx2 in relation to pRB and p53 in immortalized osteoblastic cells (human fetal osteoblasts, HFOB) and a novel osteosarcoma cell line (OS1).
Materials and Methods
To define the pathological role of Runx2 in osteosarcoma, we examined Runx2 protein levels in relation to the levels of the p53 and pRB tumour suppressors in immortalized osteoblasts and osteosarcoma cells. To evaluate the molecular characteristics of representative cell types, we examined cell lysates by western blotting to check the expression pattern of the cell growth regulatory proteins Runx2, pRB, and p53, in relation to other cell cycle markers (i.e., the p53 response CDK inhibitor p21 and cyclin D). As Runx2 is expressed at high levels in quiescent osteoblasts and mature osteoblasts but at low levels in normal actively proliferating osteoblasts, we also examined the expression of Runx2 protein at the single cell level using immunofluorescence microscopy.
We isolated cells from an osteosarcoma obtained in Singapore to develop a culture system that may be representative of the local population. While it is possible that ethnic differences may be less important than the genetic heterogeneity found in metastatic osteosarcomas, we believed that it would be prudent not to dismiss a priori that there could be ethnic differences. The human osteosarcoma cell line designated OS1 was obtained with informed consent from the tumor biopsy of a 6-year-old female patient with a diagnosis of osteosarcoma. The tissue collection protocol was approved by the National University Hospital Institutional Review Board (NHG DSRB B/00/301). Briefly, the biopsy specimen was minced after full rinsing in phosphate-buffered saline (PBS). Cells were then incubated in a medium containing a 9:1 (v/v) mixture of RPMI 1640 and Dulbecco's Modified Eagle's Medium (DMEM; Gibco, Grand Island, NY), supplemented with 15% fetal bovine serum (FBS; HyClone, Logan, UT) at 37° C in 5% CO2. One cell colony (OS1) was isolated and expanded as a cell line that was further characterized for growth properties and karyotype.
Both OS1 and HFOB 1.19 cells (SV40 T antigen transformed human fetal osteoblasts obtained from ATCC) were maintained in α-DMEM containing 10% FBS and 2 mmol/L glutamine and sub-cultured 1:4 every 3 to 4 days using 0.25% trypsin. The HFOB1.19 cells express a temperature-sensitive T-antigen and proliferate adequately at 37-degrees, albeit that T-antigen is presumably less active at this temperature. These cells are not stressed (e.g., no DNA irradiation), but express detectable levels of p53 and p21 (see Results). OS1 cells between eight and 15 passages were used in the experiments and all cells were seeded in culture plates at a density of 5000 cells/cm2. Cell morphology was analyzed using phase-contrast light microscopy. Black and white photographs of triplicates from each group were taken through an Olympus IX70 inverted phase-contrast light microscope (Olympus, Tokyo, Japan).
Upon reaching 80% confluence, cells were cultured in osteogenic medium containing ascorbic acid-2-phosphate (50 μg/mL), β-glycerophosphate (10 mmol/L), and dexamethasone (100 nmol/L) (Sigma-Aldrich, St Louis, MO) for 4-weeks. To assess whether the cells that were cultured ex-vivo possessed phenotypic properties of osteoblasts, we examined whether these cells were capable of supporting calcium deposition in their extracellular matrix, the mineralization of cultures under osteogenic inductions was evaluated using alizarin red staining. Cultures were first fixed with −20° C methanol followed by several washes with distilled water and then stained with 0.01% alizarin red S (pH 5.5) for 30-minutes. Histochemical staining of the cultures was documented on an Epson Perfection® Photo scanner (Seiko Epson Corp, Suwa, Japan). The cell layer was washed three times in PBS before disruption in a lysis buffer (5 mmol/L MgCl2, 150 mmol/L NaCl, 0.05% Nonidet® P40, 0.1% sodium dodecyl sulfate [SDS], 1% Triton® 100, pH 7.5) and protease inhibitor cocktail (Calbiochem UK Ltd, Nottingham, UK) at 4° C for 10 to 30 minutes. Protein concentrations were determined using the BCA protein assay kit (Pierce Biotechnology, Rockford, IL). Cell lysates (20–30 μg) were resolved by 10% SDS-PAGE and proteins were transferred to nitrocellulose membranes. Membranes were blocked with 5% low-fat milk in Tris-buffered saline for 1 hour and then incubated with a 1000-fold dilution of primary antibody. Mouse monoclonal antibodies against Runx2 (a gift from Dr Yoshiaki Ito and Dr Kosei Ito), pRB (Biodesign, Saco, ME), p53 and p21 (Cell Signaling Technology, Beverly, MA), and rabbit polyclonal cyclin D antibody (Santa Cruz Biotechnology, Inc, Santa Cruz, CA) were used in the experiments. Membranes were incubated with peroxide-conjugated goat anti-mouse/rabbit secondary antibodies (Santa Cruz Biotechnology) for 1-hour. Immunoreactive bands were visualized by a chemiluminescence detection kit (Pierce).
Cultures for immunoflorescence analysis were fixed in cold 3.7% paraformaldehyde (Sigma) for 30 minutes, permeabilized with 0.1% Triton® 100 in PBS for 15 minutes, and then blocked by 5% goat serum for 30 minutes. Cells were incubated at 4°C overnight with a monoclonal antibody against Runx2 (1:10,000 dilution). Cells were incubated with Alex Fluor® 594 chicken anti-mouse secondary antibody (Molecular Probes, Inc, Eugene, OR) for 30 minutes at room temperature. Nuclei were stained using 4',6-diamidino-2-phenylindole, (DAPI, Sigma) and images were recorded using a Olympus fluorescence microscope (Olympus, Tokyo, Japan).
Pixel intensities of the alkaline phosphatase stains of HFOB and OS1 cells were determined using Adobe Photoshop [17]. Luminosity of each band was determined using the histogram function. Mean luminosity and total pixels were determined for each of the three samples. Background levels for each dish were determined by measuring an unstained area on the dish. Mean luminosity and total pixels also were determined for background. Mean luminosities were multiplied by total pixels to obtain total luminosity for each dish and background. Total luminosity for each dish was divided by luminosity of the corresponding background to normalize values. Averages were taken for each sample (n =3), and standard deviation was determined. Optical density was then calculated as the inverse of the luminosity. Gray-scale TIFF files were used and quantified with Adobe Photoshop 5.0 software using the following protocol. First, an Invert command was applied to an entire image, and the intensity of staining within a selected circular image window (mean pixel value) was determined using the Image Histogram function of the software. The ratios of the staining intensities of each well were determined after subtraction of background values measured in a space between wells. A Mann-Whitney U parameteric test was used to compare the mean optical density at each time point, with a 0.05 level of significance.
Results
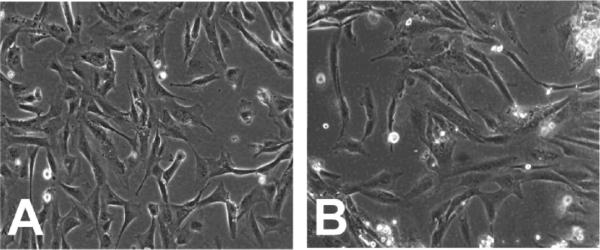
To address the hypothesis that Runx2 has an important role in osteosarcoma, we compared an established immortalized osteoblast cell line (HFOB) with a cell line (OS1) derived from an osteosarcoma from a Singaporean patient. In primary cultures, the OS1 cells demonstrated a typical spindle-like morphology and formed strand-like filopodia along the cell body. There were no distinct differences between OS1 and HFOB although the morphology of HFOB looked less spindle-like than OS1 (Fig. 1). Some cells from HFOB presented a triangular profile and formed sheet-like filopodia. Both proliferated at the same rate with a doubling time of approximately 1 to 2 days. Under osteogenic conditions (Fig. 2), the mineralization for both OS1 and HFOB started at Week 2 [Optical Density: 142.93 (SD,5.13) and 148.65 (SD, 2.49), respectively, p<0.05] and was most prominent in the extracellular matrix at Week 3 [172.72, (SD, 13.08), 174.82, (SD, 2.14), p=0.51] and Week 4 [181.88 (SD, 7.68), 181.37 (SD, 3.68), p=0.51] with no significant differences between the two cell lines (Fig. 2). For control cultures, no appreciable mineralization was observed without osteogenic induction [122.93 (SD, 5.13), 122.08 (SD, 2.79), p=0.51]. The morphologic, cell kinetic, and histochemical data suggest OS1 cells resemble immortalized HFOBs in several respects.
Fig. 1.
Both (A) HFOB and (B) OS1 cells displayed the typical spindle-like morphology. (No Stain, original magnification, ×100).
Fig. 2A–B.
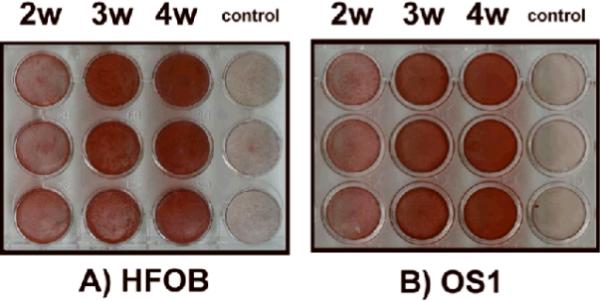
Mineralization of (A) HFOB and (B) OS1 cells was visualized by alizarin red staining up to 4-weeks. Both cell types demonstrated extensive calcium deposition in the extracellular matrix only upon osteogenic induction. Note: 2W = 2 weeks; 3W = 3 weeks; 4W = 4 weeks.
Expression of Runx2 was specifically detected in the nuclei of both OS1 and HFOB cells (Fig. 3). The western blot data (Fig. 4) show Runx2 is highly expressed in OS1, and these results corroborate the immunofluorescence microscopy data (Fig. 3). Moreover, OS1 and HFOB each express similar levels of the pocket protein pRB. The retention of pRB expression was noted with a robust expression of cyclin D1, which we assayed because expression of this cyclin is frequently deregulated in cancer.
Fig. 3.
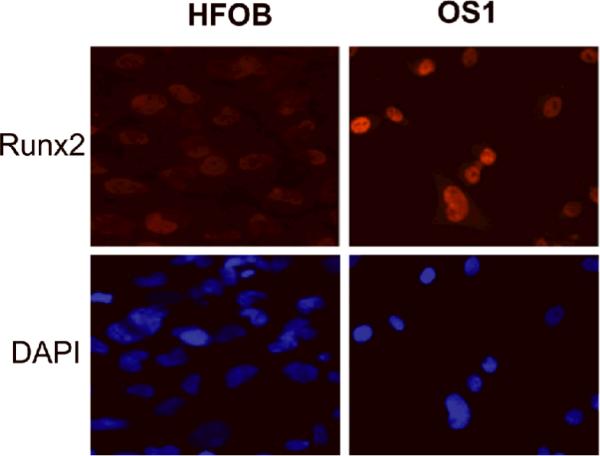
Runx2 expression detected in the nuclei by immunofluorescence microscopy. The intensity of the Runx2 signal was much more pronounced in OS1 cells than in HFOB cells (Stain, 4',6-diamidino-2-phenylindole, DAPI; original magnification, ×200).
Fig. 4.
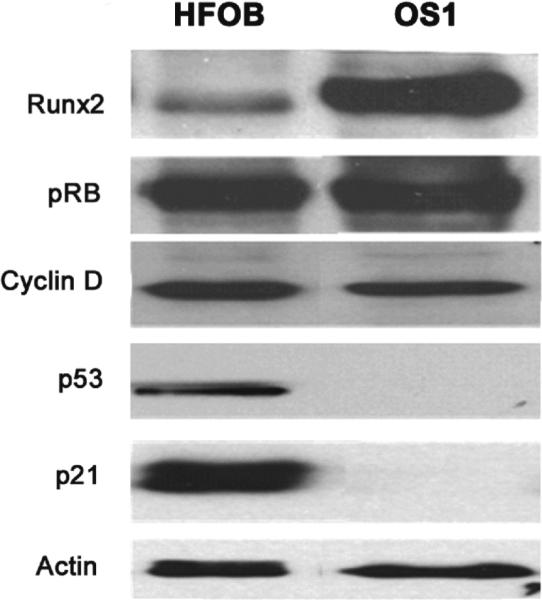
Western blots demonstrate Runx2 is highly expressed in OS1 but weakly expressed in HFOB. Pocket protein pRB was detected in OS1, but the growth suppressor p53 and the CDK inhibitor p21, which is a downstream target of p53, is not detected in OS1 cells.
Discussion
Runx2 is a key regulator of osteoblast growth and differentiation. HFOB cells are immortalized osteoblasts but retain many attributes of normal osteoblasts (e.g., growth factor-dependent and anchorage-dependent growth). We detected low levels of Runx2 in HFOB cells and only a limited number of cells revealed positive staining. These data are consistent with the low levels of Runx2 that are detected in actively proliferating immortalized MC3T3 osteoblasts that exhibit normal growth factor-dependent proliferation [12,14]. In contrast, we detected strong Runx2 signals in OS1, with over 70% of cells demonstrating positive staining, thus corroborating the interpretation that OS1 cells exhibit constitutively high levels of Runx2. Our results suggest a deregulation of the normal inverse relation between Runx2 levels and cell proliferation [12,14].
Our results show that the HFOB cell line has lower Runx2 levels than the OS1 cell line. Either confluence or reduced growth (e.g., due to loss of T-antigen activity exogenously expressed in HFOB cells) could have increased Runx2 levels [5,12], yet the levels in HFOB cells are still lower than the actively proliferating OS1 cells. Our observation that the levels of Runx2 are high in OS1 osteosarcoma cells is opposite to the findings of Thomas and colleagues who suggested that Runx2 protein is down-regulated in some osteosarcomas [19]. The question arises whether this is a novel finding or whether high Runx2 expression is a peculiarity of the OS1 cell line we characterized. In previous studies we have shown that human SAOS-2 cells and rat ROS17/2.8 cells express clearly detectable levels of Runx2 [12]. Thus, the data suggest that high Runx2 levels may be common in osteosarcoma cells.
The retention of pRB gene expression in both OS1 and HFOB cells is consistent with robust expression of cyclin D dependent kinases (i.e. CDK4 and CDK6) that can functionally inactivate pRB. Interestingly, OS1 cells did not exhibit either p53 or p21 expression and thus lack a functional p53/p21 DNA damage response pathway [9,18,19]. Taken together, the osteosarcoma cell line (OS1) we have characterized expresses two growth suppressors (Runx2 and pRB) that may have lost their suppressive potential, but exhibit loss of the p53 growth suppressor protein and its downstream target p21, as occurs frequently in osteosarcomas. While direct regulatory links between Runx2 and p53 are not evident from previous studies [20–21], Runx2 is known to repress the p53 target p21 indicating that p53 and Runx2 control at least one common pathway [22]. The absence of a functional p53/p21 DNA damage response pathway may render OS1 cells vulnerable to secondary genetic hits that may bypass the growth suppressive pathways that are controlled by pRB and Runx2 [3]. The development of OS1 as a cell culture resource may permit its use in definition of molecular mechanisms that are responsible for pathological changes of phenotypically normal osteoblast precursors into osteosarcoma cells.
One limitation of our results is that we only compare two cell lines, thus precluding generalizations at this stage and a larger sampling of osteosarcoma cells would be informative. Our findings show that OS1 is derived from an osteoblastic sarcoma, but other types of osteosarcoma exist and it would be of interest to demonstrate differences in protein expression among different osteosarcoma types [7]. Based on our results, we anticipate that at least a subset of osteosarcoma cells express elevated levels of Runx2 and this may modulate the pathogenic properties of these tumor cells.
Acknowledgments
We thank Yoshiaki Ito and Kosei Ito, Institute of Molecular and Cell Biology, Singapore, for providing the Runx2 antibody used in our studies and all the members of the laboratories participating in the Singapore Osteobiology group for stimulating discussions. In particular we thank Ren Xia Fei, Mario Galindo, Nadiya Teplyuk, Kakoli Das, David Leong, Dietmar Hutmacher, Lee Eng Hin, Wong Hee Kit, Jane Lian and Janet Stein for stimulating discussions. This study was co-supported by funding from NIH (grant R01 AR049069 to AvW and grant P01 CA082834 to GS) and Singapore Cancer Syndicate, A*STAR (grants MN-005 and MN-077, to MST)
Footnotes
Each author certifies that his or her institution has approved the human protocol for this investigation that all investigations were conducted in conformity with ethical principles of research, and that informed consent for participation in the study was obtained.
References
- 1.Kim KM, Salovaara R, Mecklin JP, Järvinen HJ, Aaltonen LA, Shibata D. PolyA deletions in hereditary nonpolyposis colorectal cancer: mutations before a gatekeeper. Am J Pathol. 2002;160:1503–6. doi: 10.1016/S0002-9440(10)62576-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibbs CP, Kukekov VG, Reith JD, Tchigrinova O, Suslov ON, Scott EW, Ghivizzani SC, Ignatova TN, Steindler DA. Stem-like cells in bone sarcomas: implications for tumorigenesis. Neoplasia. 2005;7(11):967–76. doi: 10.1593/neo.05394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat. Rev. Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 4.Kansara M, Thomas DM. Molecular pathogenesis of osteosarcoma. DNA Cell Biol. 2007;26:1–18. doi: 10.1089/dna.2006.0505. 2007. [DOI] [PubMed] [Google Scholar]
- 5.Pratap J, Javed A, Languino LR, van Wijnen AJ, Stein JL, Stein GS, Lian JB. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol. Cell. Biol. 2005;25:8581–8591. doi: 10.1128/MCB.25.19.8581-8591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas DM, Johnson SA, Sims NA, Trivett MK, Slavin JL, Rubin BP, Waring P, McArthur GA, Walkley CR, Holloway AJ, Diyagama D, Grim JE, Clurman BE, Bowtell DD, Lee JS, Gutierrez GM, Piscopo DM, Carty SA, Hinds PW. Terminal osteoblast differentiation, mediated by Runx2 and p27KIP1, is disrupted in osteosarcoma. J Cell Biol. 2004;167:925–934. doi: 10.1083/jcb.200409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol. Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- 8.Gupta S, Luong MX, Bleuming SA, Miele A, Luong M, Young D, Knudsen ES, Van Wijnen AJ, Stein JL, Stein GS. Tumor suppressor pRB functions as a co-repressor of the CCAAT displacement protein (CDP/cut) to regulate cell cycle controlled histone H4 transcription. J Cell Physiol. 2003;196:541–556. doi: 10.1002/jcp.10335. [DOI] [PubMed] [Google Scholar]
- 9.Harrington EA, Bruce JL, Harlow E, Dyson N. pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc Natl Acad Sci USA. 1998;95:11945–11950. doi: 10.1073/pnas.95.20.11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayhew CN, Bosco EE, Fox SR, Okaya T, Tarapore P, Schwemberger SJ, Babcock GF, Lentsch AB, Fukasawa K, Knudsen ES. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 2005;65:4568–4577. doi: 10.1158/0008-5472.CAN-04-4221. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs B, Pritchard DJ. Etiology of osteosarcoma. Clin Orthop Relat Res. 2002;397:40–52. doi: 10.1097/00003086-200204000-00007. [DOI] [PubMed] [Google Scholar]
- 12.Galindo M, Pratap J, Young DW, Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS, van Wijnen AJ. The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. J Biol Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit Rev Eukaryot Gene Expr. 2004;14:1–41. [PubMed] [Google Scholar]
- 14.Pratap J, Galindo M, Zaidi SK, Vradii D, Bhat BM, Robinson JA, Choi JY, Komori T, Stein JL, Lian JB, Stein GS, van Wijnen AJ. Cell growth regulatory role of Runx2 during proliferative expansion of preosteoblasts. Cancer Res. 2003;63:5357–5362. [PubMed] [Google Scholar]
- 15.Qiao M, Shapiro P, Fosbrink M, Rus H, Kumar R, Passaniti A. Cell cycle-dependent phosphorylation of the RUNX2 transcription factor by cdc2 regulates endothelial cell proliferation. J Biol Chem. 2006;281:7118–7128. doi: 10.1074/jbc.M508162200. [DOI] [PubMed] [Google Scholar]
- 16.Shen R, Wang X, Drissi H, Liu F, O'Keefe RJ, Chen D. Cyclin D1-cdk4 induces runx2 ubiquitination and degradation. J Biol Chem. 2006;281:16347–16353. doi: 10.1074/jbc.M603439200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gartel AL, Feliciano C, Tyner AL. A new method for determining the status of p53 in tumor cell lines of different origin. Oncol Res. 2003;13:405–408. doi: 10.3727/096504003108748429. [DOI] [PubMed] [Google Scholar]
- 18.Chen X, Ko LJ, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 19.Giono LE, Manfredi JJ. The p53 tumor suppressor participates in multiple cell cycle checkpoints. J Cell Physiol. 2006;209:13–20. doi: 10.1002/jcp.20689. [DOI] [PubMed] [Google Scholar]
- 20.Lengner CJ, Steinman HA, Gagnon J, Smith TW, Henderson JE, Kream BE, Stein GS, Lian JB, Jones SN. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. J Cell Biol. 2006;172(6):909–21. doi: 10.1083/jcb.200508130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Kua HY, Hu Y, Guo K, Zeng Q, Wu Q, Ng HH, Karsenty G, de Crombrugghe B, Yeh J, Li B. p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J. Cell Biol. 2006;172(1):115–25. doi: 10.1083/jcb.200507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westendorf JJ, Zaidi SK, Cascino JE, Kahler R, van Wijnen AJ, Lian JB, Yoshida M, Stein GS, Li X. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol. Cell Biol. 2002;22:7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]