Abstract
Efficient enzymatic hydrolysis of lignocellulosic material remains one of the major bottlenecks to cost-effective conversion of biomass to ethanol. Improvement of glycosylhydrolases however is limited by existing medium-throughput screening technologies. Here, we report the first high-throughput selection for cellulase catalysts. This selection was developed by adapting chemical complementation to provide a growth assay for bond cleavage reactions. First, a URA3 counter selection was adapted to link chemical dimerizer activated gene transcription to cell death. Next, the URA3 counter selection was shown to detect cellulase activity based on cleavage of a tetrasaccharide chemical dimerizer substrate and decrease in expression of the toxic URA3 reporter. Finally, the utility of the cellulase selection was assessed by isolating cellulases with improved activity from a cellulase library created by family DNA shuffling. This application provides further evidence that chemical complementation can be readily adapted to detect different enzymatic activities for important chemical transformations for which no natural selection exists. Due to the large number of enzyme variants selections can test compared to existing medium-throughput screens for cellulases, this assay has the potential to impact the discovery of improved cellulases and other glycosylhydrolases for biomass conversion from libraries of cellulases created by mutagenesis or obtained from natural biodiversity.
Introduction
Directed evolution and biodiversity mining of environmental DNA are two powerful approaches in enzyme discovery. Directed evolution is an established method for generating enzymes with improved specific activities for industrial, research, and therapeutic applications1–5. It consists of generating large numbers of protein variants, then assaying those variants en masse for the desired function. Biodiversity mining exploits the diversity found in unculturable microorganisms, which represents >99% of microorganisms6, to find novel enzymes. It consists of amplifying environmental DNA in vitro, expressing the genes in a model organism and testing them en masse for the desired function. Biodiversity mining has also been successfully applied for the discovery of industrial, research and therapeutic enzymes7. Directed evolution and biodiversity mining are limited to functional enzyme reactions that are inherently screenable or selectable, such as reactions where the product is fluorescent or an essential metabolite. To address this bottleneck we developed chemical complementation, a general high-throughput assay for enzyme catalysts that relies on the yeast three-hybrid assay to link enzyme catalysis to reporter gene transcription in vivo. This assay has been previously applied to screen for bond cleavage reactions using a beta-lactamase enzyme and lacZ as the reporter gene8. More recently, chemical complementation was adapted to select for bond formation reactions using a glycosynthase enzyme and a LEU2 reporter gene essential for cell survival9.
One of the major bottlenecks to cost-competitive conversion of biomass to fermentable sugars for biofuel production is the high enzyme cost for cellulose degradation10. Technological improvements in the breakdown of cellulose into sugars is often cited as one of the solutions to reducing cost10. Cellulose, a β-1,4-glucose polymer, represents 38–50% of the total cellulosic biomass composition11. Cellulose is hydrolyzed into glucose units by an ensemble of three enzymes: endoglucanases, also called cellulases [E.C. 3.2.1.4], exoglucanases [E.C.3.2.1.91], and glucohydrolases [E.C. 3.2.1.21]. Glucose can then be fermented to ethanol by microorganisms. Since the rate-limiting step in enzymatic cellulose depolymerization is performed by exoglucanases and endoglucanases12, discovery of improved cellulases likely is a straightforward solution to decreasing costs.
Currently, the discovery of improved endoglucanases and other glycosylhydrolase enzymes important for biomass conversion is based on medium-throughput screening technologies that rely on surrogate substrates12, 13. The two most widely used medium-throughput assays for endoglucanase activity are a halo assay carried out on petri dishes, which detects hydrolysis of carboxymethylcellulose (CMC) based on Congo red staining of carbohydrate reducing ends, and a UV assay, which detects hydrolysis of p-nitrophenyl cellobioside (pNPC)12. Because each gene in a screen must be assayed individually, even with automatation techniques only 104–106 genes can be tested, at best. A selection for cellulase catalysts would allow the search of much larger cellulase catalysts libraries (>108) because only cells expressing active enzyme variants survive. The problem is that cellulase catalysis is not inherently selectable.
Here, using chemical complementation, we have developed a URA3 counter selection for cellulose activity. To our knowledge, this is the first high-throughput selection for cellulase catalysts and should significantly increase the number of variants that can be tested over medium-throughput screens. The method was developed using the endoglucanase Cel7B from Humicola insolens that catalyzes the hydrolysis of beta-1,4-linked glucosidic bonds in cellulose. First, a URA3 counter selection was adapted to link chemical dimerizer activated gene transcription to cell death. The classic yeast URA3 5-fluoroorotic acid counter selection was chosen as the reporter gene because it is the state of the art counter selection in the two hybrid literature. Although the URA3 counter selection has been previously applied in an n-hybrid system14, 15, re-engineering of a chemical complementation URA3 counter selection strain was necessary as previously published strains were either not available or did not meet the parameters required by the chemical complementation strategy. Next, the URA3 counter selection was shown to detect cellulase activity based on cleavage of a tetrasaccharide chemical dimerizer substrate and decrease expression of the toxic URA3 reporter. Finally, the utility of the cellulose selection was assessed by isolating cellulases with improved activity from a cellulase family DNA shuffled library.
Results
Selection Scheme
In chemical complementation, enzyme catalysis of bond formation or cleavage is linked to cell survival based on covalent coupling of two small molecule ligands to reconstitute a transcriptional activator in vivo8 (Figure 2A). Bond formation is detected as activation of an essential reporter gene; bond cleavage, as repression of a toxic reporter gene. The assay is high-throughput because it can be run as a growth selection in which only cells containing the functional enzyme survive. The assay can be readily extended to new chemistry by synthesizing small molecule heterodimers with different chemical linkers as enzyme substrates.
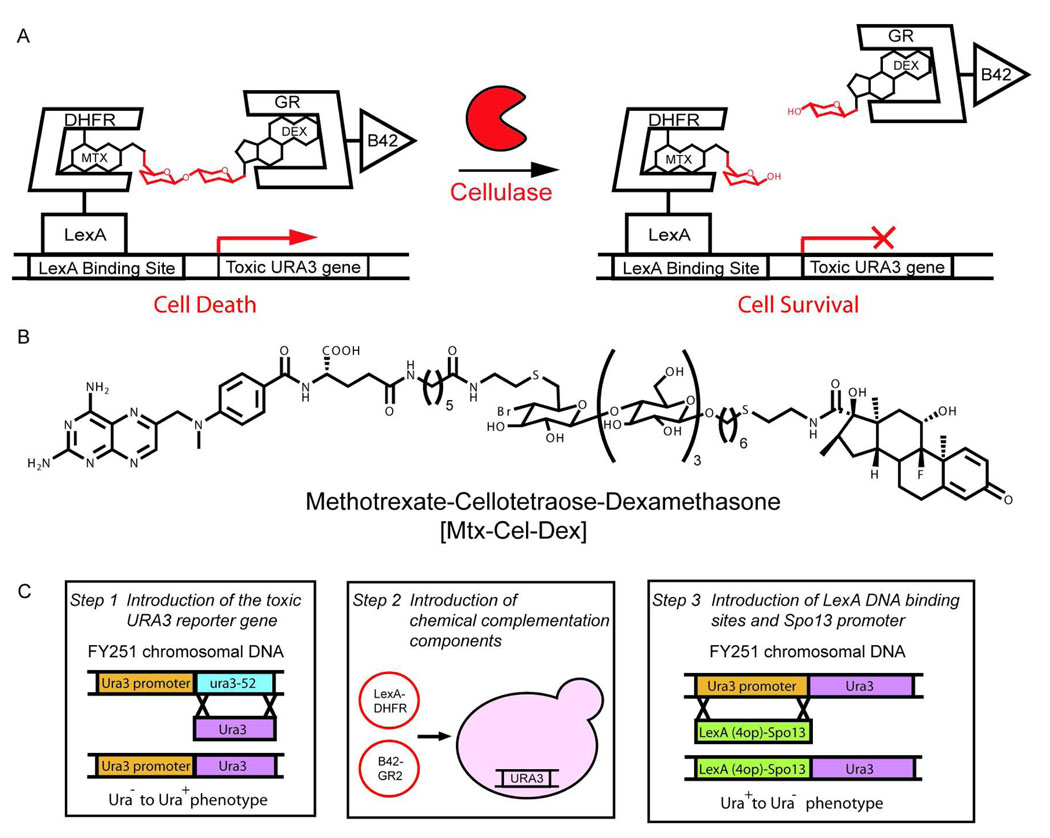
Figure 2.
Chemical complementation detects bond cleavage reactions A. In the cellulase growth selection, the Methotrexate-Cellotetraose-Dexamethasone (Mtx-Cel-Dex) heterodimer links a DNA-binding domain fused to dihydrofolate reductase (DBD-DHFR) and an activation domain fused to the glucocorticoid receptor (AD-GR), activating transcription of the toxic URA3 reporter gene, thus leading to cell death. Active cellulases cleave the β-1,4-glucosidic bond in the Mtx-Cel-Dex linker, decreasing transcription of the toxic URA3 reporter gene, and leading to cell survival. B. Structure of the Mtx-Cel-Dex chemical inducer of dimerization used for this assay. C. The URA3 counter selection strain was constructed in three steps. First, the URA3 gene was introduced at the ura3-52 locus of the chemical complementation yeast strain (FY251) via homologous recombination. Next, the AD-GR and the DBD-DHFR constructs were introduced into FY251 via transformation. Finally, the LexA DNA binding site and the tightly regulated Spo13 promoter were introduced upstream of the URA3 reporter gene via homologous recombination.
We envisioned that chemical complementation could detect cellulase activity as cleavage of a β-1,4-glucosidic bond in a Methotrexate-Cellotetraose-Dexamethasone (Mtx-Cel-Dex) substrate9, 16. Selection for bond cleavage required the construction of a chemical complementation counter selection where cleavage of the Mtx-Cel-Dex chemical dimerizer by a cellulase would relieve transcription of a toxic reporter gene. Thus, we adapted chemical complementation to incorporate the classic yeast counter selection marker URA3. In the presence of 5-fluoroorotic acid (5-FOA) the URA3 gene product, orotidine-5’-phosphate decarboxylase, produces the toxic product 5-fluorouracil (5-FU) which is misincorporated into RNA and inhibits the nucleotide synthetic enzyme thyamidylate synthase leading to cell death17. Cell survival is achieved by cleavage of the heterodimeric small molecule, disrupting expression of the URA3 gene
Design of the Mtx-Cel-Dex substrate
Since it is the chemical dimerizer substrate that imparts modularity to chemical complementation, we first synthesized a cellulase substrate carrying the Mtx and Dex handles. E. carotovora CelA, CelN, and CelV [E.C. 3.2.1.4] share 68% amino acid sequence identity with Bacillus agaradharens Cel5A. Therefore, we took advantage of the extensive crystallographic data on this cellulase to guide the design of the Mtx-Cel-Dex substrate18, 19. The high-resolution structure of B. agaradharens Cel5A predicts five subsites in the active site that accommodate five glucose units (−3, −2, −1, 1, 2). Given that in other cellulases four subsites (−2, −1, 1, 2) contribute most to the binding energy, with the fifth contributing only slightly20, the cellulase substrate was designed to have four saccharide units. Since the tetrasaccharide is embedded between Mtx and Dex, it should recapitulate the cellulase oligomer substrates. The tetrasaccharide linker is β-1,4-linked Glu-Glu-Glu-Glu. The synthesis of Mtx-Cel-Dex was carried out chemoenzymatically. Mtx-Lac-F and Dex-Cel were synthesized using published strategies from our laboratory and the linkage between the two halves was synthesized by the previously reported Cel7B:E197A glycosynthase. The full assignment of Mtx-Cel-Dex can be found in the Supplementary Information (Figure 2B, S1 and S2)9, 16.
Construction of the chemical complementation URA3 counter selection yeast strain
Guided by the construction of a previously published reverse yeast two-hybrid strain, we reduced the basal transcription of the URA3 reporter gene by placing it under control of the tightly regulated Spo13 promoter14. First, the FY251 ura3-52 chromosomal locus was replaced by a functional URA3 gene and selected for Ura+ phenotype. Next, the plasmids carrying the DNA binding domain-dihydrofolate reductase (DBD-DHFR) and the activation domain-glucocorticoid receptor (B42-GR) fusion proteins were introduced in the strain. Finally, a LexA(4op)-pSpo13 DNA fragment with four LexA binding sites was introduced upstream of the URA3 reporter gene and selected for a Ura− phenotype. This selection provided numerous chemical complementation URA3 counter selection yeast strains (Figure 2C).
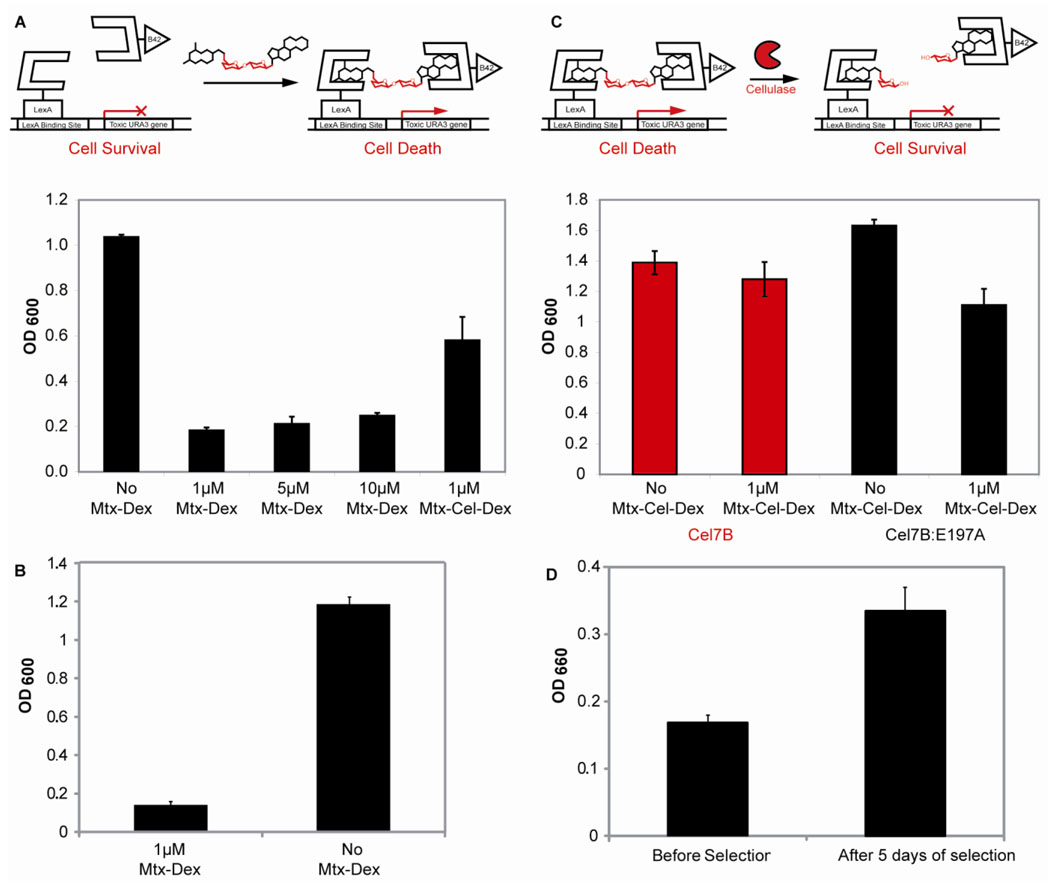
Three of these strains were used to optimize the counter selection conditions by varying the level of galactose (0.5–2%), which affects the concentration of DBD-DHFR and AD-GR in the system, and 5-FOA (0.1-0.5%), which controls the toxicity level of the reporter gene (Figure S3). Using the optimized conditions, the counter selection strain showed on-off behavior between 1–10 µM Mtx-Dex, thus all future counter selection experiments were carried out at 1 µM small molecule concentration. Next, the URA3 counter selection strain was tested with 1 µM Mtx-Cel-Dex, the small-molecule to be used in the cellulase selection, to confirm that the counter selection would also function properly with the cellulose substrate (Figure 3A). Finally, the chemical complementation URA3 counter selection yeast strain proved to be very stable after eight days under counter selection conditions. The cell growth of 93 strains under counter selection conditions was lower than the growth of the same 93 strains under noncounter selection conditions. This difference was statistically significant when analyzed with a paired one tailed Student T-test (p-value<0.0001) (Figure 3B).
Figure 3.
The chemical complementation URA3 counter selection strain. A. The URA3 reverse yeast three-hybrid system. Absence of small molecule leads to cell survival and an increase in absorption at 600nm. Presence of either 1, 5, 10 µM Mtx-Dex, or 1 µM Mtx-Cel-Dex leads to toxic URA3 reporter gene transcription, cell death, and a decrease of absorption at 600nm. The experiments were carried out in triplicate. Shown is the mean growth and the error bars represent the standard deviation from the mean. B. Reversion rate of the chemical complementation URA3 counter selection strain. Ninety three unique chemical complementation URA3 counter selection conditions (1μ M MTX-Dex) and under non-counter selection conditions (No MTX-Dex) as a control. Shown is the mean growth and the error bars represent the standard error form the mean (p-value<0.0001). C. The chemical complementation URA3 counter selection strain detects cellulase activity. Presence of 1 µM Mtx-Cel-Dex and active cellulase Cel7B leads to cell survival and an increase in absorption at 600 nm. Presence of 1 µM Mtx-Cel-Dex and inactive cellulose Cel7B:E197A leads to cell death and a decrease in absorption at 600nm. As a control, the URA3 counter selection strain was also grown in the absence of Mtx-Cel-Dex and either the presence of Cel7B or Cel7B:E197A. All experiments were carried out in triplicate. Shown is the mean growth and the error bars represent the standard deviation from the mean. D. The chemical complementation URA3 counter selection enriches for cellulase activity after five days of selection. Twenty-two cellulase variants isolated before selection and twenty-two variants isolated after five days of selection were tested for cellulase activity using carboxymethylcellulose (CMC) in crude cell extracts. Increase in aldehyde formation was detected as an increase in absorption at 660nm. Shown is the mean cellulase activity and the error bars represent the standard error form the mean (p-value<0.005).
The chemical complementation URA3 counter selection links cellulase activity to cell survival
The chemical complementation URA3 counter selection strain was adapted to detect cellulase activity by introducing the Cel7B cellulase from Humicola insolens. As figure 3C shows, expression of H. insolens Cel7B in the presence of Mtx-Cel-Dex conferred a growth advantage to the URA3 counter selection strain. Inside the cell, the Cel7B presumably cleaves the Mtx-Cel-Dex substrate, disrupting dimerization of the transcriptional activator, thus decreasing expression of the toxic URA3 reporter gene and leading to cell survival. As a negative control, we used the inactive cellulase variant Cel7B:E197A, obtained by mutation of the active-site nucleophile glutamic acid to alanine9, 21. The inactive variant lacks hydrolytic activity21, 22, thus it is unable to cleave the Mtx-Cel-Dex substrate, leading to transcription activation of the toxic URA3 reporter gene and cell death.
Assessing the utility of the cellulase selection
The utility of the cellulase selection was evaluated by isolating cellulases with improved activity from a cellulase library prepared by family DNA shuffling. First, the chemical complementation URA3 counter selection strain was confirmed to detect the cellulose activity of CelN, CelA and CelV (Figure S4). Together with H. insolens Cel7B the chemical complementation URA3 counter selection has successfully read out the four endoglucanases tested to date. The library was generated by shuffling the catalytic domains (signal peptides removed) of E. carotovora CelN, CelA, and CelV cellulases [E.C. 3.2.1.4], which share 90% sequence identity at the amino acid level and above 78–88% at the nucleotide level (Figure S5). To reach a large library size, the cellulase chimeras were introduced into the expression vector via S. cerevisiae in vivo homologous recombination23, 24 yielding a library size of 1×108 unique transformed cells. The quality of the library was determined by sequencing ten chimeras before selection. All chimeras contained sections from the three starting cellulases and an average of 10 crossovers (Figure S6).
The cellulase selection can select for cellulase variants with increased activity from a family DNA shuffled cellulase library after just five days of selection. Using a colorimetric screen based on the quantification of aldehyde formation using CMC as the substrate25, we tested if variants isolated after selection had increased cellulase activity. As figure 3D shows, the mean cellulase activity of the variants isolated after five days of selection is higher than that of variants isolated before selection. This difference is statistically significant (p-value<0.005) when analyzed using a paired two tailed Student T-test.
In Vitro Characterization of Evolved Cellulase Variants
After eight days of selection, four cellulase variants were isolated and characterized in vitro for hydrolysis of pNPC, the standard substrate for determining endoglucanase Michaelis-Menten constants. Four variants picked at random from the liquid selection after plating were overexpressed with a 6-His-tag in E. coli. Two variants had very low expression levels and no in vitro kinetics were measured. The other two variants, Cel_3.7 and Cel_5.7 showed very good expression levels (Figure S7) and 3.7-fold and 5.7-fold increases in catalytic efficiency, respectively, over the best starting cellulase, CelN (Table 1 and Figure S8).
Table 1.
Michaelis-Menten parameters of the starting cellulase genes CelN, CelA, CelV and the isolated variants using p-nitrophenyl cellobioside.
| Cellulases | Km (mM; mean ± s.d) |
kcat (s−1; mean ± s.d.) |
kcat/KM (M−1 s−1; mean ± s.d) |
|---|---|---|---|
| Cel V | 4.6 ± 0.3 | 1.10 × 103 ± 50 | 2.4 × 105 ± 1.8 × 104 |
| Cel A | 7.5 ± 0.7 | 1.45 × 103 ± 65 | 1.9 × 105 ± 2 × 104 |
| Cel N | 3.4 ± 0.2 | 1.27 × 103 ± 42 | 3.7 × 105 ± 2 × 104 |
| Cel_3.7 | 3.4 ± 0.1 | 4.5 × 103 ± 132 | 1.35 × 106 ± 7 × 104 |
| Cel_5.7 | 2.51 ± 0.08 | 5.3 × 103 ± 124 | 2.09 × 106 ± 2 ×104 |
The standard deviations were calculated from at least three independent measurements.
Sequencing of the improved cellulase variants revealed nucleotide segments from all three starting cellulases CelV, CelA and CelN (Figure S9). Cel3.7 is most similar to the parent gene CelA, while Cel5.7 is most similar to the parent gene CelN. Cel_3.7 has nine crossovers. When compared to CelA, Cel_3.7 differs by five point mutations: V86A, Q186L, L193M, R208C, and V254I. Cel_5.7 has eight crossovers. When compared to CelN, Cel_5.7 differs by eight point mutations: A86V, S126N, E154D, T172S, C208R, T262S, A267T, and A272T. To map the mutated residues onto the cellulase scaffold, homology models of Cel_3.7 and Cel_5.7 were made using the B. agaradherans Cel5A structure with a cellopentoside inhibitor26. The majority of the isolated mutations were remote from the enzyme active site.
Discussion
Together these results establish that chemical complementation provides the first high-throughput assay for cellulase catalysts, a key target in the economical conversion of cellulosic biomass to ethanol. The chemical complementation URA3 selection detected all known endoglucanases that we tested—four in total. After five days, the cellulase growth selection was shown to enrich for cellulase variants with increased activity from a family DNA shuffling library based on carboxymethylcellulose hydrolysis in cell culture. The enrichment was statistically significant, with a p-value<0.005. After eight days of selection, a cellulase variant with a six-fold increase in kcat/KM over the best parent enzyme for the hydrolysis of p-nitrophenyl cellobioside was isolated. Given that the parent cellulases are already highly active with kcat/KMs on the order of 105 M−1s−1, it is quite challenging to generate variants with improved activity. By way of comparison, it is worth noting that the most significant increase in endoglucanase activity by directed evolution in the literature is a 2.2-fold increase in activity using CMC as the substrate27. The Department of Energy’s stated goal for improvement of cellulases for biomass conversion is a 10-fold increase in activity of industrial cellulases on crystalline cellulose28.
For the first time chemical complementation has been used to detect bond cleavage as cell survival. Together with the previously reported selection for bond formation reactions9, we now have functional selections for bond cleavage and bond formation reactions. The URA3/5-FOA counter selection was chosen as the platform for the chemical complementation bond cleavage selection strain because it is the state of the art counter selection platform in the yeast two-hybrid system. Currently we are working on the design of optimized counter selections with improved dynamic ranges for biodiversity mining and directed evolution.
While applied to cellulase enzymes, the chemical complementation counter selection should be readily adapted to other glycosylhydrolases important for bioenergy production. Once the URA3 counter selection yeast strain was developed, all that was required to detect cellulase activity was the addition of the Mtx-Cel-Dex substrate and a cellulase. With the same ease, chemical complementation could be adapted to select for other hydrolase catalysts important for biomass conversion, such as hydrolases able to degrade pectin, hemicellulose, and even lignin. The generality of the chemical complementation selection for bond cleavage reactions should also allow testing other glycosylhydrolases not involved in bioenergy.
Ultimately, we envision using the chemical complementation high-throughput selection followed by a low-throughput assay on crystalline cellulose that mimics industrial bioreactor conditions to discover new, industrial useful cellulases. The chemical complementation assay could be used to search environmental DNA libraries to discover new cellulases, or for directed evolution to increase the activity of known cellulases. A low-throughput assay could then evaluate which of these cellulases had improved activity under industrial conditions. While the chemical complementation assay cannot mimic the natural polymer, crystalline cellulose substrate, notably the assay does successfully read out the four endoglucanases we have tested to date. With library sizes of ca. 108 achievable in yeast, chemical complementation has the potential to increase by several orders of magnitude the number of variants that can be initially tested, thus increasing the likelihood of discovering improved variants.
Experimental Section
URA3 counter selection yeast strain construction
Standard methods for molecular biology in S. cerevisiae and E. coli were used29, 30. The URA3 gene from pMW112 was recombined at the ura3-52 locus of FY251 (MATa trp1Δ63 his3Δ200 ura3-52 leu2Δ 1 Gal+) and selected on plates lacking uracil to give strain VC2169Y. Vectors pBC398 and pKB521 carrying the GR-B42 and LexA-DHFR constructs were transformed into VC2169Y to give strain VC2291Y. The LexA(4op)-Spo13 reporter construct was obtained by fusion PCR of the LexA(4op) from pMW112 and the Spo13-promoter from MaV9514. The reporter construct was amplified with oligos carrying 30bp homology to the URA3 promoter and the URA3 gene, recombined at the URA3 locus of VC2291Y, and selected on 5-FOA plates to give the URA3 counter selection strain VC2240Y.
Cellulase mutagenesis
The expression vector pPPY2148 was constructed by digestion of p425Met25 (ATCC 87323) with HindIII and PstI and introduction of an 800bp stuffer flanked by SfiI sites. A C-terminal 6-Histag was introduced to the catalytic domains, signal peptides removed, of the cellulases CelN31, CelA32, and CelV33, and subcloned into pPPY2148 to generate vectors pPPY2230, pPPY2234, and pPPY2236, respectively. The cellulase genes were shuffled and amplified with primers incorporating 30bp homology to the promoter and terminator of pPPY2148 and subcloned into pPPY2148 via S. cerevisiae in vivo homologous recombination.
Yeast transformation and cellulase selection
VC2240Y was transformed with the 12.5 µg of cellulase chimeras and 3.6 µg of linear pPPY2148 via electroporation using high transformation efficiency procedures with slight variations to give a library size of 108 unique transformants. After recovery, 10 µL were used to determine the library size. The rest was washed, resuspended in water and placed at 4°C for three days to eliminate any residual uracil inside the cell. Once the library was determine to be >107, it was resuspended in counter selection media (SC(HTL−) 2% gal, 2% raf, 0.2% 5-FOA, and 1 µM Mtx-Cel-Dex, at pH=5) to a final volume of 2ml. The selection was run for eight days, and samples were taken on days 5 and 8. To isolate selected variants, samples of the selection were diluted and plated under non-selective conditions.
CMC cellulase secondary screen
Cellulase activity was detected using a carboxymethylcellulose colorimetric secondary assay. This secondary screen for aldehyde formation using CMC was adapted to be carried out in a 96-well plate format25. The 22 colonies isolated before selection and the 22 colonies isolated after 5 days of selection were grown in 10mL synthetic media lacking leucine for two days. The same number of cells (2×107) from each cell culture were arrayed on a 96-well plate and lysed using 100 µL YPER (Novagen). The cell extract (80 µL) was transferred onto a second 96-well plate containing 120 µL 2% CMC (low viscosity, Sigma) in 0.1M NaOAc buffer pH 5, and the reaction was incubated at 37°C for 24hrs. The next day, 10 µL of the reaction mixture were incubated with 90µl of 0.1% tetrazolium blue (0.5M potassium sodium tartrate, 0.05M NaOH) and heated at 98°C for 10min. The reaction was cooled to room temperature and absorption at 660nm was taken immediately.
In vitro characterization of cellulases on pNPC
The three starting cellulases and the improved cellulase variants were subcloned into the E. coli T7 vector, pAED4, between NdeI and HindIII sites to give vectors pPPY2256 (CelN), pPPY2258 (CelA), pPPY2257 (CelV), pPPY2291 (Cel_3.7), and pPPY2292 (Cel_5.7). The cellulases were purified using Quick spin Ni-NTA affinity column (Qiagen). Protein concentration was determined using U.V. absorption (ε=76890, A280). To determine the kinetic constants for the hydrolysis of pNPC, the cellulase chimeras were incubated at room temperature in phosphate buffer (25mM K2HPO4, 100mM NaCl, pH 7) containing seven different concentrations of pNPC ranging from 12 mM to 0.185 mM. The release of p-nitrophenol (εpH7= 4000M−1cm−1, A420) was recorded continuously in a SpectraMax Plus 384 spectrophotometer at 420nm.
Supplementary Material
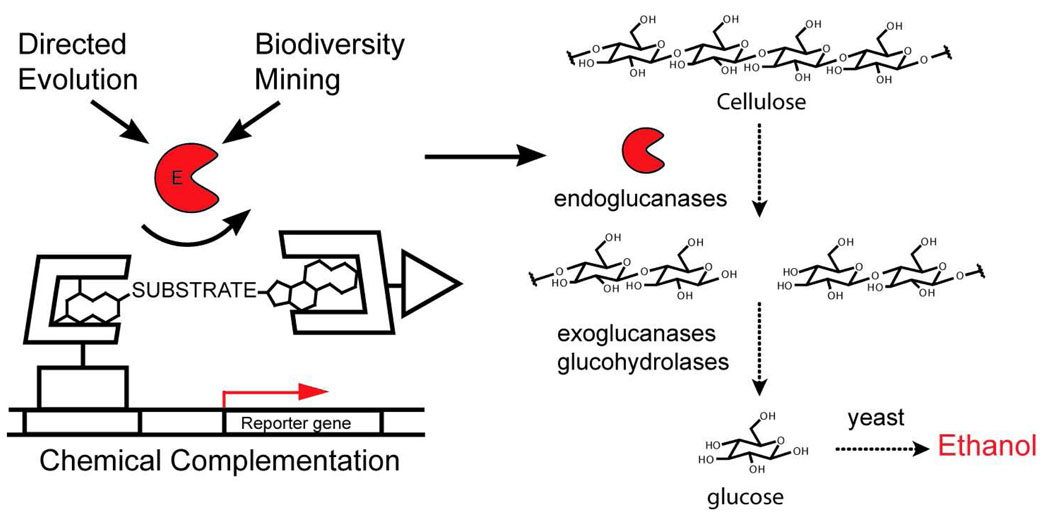
Figure 1.
Chemical complementation provides a high-throughput selection for cellulase catalysts. The discovery of improved cellulases, and other hydrolase enzymes important for biomass conversion is limited by existing medium-throughput screening technologies. Chemical complementation has been adapted to provide a high-throughput selection for bond cleavage reactions and specifically cellulose catalysts. Chemical complementation detects enzyme catalysis of bond formation or cleavage reactions based on covalent coupling of two small molecule ligands. The heterodimeric small molecule reconstitutes a transcriptional activator, turning on the transcription of a downstream reporter gene. Here, a dexamethasone (Dex)-methotrexate (Mtx) yeast three-hybrid system is used. Endoglucanase activity is detected as cleavage of a glycosidic linkage between Mtx and Dex. A cellulase selection could be used as an initial high-throughput assay to evaluate very large libraries of cellulase variants generated either by mutagenesis or isolated from biodiversity sources. Active cellulases emerging from the high-throughput assay could be then tested using low-throughput assays using plant material that more closely mimic industrial conditions.
Acknowledgment
The authors thank Prof. O. Olsen at the Carlsberg Research Center for providing the E. carotovora CelN gene, Prof. H.D. Yun at Gyenongsang National University for providing the E. carotovora CelA gene, and Prof. I. Toth at Cambridge University for providing E. carotovora subspecies atroceptica genomic DNA from which CelV gene was cloned. We also thank Prof. A. Mitchell for helpful advice on S. cerevisiae genetics and Prof. J. Ju at the Columbia Genome Center for assistance with high-throughput sequencing. We are grateful for financial support from the National Science Foundation (CHE-0350183) and the National Institutes of Health (GM62867).
Footnotes
Supporting Information Available. A more complete description of the general methods for molecular biology, strain and vector construction, DNA family shuffling, yeast transformation, secondary screening, and enzyme purification is given in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Tao H, Cornish VW. Curr Opin Chem Biol. 2002;6(6):858–864. doi: 10.1016/s1367-5931(02)00396-4. [DOI] [PubMed] [Google Scholar]
- 2.Aharoni A, Griffiths AD, Tawfik DS. Curr. Opin. Chem. Biol. 2005;9(2) doi: 10.1016/j.cbpa.2005.02.002. 210-206. [DOI] [PubMed] [Google Scholar]
- 3.Bloom JD, Meyer MM, Meinhold P, Otey CR, MacMillan D, Arnold FH. Curr. Opin. Struct. Biol. 2005;15(4):447–452. doi: 10.1016/j.sbi.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Johannes TW, Zhao H. Curr. Opin. Microbiol. 2006:261–267. doi: 10.1016/j.mib.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Kurtzman AL, Govindarajan S, Vahle K, Jones JT, Heinrichs V, Patten PA. Curr. Opin. Biotechnol. 2001;12(4):361–370. doi: 10.1016/s0958-1669(00)00228-7. [DOI] [PubMed] [Google Scholar]
- 6.Ferrer M, Martinez-Abarca F, Golyshin PN. Curr Opin Biotechnol. 2005;16(6):588–593. doi: 10.1016/j.copbio.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Cowan D, Meyer Q, Stafford W, Muyanga S, Cameron R, Wittwer P. Trends Biotechnol. 2005;23(6):321–329. doi: 10.1016/j.tibtech.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Baker K, Bleczinski C, Lin H, Salazar-Jimenez G, Sengupta D, Krane S, Cornish VW. Proc. Natl. Acad. Sci. U S A. 2002;99(26):16537–16542. doi: 10.1073/pnas.262420099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin H, Tao H, Cornish VW. J. Am. Chem. Soc. 2004;126(46):15051–15059. doi: 10.1021/ja046238v. [DOI] [PubMed] [Google Scholar]
- 10.Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. Science. 2007;315(5813):804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 11.Gray KA, Zhao LS, Emptage M. Curr. Opin. Chem. Biol. 2006;10(2):141–146. doi: 10.1016/j.cbpa.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 12.Percival Zhang YHP, Himmel ME, Mielenz JR. Biotechnol. Adv. 2006;24(5):452–481. doi: 10.1016/j.biotechadv.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Himmel ME, Ruth MF, Wyman CE. Curr Opin Biotechnol. 1999;10(4):358–364. doi: 10.1016/S0958-1669(99)80065-2. [DOI] [PubMed] [Google Scholar]
- 14.Vidal M, Brachmann RK, Fattaey A, Harlow E, Boeke JD. Proc. Natl. Acad. Sci. U S A. 1996;93(19):10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J, Schreiber SL. Proc. Natl. Acad. Sci. U S A. 1997;94(25):13396–13401. doi: 10.1073/pnas.94.25.13396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tao H, Peralta-Yahya P, Lin H, Cornish VW. Bioorg. Med. Chem. 2006:6940–6953. doi: 10.1016/j.bmc.2006.06.034. [DOI] [PubMed] [Google Scholar]
- 17.Longley DB, Harkin DP, Johnston PG. Nat Rev Cancer. 2003;3(5):330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 18.Varrot A, Schulein M, Fruchard S, Driguez H, Davies GJ. Acta Crystallogr. D Biol. Crystallogr. 2001;57(Pt 11):1739–1742. doi: 10.1107/s0907444901013993. [DOI] [PubMed] [Google Scholar]
- 19.Varrot A, Davies GJ. Acta Crystallogr. D Biol. Crystallogr. 2003;59(Pt 3):447–452. doi: 10.1107/s0907444902023405. [DOI] [PubMed] [Google Scholar]
- 20.Schou C, Rasmussen G, Kaltoft MB, Henrissat B, Schulein M. Eur. J. Biochem. 1993;217(3):947–953. doi: 10.1111/j.1432-1033.1993.tb18325.x. [DOI] [PubMed] [Google Scholar]
- 21.Ducros VMA, Tarling CA, Zechel DL, Brzozowski AM, Frandsen TP, von Ossowski I, Schulein M, Withers SG, Davies GJ. Chemistry & Biology. 2003;10(7):619–628. doi: 10.1016/s1074-5521(03)00143-1. [DOI] [PubMed] [Google Scholar]
- 22.Wang QP, Graham RW, Trimbur D, Warren RAJ, Withers SG. Journal of the American Chemical Society. 1994;116(25):11594–11595. [Google Scholar]
- 23.Cherry JR, Lamsa MH, Schneider P, Vind J, Svendsen A, Jones A, Pedersen AH. Nature Biotechnology. 1999;17(4):379–384. doi: 10.1038/7939. [DOI] [PubMed] [Google Scholar]
- 24.Colby DW, Kellogg BA, Graff CP, Yeung YA, Swers JS, Wittrup KD. Methods Enzymol. 2004;388:348–358. doi: 10.1016/S0076-6879(04)88027-3. [DOI] [PubMed] [Google Scholar]
- 25.Jue CK, Lipke PN. J. Biochem. Biophys. Methods. 1985;11(2–3):109–115. doi: 10.1016/0165-022x(85)90046-6. [DOI] [PubMed] [Google Scholar]
- 26.Schwede T, Kopp J, Guex N, Peitsch MC. Nucleic. Acids. Res. 2003;31(13):3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y-S, Jung H-C, Pan F-G. Appl. and Environ. Microbiol. 2000;66(2):788–793. doi: 10.1128/aem.66.2.788-793.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cellulose Cost Reduction Contracts. Department of Energy. 2007 [Google Scholar]
- 29.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 30.Adams A, Gottschling D, Kaiser C, Stearns T. Methods in Yeast Genetics. Plainview, New York: Cold Spring Harbor Lab Press; 1998. [Google Scholar]
- 31.Olsen O, Thomsen KK, Weber J, Duus JO, Svendsen I, Wegener C, von Wettstein D. Biotechnology (N Y) 1996;14(1):71–76. doi: 10.1038/nbt0196-71. [DOI] [PubMed] [Google Scholar]
- 32.Park YW, Lim ST, Cho SJ, Yun HD. Biochem. Biophys. Res. Commun. 1997;241(3):636–641. doi: 10.1006/bbrc.1997.7747. [DOI] [PubMed] [Google Scholar]
- 33.Bell KS, et al. Proc. Natl. Acad. Sci. U S A. 2004;101(30):11105–11110. doi: 10.1073/pnas.0402424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.