Abstract
Non-natural oligomers have recently shown promise as functional analogues of lung surfactant proteins B and C (SP-B and SP-C), two helical and amphiphilic proteins that are critical for normal respiration. The generation of non-natural mimics of SP-B and SP-C has previously been restricted to step-by-step, sequence-specific synthesis, which results in discrete oligomers that are intended to manifest specific structural attributes. Here we present an alternative approach to SP-B mimicry that is based on sequence-random copolymers containing cationic and lipophilic subunits. These materials, members of the nylon-3 family, are prepared by ring-opening polymerization of β-lactams. The best of the nylon-3 polymers display promising in vitro surfactant activities in a mixed lipid film. Pulsating bubble surfactometry data indicate that films containing the most surface-active polymers attain adsorptive and dynamic-cycling properties that surpass those of discrete peptides intended to mimic SP-B. Attachment of an N-terminal octadecanoyl unit to the nylon-3 copolymers – inspired by the post-translational modifications found in SP-C – affords further improvements by reducing the percent surface area compression to reach low minimum surface tension. Cytotoxic effects of the copolymers are diminished relative to that of an SP-B-derived peptide and a peptoid-based mimic. The current study provides evidence that sequence-random copolymers can mimic the in vitro surface-active behavior of lung surfactant proteins in a mixed lipid film. These findings raise the possibility that random copolymers might be useful for developing a lung surfactant replacement, which is an attractive prospect given that such polymers are easier to prepare than are sequence-specific oligomers.
INTRODUCTION
Lung surfactant proteins SP-B and SP-C are required for the biophysical activity of lung surfactant (LS), the complex lipid-protein mixture that coats the internal air-liquid (a/l) interface of the vertebrate lung and reduces the work of breathing.1-2 Both proteins (~ 1.5-2.0 combined weight percent of natural LS) contain a high proportion of lipophilic residues, and they adopt amphiphilic and helical conformations. The main function of LS is to regulate surface tension (γ, mN m−1) in the alveoli, tiny sacs that mediate gas exchange between the blood and air spaces of the lung, by optimizing available surface area, maximizing lung compliance, and stabilizing the alveolar network against collapse.3 Three crucial functional characteristics of LS are: (1) rapid adsorption to the a/l interface, (2) near-zero γ upon film compression, and (3) efficient re-spreading of material and minimal loss to the subphase through multiple breathing cycles.4
Deficient or dysfunctional LS results in infant or acute respiratory distress syndromes (IRDS or ARDS, respectively).5-6 Although no current exogenous treatment for ARDS exists,7 IRDS can be successfully treated with porcine- or bovine-derived surfactant replacement therapies (SRTs).8 Use of animal-derived substances, however, is non-ideal because of the risk of zoonotic infection and the high cost of large-scale extraction, isolation, and purification. To eliminate dependence on animal-derived material, many groups have sought to develop biomimetic LS replacements based on synthetic surfactant protein analogues.4 This approach could lead to a safe and bioavailable alternative to SRTs that may be able to treat or mitigate both IRDS and ARDS.
SP-B in monomeric form is a 79-residue protein (8.7 kDa) with a net cationic charge, and is postulated to contain four or five facially amphiphilic helices. SP-B forms four disulfide bonds: three intramolecular connections that presumably constrain conformational flexibility, and one intermolecular bond that results in homodimerization.9-12 SP-C contains just 35 residues and forms a single helix.13 This protein has two palmitoylation points (positions 5 and 6), two cationic residues (11 and 12), and an extremely lipophilic poly-valine helix that approximates the length necessary for spanning a lipid bilayer.12,14-17 The sequences and structural attributes of both proteins are highly conserved across mammalian species, implying that these features are necessary for their ability to organize and regulate lipid film formation, and to anchor the film to the a/l interface.3,18 Unfortunately, these attributes render the proteins very troublesome to obtain on a large scale by extraction or chemical synthesis; efforts to synthesize SP-B or SP-C or fragments thereof are often hampered by misfolding or aggregation.4
The main approaches toward functional mimicry of surfactant proteins have involved peptide fragment synthesis, limited dimerization of SP-B and its analogues, recombinant protein expression, and more recently, peptoid synthesis, the subject of our recent contributions in this field.19-24 Although a recombinant form of SP-C is available,25 it is not palmitoylated, and no recombinant form of SP-B has yet been reported. Chemically synthesized, surface-active peptide fragments of SP-B such as SP-B1-2526-27 and the dimeric constructs dSP-B1-2528 and “Mini-B”,29 have demonstrated in vitro and in vivo success, but the challenge of generating these materials on a large scale is a stumbling block to pharmaceutical development. The chronic problem of achieving the desired extent of dimerization and multiple amphiphilic helices when mimicking SP-B has prompted recent endeavors to determine whether the incorporation of dimerization points can be circumvented while retaining good surfactant activity.24,30
The high cost of large-scale, step-wise synthesis and purification represents a significant barrier to the development of peptide-based drugs and has generated an interest in alternatives for use in an LS replacement.4 Non-natural oligomers, such as peptoids,31 β-peptides32-33 and α/β-peptides34-35 can circumvent some peptide-associated problems, including irreversible aggregation and protease susceptibility; however, step-wise synthesis is required for preparation of these sequence-specific oligomers, and reversed-phase high performance liquid chromatography (RP-HPLC) is necessary for their purification.32,36 Therefore, although these types of peptide mimics can display impressive biological activities and thereby shed light on relationships between molecular structure and resultant biophysical activities, sequence-specific non-natural oligomers do not alleviate the production cost problem.
It is generally assumed that the function of a protein depends upon the sequence of amino acid residues and the three-dimensional arrangement of amino acid side chains (or a subset thereof) that results from adoption of a specific secondary or tertiary structure. Recently, however, we proposed that discrete sequences and folding patterns may not be necessary for mimicry of the cell type-selective toxicity of host-defense peptides.37-38 Many of these natural antimicrobial peptides adopt helical conformations upon interaction with bacterial membranes and ultimately compromise the barrier function of the membrane; these peptides are generally selective as membrane-disrupting agents, acting on bacteria in preference to eukaryotic cells.39 We have shown that nylon-3 copolymers (poly-β-peptides) containing sequence-random mixtures of lipophilic and cationic subunits can mimic the selective antibacterial activity of helical host-defense peptides.40-42 Based on this finding, we hypothesized that nylon-3 copolymers might be able to mimic lung surfactant proteins SP-B and SP-C, the function of which depends upon interaction with lipids,18 as is true of natural antimicrobial peptides.
Here we describe flexible, sequence-random nylon-3 copolymers that are intended to mimic lung surfactant protein B. The efficacy of the designs was estimated by in vitro surfactant behavior in a mixed lipid film. Surface activity has been evaluated by pulsating bubble surfactometry (PBS)43-44 of copolymers in a Tanaka lipid (TL) film45 (1,2-diacyl-sn-glycero-3-phosphocholine (DPPC): 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (POPG): palmitic acid (PA) 68:22:9 by weight). Our materials, which were synthesized via anionic ring-opening polymerization of β-lactams,42,46 display increased surface-active behavior relative to known peptide- and peptoid-based SP-B mimics, with values approaching that of porcine-derived SP-B in the TL film.30 These nylon-3 copolymers display lower toxicity toward mammalian cells than do an SP-B-derived peptide and a peptoid-based mimic. Our polymers are heterochiral because they were prepared from racemic β-lactams; therefore, these nylon-3 copolymers presumably cannot adopt specific, regular, or patterned conformations. Thus, our results challenge the notion that a helix or other regular conformation is strictly required if a molecule is to achieve global amphiphilicity (i.e., global segregation of lipophilic and hydrophilic subunits), a property that is thought to be necessary for SP-B-like activity. We postulate that the nylon-3 copolymers are able to achieve global amphiphilicity in irregular conformations after association with lipids, which facilitate their surfactant behavior.
RESULTS
Copolymer Design, Synthesis, and Characterization
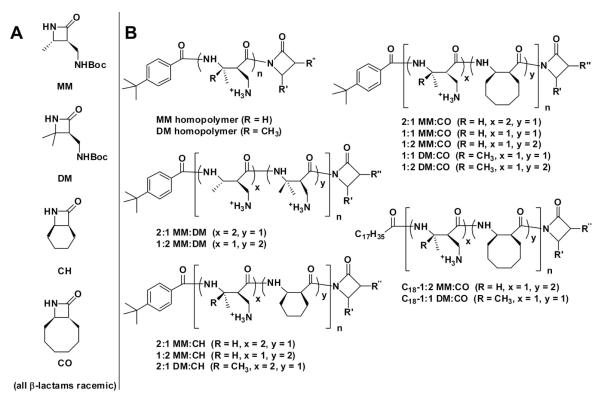
All monomers and polymers described here were prepared using previously reported procedures;40,42,46 their chemical structures are presented in Figure 1. The main variables in copolymer design were N-terminal modification and subunit composition. The β-lactams we employed gave rise both to lipophilic subunits, CH (for “cyclohexyl”) or CO (“cyclooctyl”), and to cationic subunits, MM (“monomethyl”) and DM (“dimethyl”). Incorporation of these subunits ensured that our polymers bear a net positive charge in aqueous solution and are amphiphilic, as is true of SP-B itself. However, in contrast to SP-B, our polymers presumably cannot achieve global segregation of lipophilic and cationic side chains by adopting a specific, regular, or patterned conformation, because the sequence of lipophilic and cationic subunits varies among polymer molecules. In addition, because the β-lactam monomers are racemic, stereochemistry varies among polymer chains.
Figure 1. Monomers (A) and polymers (B) examined in this work.
The copolymers are sequence-random. Because the β-lactams are racemic, the polymers are heterochiral and stereorandom. The C-terminal imide unit is derived from the β-lactam(s) used in the polymerization reaction. Thus, for each copolymer, R’ and R” could correspond to either of the β-lactam precursors. All polymers are cationic and were isolated as trifluoroacetate salts. Nomenclature, characterization, and synthesis details are presented in Supporting Information (Table S1).
The CH and CO subunits in our nylon-3 copolymers were intended to mimic the roles of lipophilic side chains and cyclically constrained residues in SP-B. The so-called “insertion region” of human SP-B, residues 1-9, contains a high proportion of residues that are aromatic and lipophilic (Phe1, Tyr5, and Trp8) or that are cyclically constrained (Pro2, Pro4, and Pro6). In previous work, this region was shown to be critical for the γ-reducing behavior of SP-B.47-48 Furthermore, when aromatic residues or prolines were substituted with alanine, the resulting peptides showed significantly decreased surface activity.48 It has been hypothesized that the N-terminal region of SP-B inserts transiently into the lipid layer with Trp as an anchor,49 thus allowing the protein to function overall as a lipid organizer and transporter.
The designations we employed for the nylon-3 polymers (Figure 1) indicate the subunit identities and proportions, the latter determined by the ratio of β-lactams used in the polymerization reaction. The subunit sequence is random within the copolymers, and because the β-lactams were racemic, each polymer sample contains a mixture of backbone configurations. Most of the polymers have a p-(tert-butyl)benzoyl group at the N-terminus, but two of the copolymers were prepared with an octadecanoyl group at the N-terminus in an attempt to introduce an SP-C-like lipophilic tail into our SP-B mimetic polymers. We previously examined peptoid-based SP-C analogues that bear one or two octadecyl groups at the N-terminus.50 The favorable impact of the lipophilic tail(s) on surface activity prompted us to examine similar modifications of peptoid-based SP-B analogues, and again we observed a marked improvement in surfactant activities relative to unalkylated analogues.30 These findings with sequence-specific surfactant protein mimics led us to explore analogous N-terminal modifications of nylon-3 copolymers.
The β-lactams required for nylon-3 copolymer synthesis were prepared by previously published methods involving the [2+2] cycloaddition of chlorosulfonyl isothiocyanate (CSI) to alkenes. Anionic ring-opening copolymerization of the β-lactams51-52 followed by acid-mediated removal of the t-butylcarbamate protecting groups yielded nylon-3 materials that are cationic at neutral pH.40 Gel permeation chromatography (GPC) performed before deprotection indicated polydispersity indices (PDI) in the range 1.04-1.18 (see Supporting Information, SI). We were unable to measure polydispersity after side chain deprotection, but we assume that this process does not alter polydispersity. NMR measurements suggested that deprotection reactions proceeded to completion (see SI).
Copolymers identified as promising in early PBS screenings (1:2 and 2:1 MM:CO, and 1:1 and 2:1 DM:CO) were re-synthesized to establish the reproducibility of the polymer preparation protocol, as manifested in surface activity and cytotoxicity. For these polymers, different batches are denoted as a, b and c.
Pulsating Bubble Surfactometry: Static-Bubble Mode
The immediate adsorptive, γ-reducing effect, and stability of the films over time at a bubble a/l interface were assessed via PBS in static-bubble mode. Polymers dried with lipids (Tanaka Lipids, TL, DPPC:POPG:PA 68:22:9 [wt:wt:wt]) were suspended in aqueous buffer (150 mM NaCl, 10 mM HEPES, 5 mM CaCl2, pH 6.9) at 37 °C and allowed to adsorb to the interface of a 0.40 mm radius bubble for 20 minutes, yielding γ as a function of time. In this study, it is crucial for surface-active films to both adsorb rapidly and reach a low ‘equilibrium’ or final γ in the time tested. Therefore, a mimic is considered very surface-active in the lipid film if it adsorbs to ~ 25 mN m−1 within ~ 1-5 minutes; for instance, Infasurf®, an animal-derived SRT, attains a low γ ~ 23 mN m−1 within ~ 1-2 minutes on the PBS,44 and TL + porcine-derived SP-B films reach ~ 26 mN m−1at 5 minutes.30 Select polymer adsorption traces are depicted in Figure 2, while the adsorbed γ of all the lipid films containing polymers and positive controls, including lipids alone (TL), the peptides SP-B1-25 and KL4 (the latter is a peptide-based SP-B mimic with broad, biomimetic cationic residue patterning),53 and aromatic-rich Peptoid B1,20 are presented at 5 minutes adsorption in Figure 3, Panels A-D. Mean adsorptions (γ) ± standard deviation (σ) at selected time intervals for all films are available in the Supporting Information (Tables S2 and S3). For static-bubble experiments, relative closeness of γ values for different lipid-polymer films was largely determined by comparing the σ of the mean γ for the films; however, as a guideline, we consider that a difference of ≥ 2 mN m−1 among adsorptive γ values of different films is significant.
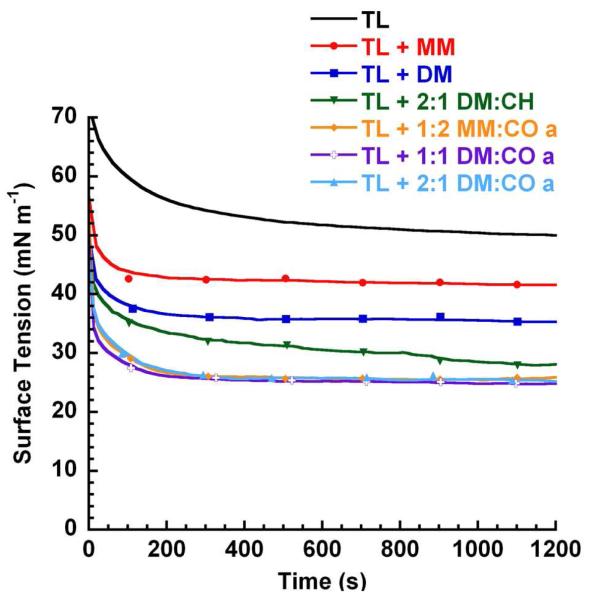
Figure 2. PBS Adsorption Plot for Lipid-Polymer Films in Static-Bubble Mode at 37 °C.
Representative static-bubble adsorption traces for Tanaka lipids alone (TL) and TL + 10 relative weight % of each mimic in an aqueous buffer (150 mM NaCl, 10 mM HEPES, 5 mM CaCl2, pH 6.9) suspension at 37 °C.
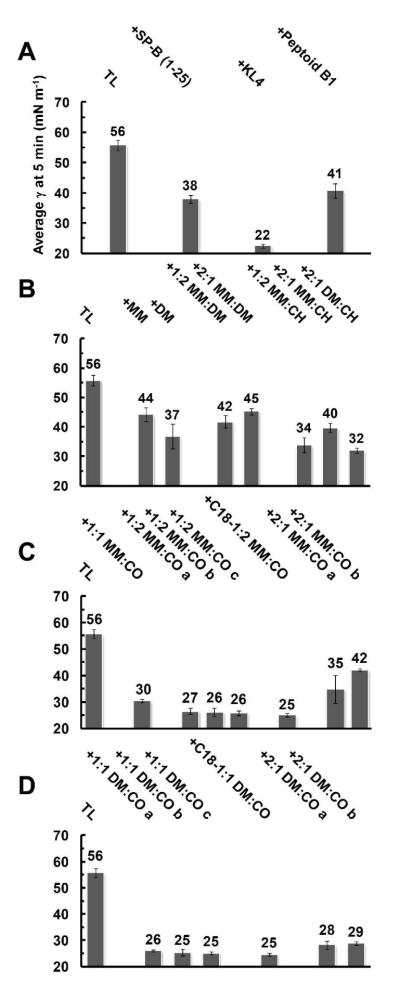
Figure 3. PBS Adsorption Data for Lipid-Polymer Films in Static-Bubble Mode at 37 °C.
Mean γ (mN m−1) at 5 minutes adsorption are presented for all films including polymers or positive controls in Panels A-D. Error bars are the standard deviation of the mean (σ). See SI (Tables S2-S3) for mean adsorption γ data ± σ at selected time intervals.
Any additive to the lipid film significantly improved adsorptive characteristics relative to TL alone, with TL + KL4 exhibiting the most positive surfactant activity among the positive controls, reaching 22 mN m−1 at 5 minutes (Figure 3, Panel A). Control TL + SP-B1-25 was more active than TL + B1, but both were less active than TL + KL4 (see adsorptive traces for positive control films in SI, Figure S29). All of the nylon-3 polymers displayed surface activity in this assay. Among the nylon-3 homopolymers, TL + DM yielded a lower γ than TL + MM, and an analogous trend was evident among the films containing MM:DM copolymers. The introduction of lipophilic CH subunits to polymers in the lipid films moderately improved the γ’s reached (Figure 3, Panel B, and SI, Table S2), but impeded the adsorption rate relative to films containing polymers with entirely cationic subunits (MM homopolymer, DM homopolymer, or MM:DM copolymers). Among CH-containing copolymer-lipid films, an increasing proportion of lipophilic CH subunits (1:2 MM:CH vs. 2:1 MM:CH) or replacement of MM subunits with slightly more lipophilic DM subunits (2:1 DM:CH vs. 2:1 MM:CH) also resulted in lower γ’s during adsorption (Figure 3, Panel B, and SI, Table S2).
The best surface activities were observed among films with nylon-3 copolymers containing CO subunits. Both TL + 1:1 DM:CO and TL + 1:2 DM:CO displayed excellent activities, with γ reaching 26-28 mN m−1 after as little as 2.5 minutes of adsorption (Figure 2; Table S2). Replacing the p-(tert-butyl)benzoyl group at the N-terminus with an octadecanoyl group did not affect PBS static-mode surface activity (films containing 1:1 DM:CO copolymers). Raising the cationic subunit proportion to 67% led to a significant decrease in surface activity, and among these polymers, the use of the DM subunit provided improvement relative to use of the MM subunit (2:1 DM:CO vs. 2:1 MM:CO). In addition, the adsorptive activities of the nylon-3 copolymers were reproduced in subsequent batches (Figure 3, Panels A-D, also see SI, Tables S2-S3).
Pulsating Bubble Surfactometry: Dynamic-Bubble Mode
The dynamic surfactant activity during changes in volume or film surface area provides an indication of film sustainability over time. Bubble pulsation at the approximate adult respiratory rate of 20 cycles per minute (cpm) in PBS dynamic-bubble mode at 37 °C permits a simplified evaluation of such dynamic film behavior.44 In dynamic mode, after static-bubble adsorption, the bubble was subsequently pulsed for 10 minutes and γ was recorded with respect to surface area. In Figure 4, Panels A-D depict the attained maximum and minimum γ (γmax/min) at 5 minutes of cycling for all lipid films containing polymers or positive controls. Mean γmax/min’s ± σ at selected time intervals for up to 10 minutes of cycling are located in SI (Tables S4 and S5).
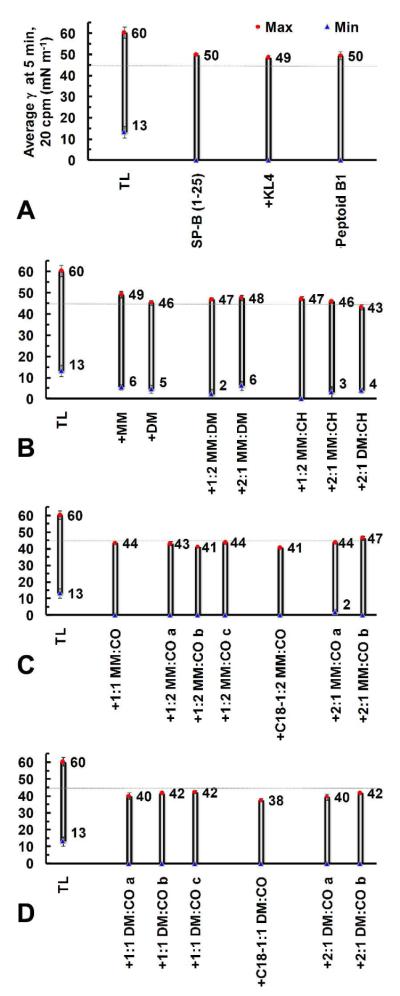
Figure 4. PBS Data for Lipid-Polymer Films in Dynamic-Bubble Mode at 5 Minutes, 37 °C.
Mean maximum (γmax, red circles) and minimum (γmin, blue triangles) surface tensions (γ, mN m−1) at 5 minutes of dynamic-bubble pulsation, 20 cycles per minute (cpm), are presented for all lipid films containing polymers or positive controls in an aqueous buffer (150 mM NaCl, 10 mM HEPES, 5 mM CaCl2, pH 6.9) suspension at 37 °C (Panels A-D). The faint line at 45 mN m−1 represents the defined activity threshold for a very surface-active mimic. See SI (Tables S4-S5) for mean dynamic γ data ± σ at selected time intervals.
Representative single pulsation γ-surface area (SA) bubble hysteresis loops at 5 minutes cycling for select lipid-polymer films are presented in Figure 4, Panels A-D. Bubble expansion corresponds to a clockwise loop direction, and vice versa for compression. The absence of low-γ data in some loops is caused by the limited ability of the image analysis system to trace the bubble shape in this regime.44 The highly compressed state of the film, which enables it to reach near-zero γ, often causes significant bubble shape deformation. This deformation may prevent the level of bubble tracing needed to obtain SA and to calculate γ via the ellipsoidal Laplace equation.44 However, visual, real-time bubble inspection during the experiment confirmed that γ reached near-zero in these films.44 In addition, lipid-polymer films that did not reach < 1 mN m−1 could be accurately traced to the minimum value reported and never exhibited significant bubble deformation. Although bubble size was not uniform for every experiment, differences in x-axis positioning (SA) had no appreciable effect on γ (data not shown).
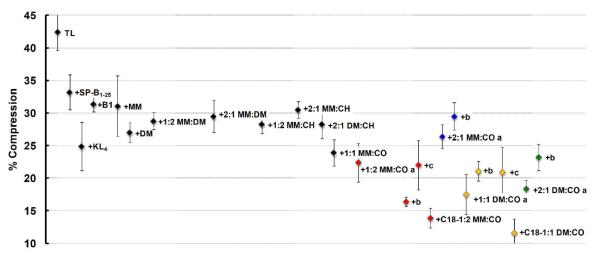
The key features for good surfactant activity of a film in dynamic-mode are a reduced γmax and a γmin near zero, with the latter attribute absolutely essential for normal respiration. The PBS cycling loop for Infasurf® maintains a γmax of ~ 35 mN m−1 and a γmin near-zero,44 while the TL + porcine-derived SP-B film exhibits a γmax of ~ 36 mN m−1 and near-zero γmin.30 To enable normal respiration and minimize the work of breathing, a near-zero γmin should occur upon compression of the first cycle, and reduced γmax/min values should remain throughout the time tested with minimal SA compression to reach near-zero γmin. The latter parameter is monitored as percent SA compression to reach 20 mN m−1, and is graphically represented in Figure 5. Tabulated data are available in the SI (Table S6). The TL + SP-B film displayed ~ 21% compression to reach 20 mN m−1.30
Figure 5. PBS Percent Compression Data for Films in Dynamic-Bubble Mode at 5 Minutes, 37 °C.
The corresponding mean percent surface area (SA) compression (percent compression) to reach 20 mN m−1 at 5 minutes pulsation is depicted. Percent compression is defined here as 100*[(SAmax – SA20)/(SAmax)], where SAmax was the maximum bubble surface area (SA) at expansion, and SA20 was the SA at which γ first reaches 20 mN m−1 upon compression at 5 minutes pulsation. Error bars are the standard deviation of the mean (σ). See SI (Tables S4-S6) for mean dynamic γ data ± σ at selected time intervals and tabulated % Compression data ± σ.
Although the significance of γmax is debated with regard to the efficacy of an additive (peptide, peptoid, etc.) as a lung surfactant protein mimic, and no specific criteria exist for estimating the ideal value of γmax, it has been established that γmin must reach < 2 mN m−1 in an lipid-additive film if the additive is to be considered an effective mimic.18 In light of the high TL γmax of ~ 60 mN m−1 and the low TL + SP-B γmax of ~ 36 mN m−1, we conclude that a very surface-active mimic should exhibit a γmax of ≤ 45 mN m−1 and a γmin of < 2 mN m−1. It should be noted that the former criterion is ~ 5 mN m−1 below the γmax of all peptide- and peptoid-based positive controls. Relative closeness of γ values for different lipid-polymer films were largely determined by comparing the σ of the mean γ for the films. We consider a difference in γmax of > 3 mN m−1 and a difference in γmin of ≥ 2 mN m−1 between films to be significant.
As in static-bubble mode, the addition of any mimic to the TL film in dynamic-bubble mode significantly reduced γmax/min values, demonstrating surfactant activity (Figure 4). The TL film manifested poor surface activity, with a high γmax ~ 60 mN m−1, a γmin ~ 13 mN m−1, and ~ 43% SA compression to reach 20 mN m−1 (Figure 4, Panel A, and Figure 5). Among the films containing positive control compounds, addition of SP-B1-25, KL4, or Peptoid B1 to the film reduced γmax to ~ 50 mN m−1 and γmin to near zero, with TL + KL4 displaying the lowest percent SA compression of the positive controls, at ~ 25%. Lipid films containing MM homopolymer, DM homopolymer, or MM:DM copolymers displayed slightly reduced γmax relative to the controls, but none of these purely cationic polymers was able to achieve γmin < 2 mN m−1, and all displayed percent SA compressions in the range between films containing SP-B1-25 and KL4 (Figure 4, Panel B, and Figure 5).
The inclusion of CH in nylon-3 copolymers slightly reduced the γmax relative to lipid-MM and lipid-DM films; the lowest γmax observed among these polymers was for TL + 2:1 DM:CH at ~ 43 mN m−1, but the γmin only reached ~ 4 mN m−1 in this case (Figure 4, Panel B). TL + 1:2 MM:CH reached a near-zero γmin, which was not achieved among lipid-polymer films containing purely cationic polymers. However, the CH-containing copolymers exhibited percent SA compressions in the realm of peptide-based controls (Figure 5).
Lipid films containing nylon-3 copolymers with CO subunits displayed surfactant activity that was remarkably enhanced relative to all other copolymers, with significantly lower percent SA compression values, near-zero γmin in most cases, and consistently low γmax (~ 44 mN m−1 for TL + 1:1 MM:CO and TL + 2:1 MM:CO a, ~ 43 mN m−1 for TL + 1:2 MM:CO a, and ~ 40 mN m−1 for TL + 1:1 DM:CO a and TL + 2:1 DM:CO a (Figure 4, Panels C-D, and Figure 5)). TL +2:1 MM:CO a did not quite reach γmin = 0 (~ 1.8 mN m−1 ± 1.6 mN m−1) and demonstrated a slightly higher percent compression (~ 26%) than other CO-containing polymers (~ 18-24%, Figure 5). As in static-bubble mode, the dynamic activities of the nylon-3 copolymers were reproduced in subsequent batches (Figures 4-5, also see SI, Tables S4-S5).
Inclusion of copolymers with an N-terminal octadecanoyl group, C18-1:1 DM:CO and C18-1:2 MM:CO, resulted in the greatest surface activity improvements to the lipid film. The TL + C18-1:2 MM:CO film reached γmax ~ 41 mN m−1, which matched that of the most surface-active batch of TL + 1:2 MM:CO (b), and the sample containing the octadecanoyl end group displayed a percent compression roughly ~ 2% lower (~ 14% vs. ~ 16% for TL + 1:2 MM:CO b). TL + C18-1:1 DM:CO demonstrated the highest surface activity, with γmax ~ 38 mN m−1 and a low ~ 12% SA compression.
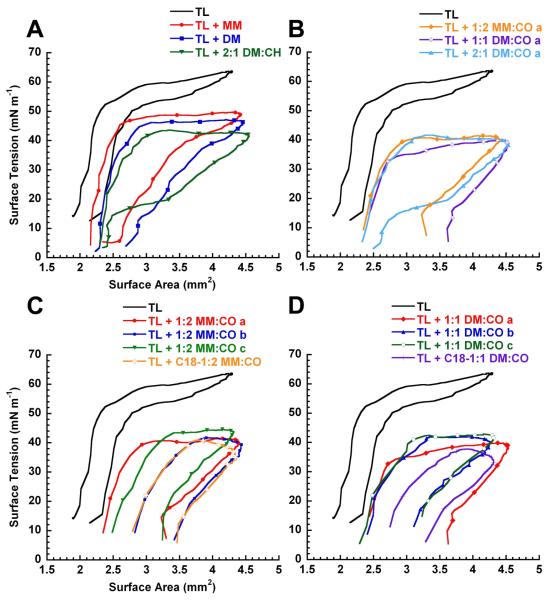
In addition to low γmax/min and percent SA compression values, a loop shape during dynamic cycling that is qualitatively similar to natural LS or TL + SP-B is considered desirable and an indication of high surfactant activity. Although the significance of loop shape and hysteresis have yet to be clearly defined, it has been observed that Infasurf®44 and the TL + SP-B30 film exhibit ‘knob-like’ loop shapes that have a small extent of hysteresis and no dramatic changes in slope or shape upon expansion or compression. In contrast, the TL film had a significantly different but characteristic loop shape that resulted in a high percent SA compression to reach the γmin (Figure 5, and Figure 6, Panels A-D).
Figure 6. PBS Bubble Pulsation Hysteresis Loops for Lipid-Polymer Films in Dynamic-Bubble Mode, 5 Minutes, 37 °C.
Representative loops for lipid-polymer films after 5 minutes of pulsation (20 cpm) in dynamic-bubble mode on the PBS (Panels A-D) in an aqueous buffer (150 mM NaCl, 10 mM HEPES, 5 mM CaCl2, pH 6.9) suspension at 37 °C. In dynamic cycling, bubble surface area expansion is clockwise from left to right, and vice versa for compression. See SI (Figure S30) for TL + 2MM:1CO a and b loops. Also see SI (Tables S4-S6) for mean dynamic γ data ± σ at selected time intervals at tabulated % Compression data ± σ.
The loop shapes of TL + MM homopolymer or TL + DM homopolymer were very similar to one other, as can be seen in Figure 6, Panel A; a similar loop shape was also observed for TL + SP-B1-25, + KL4, and + Peptoid B1 (SI, Figure S29). TL + 2:1 DM:CH also had a similar loop shape, but a distinctive feature was present upon compression, starting at ~ 20 mN m−1, wherein large changes in SA occurred with relatively small changes in γ. The loop features remained consistent for films containing MM:CO and DM:CO copolymers, aside from a lower γmax, with the distinctive compression feature observed for TL + 2:1 DM:1CO a (Figure 6, Panel B).
The loop shapes remained fairly consistent among different batches of MM:CO and DM:CO copolymers, although there were some inter-batch variations in hysteresis (TL + 2:1 DM:CO a vs. b; Figure S30). Small variations in hysteresis are not uncommon between lipid-only batches or films containing different batches of peptides or peptoids. Interestingly, in the case of TL + 1:2 MM:CO b, TL + C18-1:2 MM:CO, and TL + C18-1:1 DM:CO, the loop shape closely resembled that seen for Infasurf®44 and TL + SP-B films (Figure 6, Panels A & B).30 The ‘knob-like’ shape at expansion and low amount of hysteresis are very characteristic for Infasurf® and TL + SP-B films; the presence of these features in some TL + polymer films suggests that these materials may be very effective LS protein mimics.
Cytotoxicity Assay Against NIH 3T3 Fibroblasts
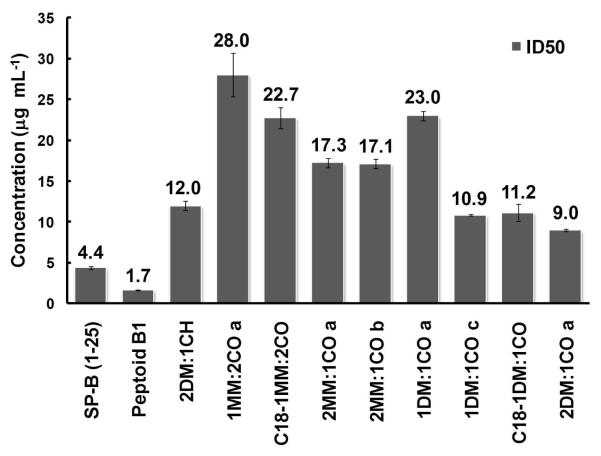
The biocompatibility of select nylon-3 copolymers was evaluated with NIH 3T3 fibroblasts using an MTS colorimetric assay. Figure 7 shows that the 50% metabolic inhibitory dose (ID50) of all tested copolymers compared favorably to that of the SP-B1-25 peptide and Peptoid B1. Copolymers 1:2 MM:CO a, C18-1:2 MM:CO, and 1:1 DM:CO a were the least toxic materials, with ID50 values more than five-fold higher than that of SP-B1-25, and nearly 15-fold higher than that of Peptoid B1. Interestingly, an increased proportion of charged subunits correlated with increased cytotoxicity (2:1 MM:CO vs. 1:2 MM:CO, and 2:1 DM:CO vs. 1:1 DM:CO). Polymers bearing an N-terminal octadecanoyl group exhibited equal or greater cytoxicity relative to analogous polymers bearing an N-terminal p-(tert-butyl)benzoyl group. This trend may result from the increased hydrophobicity of the octadecanoyl group relative to the p-(tert-butyl)benzoyl group, which should lead to enhanced lipid binding for the former relative to the latter. The highest toxicities (lowest ID50 values) were observed for copolymers with a high proportion of DM subunit, 2:1 DM:CH (~ 12 μg mL−1) and 2:1 DM:CO (~ 9 μg mL−1). Polymers with a comparable proportion of MM were not as toxic (2:1 MM:CO vs. 2:1 DM:CO). This trend may arise from the increased hydrophobicity of DM subunit relative to the MM subunit.
Figure 7. Cytotoxicity Assay Data for Polymers and Peptide or Peptoid Positive Controls.
Cytotoxicity data for select polymers, the SP-B1-25 peptide, and Peptoid B1 against NIH/3T3 fibroblast cells. Error bars represent the standard error of the mean (SEM).
DISCUSSION
The current study demonstrates that nylon-3 copolymers with random sequences of cationic and lipophilic subunits and random backbone stereochemistry can display in vitro surfactant activity in mixed lipid films analogous to that of SP-B or of a sequence-specific peptide- or peptoid-based surfactant protein B mimic. In many cases, the surface activities of the polymers match or exceed those of peptide- and peptoid-based positive controls. The cytotoxicities of the nylon-3 copolymers toward mammalian cells are lower than those of a well-studied peptide fragment of SP-B and Peptoid B1. These results are highly significant because the polymers presumably cannot adopt a regular conformation that would lead to global segregation of lipophilic and cationic side chains. In addition, the copolymers are much less costly to prepare than are sequence-specific oligomers such as peptides.
The polymer synthesis method we employ46 enables facile variation of subunit identities and the N-terminal group, and we took advantage of this capability to explore a small set of nylon-3 materials as LS protein mimics. Variations in polymer composition led to clear trends in terms of in vitro surface activity. In adsorption experiments, copolymers containing both lipophilic and cationic subunits display the most promising behavior. We examined two different lipophilic subunits, CH and CO. Incorporation of CO subunits (Figure 1) was required to achieve properties similar to those of SP-B in the same lipid film (these properties of SP-B are described in a submitted manuscript).24 The best properties were observed for copolymers containing > 50% CO subunits, with the remainder of the subunits cationic (MM or DM). Variation in the N-terminal group, p-(tert-butyl)benzoyl vs. octadecanoyl, had little impact on the static measurements. However, in the dynamic cycling measurements, the octadecanoyl end group proved to be superior.
The structural difference between the two cationic subunits we employed, DM and MM, is relatively subtle: DM contains an additional CH2 unit relative to MM. Nevertheless, favorable effects of DM vs. MM are noticeable in the dynamic measurements. The slower adsorption of CH-containing polymers, despite reaching lower surface tensions relative to purely cationic polymers (MM, DM, and MM/DM), could be attributed to the ability of the purely cationic polymers to increase the lipid interfacial adsorption rate through primarily Coulombic interactions between the charged lipid head groups and the cationic subunits. However, the purely cationic polymers result in undesirably high surface tensions relative to CH-containing polymers because they lack of a strong hydrophobic component. In the case of CH vs. CO-containing polymers, the larger, more hydrophobic CO subunit may enhance the adsorption rate as well as lower surface tension values through stronger hydrophobic associations between the CO-containing subunits and the lipid acyl chains, relative to CH-containing polymers.
Films containing DM:CO copolymers reached a slightly lower γmax, more consistently attained a near-zero γmin, and exhibited significantly less percent surface area compression to reach a surface tension of 20 mN m−1 relative to films containing MM:CO copolymers. In these two copolymer families, placement of an octadecanoyl group at the N-terminus was quite beneficial, leading to the lowest γmax/min values and the lowest percent surface area compression to reach 20 mN m−1 of all the lipid-polymer films we examined. In addition, for the N-terminal octadecanoyl polymers, the loop shape manifested by the lipid-copolymer films is substantially more like that of Infasurf®44 or of SP-B in a lipid film (submitted manuscript),30 exhibiting significantly less hysteresis and more uniform loop shape, relative to the N-terminal p-(tert-butyl)benzoyl polymers.
Protein lipidation as a natural modification enhances protein-membrane interactions and lipid-associated functioning,54 and examples of designed peptide-fatty acid conjugation to enhance biological activity are present in the literature.55 In our nylon-3 copolymers, replacing the p-(tert-butyl)benzoyl end group with a more lipophilic octadecanoyl end group probably enhances lipid-polymer interactions, especially in highly compressed surfactant states, a situation in which a polymer lacking the highly lipophilic end group might otherwise be excluded from (or “squeezed-out” of) the lipid film. In addition, incorporation of the octadecanoyl group may promote polymer-polymer association through hydrophobic interactions of the hydrocarbon chains.42
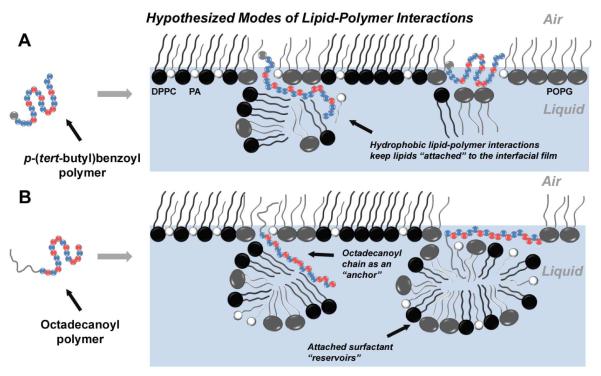
Figure 8 proposes distinct modes of lipid-polymer interaction for the copolymers bearing an N-terminal p-(tert-butyl)benzoyl group (small gray lipophilic unit; hypothesis A) vs. copolymers bearing an N-terminal octadecanoyl group (large gray lipophilic unit; hypothesis B). Panels A and B each represent DPPC:POPG:PA lipid monolayers at the air-liquid interface. At physiological temperature in the relevant surface tension regime assayed by the PBS (~ 0-40 mN/m surface tension), the lipid monolayer is composed of lipid-ordered regions, consisting of tightly packed saturated lipids DPPC:PA, and lipid-disordered regions that largely comprise unsaturated POPG.56 From previous work, it is known that natural SP-B and SP-C, as well as peptide- and peptoid-based surfactant protein mimics, reside in the fluid phase of the film, and we therefore assume that our polymers would likely occupy the same region.30,57 In A, the polymer adopts a lipid-associated amphiphilic conformation and ‘inserts’ into the lipid film. The cationic subunits of the polymer could be favorably solvated by the aqueous buffered subphase or be attracted Coulombically to the anionic lipid head groups of POPG and PA, while the lipophilic subunits could facilitate interactions with the lipid acyl chains in or below the interfacial film. In B, on the other hand, the large lipophilic end group could enable a polymer molecule to behave as a lipid “anchor,” facilitating deeper insertion into the acyl chains of the interfacial lipid monolayer, and sustaining a “surfactant reservoir” of lipids attached to the interfacial lipid film via favorable interactions between the polymer and lipids in both the interfacial film and the reservoir. Such reservoirs are known to occur in natural LS and are believed to be responsible for increasing interfacial adsorption, reducing γ, and allowing efficient re-spreading of the interfacial film upon inspiration.58 This behavior is postulated to occur more substantially with SP-C than with SP-B, where the highly lipophilic helix of SP-C is capable of spanning the length of a lipid bilayer;59 thus, it is possible that our octadecanoyl polymers emulate some surface-active attributes of both SP-B and SP-C.
Figure 8. Hypothesized Modes of Lipid-Polymer Interactions.
The hypothesized lipid-polymer interaction(s) contributing to enhanced surface activity. For the polymers, blue spheres represent the lipophilic units while red spheres indicate cationic units. The gray sphere in the ‘polymer sequence’ of A represents the p-(tert-butyl)benzoyl end group, while the gray line extending from the ‘polymer sequence’ in B represents the octadecanoyl chain. Therefore, the picture outlines two possibilities of modes of action: one for polymers without an octadecanoyl chain (A), and one for octadecanoylated polymers (B). Panels A and B each represent a Tanaka lipid (TL, DPPC:POPG:PA) monolayer at the air-liquid interface, with black spheres representing DPPC, gray spheres representing POPG, and white spheres representing PA. Copolymers with a p-(tert-butyl)benzoyl end group (A) adopt an amphiphilic conformation and insert into the lipid film, where they are able to retain attached lipids via Coulombic interactions between cationic subunits and charged lipid head groups, or hydrophobic interactions between lipophilic subunits and the lipid acyl chains. However, octadecanoylated copolymers (B) could act as lipid “anchors” by increasing the degree of insertion into the lipid acyl chains through hydrophobic hydrocarbon chain-chain interactions. These polymers may also be able to sustain sublayer lipid structures by either adopting a lipid-associated amphiphilic conformation, or utilizing the octadecanoyl chain to retain pockets of lipid material.
Studies with NIH 3T3 cells indicate that all of the nylon-3 polymers exhibit some toxicity, but in every case, the toxicity is significantly lower than that observed for Peptoid B1 and SP-B1-25, a synthetic peptide fragment derived from the N-terminal segment of natural SP-B. Note that to the best of our knowledge, mammalian cell toxicity for SP-B1-25 has not previously been reported; in addition, similar peptide mimics KL4 (in Surfaxin®, pending FDA approval), dSP-B1-25, and “Mini B” also all lack published cytotoxicity profiles, yet are actively being pursued for in vivo efficacy and possible pharmaceutical development. However, a great deal of additional research would be necessary to explore whether nylon-3 copolymers could be sufficiently non-toxic to support therapeutic applications. Since the identities and proportions of subunits and the nature of the N-terminal group can be easily varied, along with such features as average chain length and subunit stereochemistry, it may be possible to tune polymer properties in ways that enhance surfactant protein mimicry and diminish toxicity. It should be noted that the toxicities of these copolymers are likely to be substantially reduced in a lipid-rich environment, such as that found in lung surfactant. Indeed, the hemolytic activity for SP-B is considerably higher in the absence of lipids than in their presence.60
It is widely believed that the global segregation of cationic and lipophilic side chains on the surface of SP-B, i.e., the protein’s global amphiphilicity, is critical to the interactions of this protein with lipids and the overall surfactant function.29,61 This global amphiphilicity, in turn, depends upon adoption of a specific folding pattern. Most mimics of surfactant proteins examined to date have been sequence-specific oligomers, either peptides or peptoids;4 these oligomers have been designed to adopt a specific conformation, usually a helix, that is globally amphiphilic by virtue of the amino acid sequence.
If one assumes that a specific globally amphiphilic conformation must be adopted in order for an oligomer or polymer to display surfactant protein-like behavior, then it might seem pointless to explore sequence- and stereo-random copolymers in this regard, despite the profound advantages offered by such copolymers in terms of synthesis, relative to sequence-specific oligomers. However, we have recently discovered that sequence- and stereo-random nylon-3 copolymers can mimic the antibacterial function of sequence-specific host-defense peptides,40-42 and this precedent encouraged us to undertake the studies reported here. The favorable in vitro properties we observed for some of the nylon-3 copolymers suggest that the adoption of a specific conformation may not be required for effective lung surfactant protein B biophysical mimicry. We propose that the nylon-3 copolymers can achieve global amphiphilicity via irregular conformations that lead to induced segregation of cationic and lipophilic side chains in a lipid-buffer environment. Further testing of this possibility will be necessary, including in vitro studies with varying lipid composition22 and formulation, as well as surface activity assessment using techniques such as captive bubble surfactometry, surface balance studies, and lipid vesicle imaging. Ultimately, in vivo testing may be warranted. A convincing demonstration that sequence- and stereo-random copolymers could mimic natural lung surfactant protein behavior would be quite significant from a fundamental perspective, since this finding would broaden our understanding of sequence-activity relationships in this complex functional arena. Moreover, such results could be of practical importance, since the nylon-3 copolymers should be much less expensive to manufacture than are sequence-specific peptide, peptoids or other oligomers.
CONCLUSIONS
The current study provides the first evidence that sequence-random copolymers can mimic the behavior of surfactant protein B in terms of in vitro surface activity in a mixed lipid film. These polymers might represent a cost-effective strategy for generating lung surfactant protein mimics that does not rely on step-wise synthesis, extensive purification, or generating complex structural motifs through more complex chemistries. This work has effectively demonstrated that 1) sequence-random copolymers positively interact with a lipid film to dramatically enhance surface activity, 2) cyclooctyl subunit inclusion in the sequence substantially lowers the surface tension of the interfacial film, and 3) N-terminus octadecanoylation provides additional surface activity improvement, furthering the possibility that both SP-B and SP-C could be effectively mimicked with one molecule. We have challenged the notion that helicity is a strict requirement for amphiphilicity where lipid-bound molecules are concerned. Instead, we postulate that the copolymers can adopt an irregular conformation that nevertheless is globally amphiphilic and surface-active. The results of this work widen the biomimicry possibilities for polymers and have direct relevance to researchers involved in the design, synthesis, and characterization of peptidomimetics in many areas of chemistry and biology.
MATERIALS AND METHODS
Materials
All commercially available compounds were purchased from Sigma-Aldrich (Milwaukee, WI) or ACROS (Geel, Belgium) and used as received unless otherwise noted. Peptide and peptoid synthesis reagents and supplies were obtained from Applied Biosystems (ABI) (Foster City, CA) and Aldrich. Fmoc-protected amino acids and resins were purchased from EMD Biosciences (NovaBiochem, San Diego, CA). Primary amines for peptoid synthesis, highest percent purity and enantiomeric excess available, di-tert-butyl dicarbonate (Boc), and palmitic acid (PA) were obtained from Aldrich. All buffer salts, and solvents acetonitrile, chloroform, methanol, and trifluoroacetic acid (TFA), HPLC grade or better, were purchased from Fisher Scientific (Pittsburgh, PA). CDCl3 and D2O for NMR were purchased from Aldrich (see SI). DPPC and POPG were obtained from Avanti Polar Lipids (Alabaster, AL) and used as received. MTS reagent was purchased from Promega (Madison, WI), Hank’s Balanced Salt Solution (HBSS) from Lonza (Basel, Switzerland), Dulbecco’s Modified Eagle’s Media (DMEM) from ATCC or Invitrogen (Carlsbad, CA), and the microplate reader from Molecular Devices (Sunnyvale, CA). Water was Milli-Q 18.2 mΩ·cm quality.
Peptide and Peptoid Synthesis and Purification
The modified peptide SP-B1-25 (Cys8,11 → Ala)20-21,26-27 and the KL4 peptide20-21,53 were synthesized by standard SPPS62 Fmoc chemistry on a 0.25 mmol scale using preloaded Wang resin and an ABI 433A automated peptide synthesizer. Peptoid B120-21 was synthesized by the submonomer method36 using Rink amide resin on a 0.25 mmol scale, and an ABI 433A, with Boc protection of N-(4-aminobutyl)glycine (NLys). SP-B1-25, KL4, and Peptoid B1 were cleaved from their respective resins by agitation in 90-95% TFA/water [v/v], along with the appropriate scavengers, for 10 minutes up to 1 hour. Crude product for purification was obtained by immediate resin filtration of the mixture, dilution with ACN/water, repeated lyophilization, and re-dissolution in ACN/water. SP-B1-25, KL4, and Peptoid B1 were purified on a Waters (Waters Corp., Milford, MA) RP-HPLC system with a Grace Vydac (Deerfield, IL) C4-silica column. All SPPS-based molecules were purified using a linear gradient of percent solvent B in percent solvent A over a selected time period (solvent A is 0.1% TFA in water [v/v] and solvent B is 0.1% TFA in ACN [v/v]), using standard purification techniques. Final purities were confirmed to be > 97% by analytical RP-HPLC and molecular weights were obtained by either electrospray ionization mass spectrometry (ESI/MS) or matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) (MW (Da) calc:found as follows: SP-B1-25, 2865.55:2865.30; KL4, 2469.40:2469.70; Peptoid B1, 2592.40:2592.50).
Polymer Synthesis and Characterization
Polymers were synthesized using previously reported procedures,40,46 and detailed polymer characterization (MALDI-TOF/MS, GPC, and 1H NMR) are available in the SI.
Surfactant Sample Preparation
The lipids DPPC, POPG, and PA were individually dissolved in a chloroform/methanol solution (3/1 [v/v]) to ~ 2 or 4 mg mL−1. Single-lipid solutions were then combined by volume at the ratio of DPPC:POPG:PA, 68:22:9 [wt:wt:wt] and to ~ 2 mg lipid mL−1. This well-characterized and well-known lipid formulation is considered an adequate mimic of the non-protein (lipid) fraction of LS.45 The peptides, peptoid, and copolymers were individually dissolved in methanol from a lyophilized powder to ~ 1-2 mg mL−1. For the PBS studies, the peptides, peptoids, and polymers were ‘spiked’ into the lipid mixture at ~ 10 wt% relative to the total lipid content (~ 9 absolute wt%), and to a final concentration of ~ 1 mg lipid mL−1. For comparative purposes, the inclusion of peptide/peptoid/copolymer at ~ 10 wt% corresponds to ~ 10 wt% SP-B1-25 (~ 9 absolute wt%) relative to the total lipid content. The total combined protein fraction in natural LS is estimated to be ~ 10 wt%.18 For PBS experiments, all σ values reported is the standard deviation of the mean.
Pulsating Bubble Surfactometry
A commercial PBS instrument (General Transco, Largo, FL), modified with a direct, real-time imaging system, which has been previously described and validated in detail,44 was utilized to obtain both static-mode and dynamic-mode data. Samples were dried from chloroform/methanol 3/1 [v/v] in Eppendorf tubes using a DNA 120 speedvac (Thermo Electron, Holbrook, NY), forming a pellet. The pellet was suspended in buffer (150 mM NaCl, 10 mM HEPES, 5 mM CaCl2, pH 6.9) to ~ 1 mg lipid mL−1, with a final volume of ~ 70 μL. The samples were then mixed with a pipette 20 times, sonicated with a Fisher Model 60 probe sonicator for two 15 second spurts, and then mixed again 20 times to form a dispersed suspension. Samples were then loaded into a small plastic sample chamber (General Transco) using a modified leak-free methodology.43-44 The sample chamber was then placed in the instrument, surrounded by a water bath held at 37 °C. A bubble with a radius of 0.4 mm was then formed, and surface area was monitored throughout the experiment. Bubble size gradually increased in both data collection modes, but had a negligible effect on γ.
Static-mode adsorption data were collected for 20 minutes, where the suspension was allowed to adsorb to the bubble surface over time. Adsorption data were smooth fit to a curve in the Kaleidagraph program by applying a Stineman function to the data, where the output of this function then had a geometric weight applied to the current point and ± 10% of the data range to arrive at the smoothed curve (Figure 2). Dynamic-mode data were then subsequently obtained for each sample at the adult respiratory cycle frequency of 20 cpm for 10 minutes, with a ~ 50% reduction in surface area per pulsation cycle. PBS experiments were repeated three to six times for each sample to ensure repeatability. Representative PBS loops are presented at five minutes of cycling, and indicate clockwise bubble expansion and counterclockwise compression (Figure 6, Panels A-D). Average γ ± σ at selected time intervals are listed in SI for both data collection modes. Percent compression is defined here as 100*[(SAmax – SA20)/(SAmax)], where SAmax was the maximum SA value at expansion, and SA20 was the SA at which γ first reaches 20 mN m−1 upon compression (Figure 5).
Cytotoxicity (MTS) Assay
The cytotoxicities of SP-B1-25, Peptoid B1, and select copolymers against NIH/3T3 fibroblast cells were evaluated using a 3-(4,5-diemethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt (MTS) (Promega) colorimetric assay. NIH/3T3 fibroblast cells (ATCC, Manassas, VA) were cultured at 37 °C and 5% CO2 in Dulbecco’s Modified Eagle’s Media (DMEM) (ATCC or Invitrogen) supplemented with 1% sodium pyruvate, 1% penicillin-streptomycin, 1.5 g L−1 NaHCO3, and 10% fetal bovine serum (cDMEM). Cells were seeded at a density of 5,000 cells per well for NIH/3T3 cells in 96-well plates (100 μL total volume). A SP-B1-25 peptide, Peptoid B1, or copolymer solution plate (100 μl per well) was prepared by serial dilution of aqueous peptide or copolymer stock solutions of known concentration (~ 5 mg mL−1) in Hank’s Balanced Salt Solution (HBSS) (Lonza). A day-old monolayer of cells plated at 5000 cells per well (100 μl) was washed once, and the media was replaced with HBSS to minimize interference with absorbance readings. Peptide, peptoid, or copolymer solutions were then transferred to corresponding wells of the cell plate. 40 μl MTS reagent (Promega) was then added to each well, and the plate was subsequently incubated at 37 °C for 3 h, after which UV/Vis (Molecular Probes) absorbance measurements at λ ~ 490 nm were recorded. Percent inhibition = [1 - (A - Atest blank)/(Acontrol - Ablank)] * 100, where A is the UV/Vis absorbance of the test well, and Acontrol is the average absorbance of wells with cells exposed to media and MTS (no peptide or copolymer). Atest blank (media, MTS, and peptide or copolymer) and Ablank (media and MTS) were background absorbances measured in the absence of cells. Results are reported as the inhibitory dose (μg mL−1) at which 50% of cells experienced metabolic inhibition (ID50). The average of six replicates is reported, where error is the standard error of the mean (SEM) (Figure 7).
Supplementary Material
ACKNOWLEDGMENT
AEB and MTD gratefully acknowledge Mark Johnson for PBS use. AMC and MTD thank Jennifer Cruz Rea for assistance with cytotoxicity experiments. BPM would like to thank Jihua Zhang for assistance with GPC measurements. The purchase of the REFLEX II™ was partially funded by an NSF Award (#CHE-9520868) to the UW-Madison Department of Chemistry. This work was supported by the US National Institutes of Health (Grant 2 R01 HL67984) and the US National Science Foundation (Grant BES-0101195 and Collaborative Research in Chemistry Grant CHE-0404704).
Footnotes
SUPPORTING INFORMATION AVAILABLE. Detailed polymer characterization (MALDI/MS, GPC, and 1H NMR) and additional tabulated PBS data and hysteresis loops.
REFERENCES
- (1).Creuwels L, vanGolde LMG, Haagsman HP. Lung. 1997;175:1–39. doi: 10.1007/PL00007554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Notter RH. Lung Surfactants: Basic Science and Clinical Applications. Marcel Dekker; New York: 2000. [Google Scholar]
- (3).Orgeig S, Bernhard W, Biswas SC, Daniels CB, Hall SB, Hetz SK, Lang CJ, Maina JN, Panda AK, Perez-Gil J, Possmayer F, Veldhuizen RA, Yan W. Integr. Comp. Biol. 2007;47:610–627. doi: 10.1093/icb/icm079. [DOI] [PubMed] [Google Scholar]
- (4).Mingarro I, Lukovic D, Vilar M, Perez-Gil J. Curr. Med. Chem. 2008;15:393–403. doi: 10.2174/092986708783497364. [DOI] [PubMed] [Google Scholar]
- (5).Avery ME, Mead J. Am. J. Dis. Child. 1959;97:517–523. doi: 10.1001/archpedi.1959.02070010519001. [DOI] [PubMed] [Google Scholar]
- (6).Pison U, Seeger W, Buchhorn R, Joka T, Brand M, Obertacke U, Neuhof H, Schmit-Nauerburg KP. Am. Rev. Respir. Dis. 1989;140:1033–1039. doi: 10.1164/ajrccm/140.4.1033. [DOI] [PubMed] [Google Scholar]
- (7).Lewis JE, Jobe AH. Am. Rev. Respir. Dis. 1993;147:218–233. doi: 10.1164/ajrccm/147.1.218. [DOI] [PubMed] [Google Scholar]
- (8).Moya FR, Maturana A. Clin. Perinatol. 2007;34:145–177. doi: 10.1016/j.clp.2006.12.005. [DOI] [PubMed] [Google Scholar]
- (9).Hawgood S, Schiffer K. Annu. Rev. Physiol. 1991;53:375–394. doi: 10.1146/annurev.ph.53.030191.002111. [DOI] [PubMed] [Google Scholar]
- (10).Vandenbussche G, Clercx A, Clercx M, Curstedt T, Johansson J, Jornvall H, Ruysschaert JM. Biochemistry. 1992;31:9169–9176. doi: 10.1021/bi00153a008. [DOI] [PubMed] [Google Scholar]
- (11).Beck DC, Ikegami M, Na CL, Zaltash S, Johansson J, Whitsett JA, Weaver TE. J. Biol. Chem. 2000;275:3365–3370. doi: 10.1074/jbc.275.5.3365. [DOI] [PubMed] [Google Scholar]
- (12).Haagsman HP, Diemel RV. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2001;129:91–108. doi: 10.1016/s1095-6433(01)00308-7. [DOI] [PubMed] [Google Scholar]
- (13).Johansson J, Szyperski T, Curstedt T, Wuthrich K. Biochemistry. 1994;33 doi: 10.1021/bi00185a042. [DOI] [PubMed] [Google Scholar]
- (14).Johansson J, Curstedt T, Robertson B. Eur. Respir. J. 1994;7:372–391. doi: 10.1183/09031936.94.07020372. [DOI] [PubMed] [Google Scholar]
- (15).Creuwels LA, Boer EH, Demel RA, van Golde LMG, Haagsman HP. J. Biol. Chem. 1995;270:16225–16229. doi: 10.1074/jbc.270.27.16225. [DOI] [PubMed] [Google Scholar]
- (16).Kramer A, Wintergalen A, Sieber M, Galla HJ, Amrein M, Guckenberger R. Biophys. J. 2000;78:458–465. doi: 10.1016/S0006-3495(00)76608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Bi XH, Flach CR, Perez-Gil J, Plasencia I, Andreu D, Oliveira E, Mendelsohn R. Biochemistry. 2002;41:8385–8395. doi: 10.1021/bi020129g. [DOI] [PubMed] [Google Scholar]
- (18).Perez-Gil J. Biochim. Biophys. Acta. 2008;1778:1676–1695. doi: 10.1016/j.bbamem.2008.05.003. [DOI] [PubMed] [Google Scholar]
- (19).Wu CW, Seurynck SL, Lee KYC, Barron AE. Chem. Biol. 2003;10:1057–1063. doi: 10.1016/j.chembiol.2003.10.008. [DOI] [PubMed] [Google Scholar]
- (20).Seurynck SL, Patch JA, Barron AE. Chem. Biol. 2005;12:77–88. doi: 10.1016/j.chembiol.2004.10.014. [DOI] [PubMed] [Google Scholar]
- (21).Seurynck-Servoss SL, Dohm MT, Barron AE. Biochemistry. 2006;45:11809–11818. doi: 10.1021/bi060617e. [DOI] [PubMed] [Google Scholar]
- (22).Seurynck-Servoss SL, Brown NJ, Dohm MT, Wu CW, Barron AE. Coll. Surf. B. Biointerfaces. 2007;57:37–55. doi: 10.1016/j.colsurfb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- (23).Brown NJ, Wu CW, Seurynck-Servoss SL, Barron AE. Biochemistry. 2008;47:1808–1818. doi: 10.1021/bi7021975. [DOI] [PubMed] [Google Scholar]
- (24).Dohm MT, Seurynck-Servoss SL, Seo J, Zuckermann RN, Barron AE. Biopolymers (Peptide Sci.) 2009;92:538–553. doi: 10.1002/bip.21309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Hawgood S, Ogawa A, Yukitake K, Schlueter M, Brown C, White T, Buckley D, Lesikar D, Benson BJ. Am. J. Respir. Crit. Care Med. 1996;154:484–490. doi: 10.1164/ajrccm.154.2.8756826. [DOI] [PubMed] [Google Scholar]
- (26).Waring A, Taeusch HW, Bruni R, Amirkhanian JD, Fan BR, Stevens R, Young J. Pept. Res. 1989;2:308–313. [PubMed] [Google Scholar]
- (27).Bruni R, Taeusch HW, Waring AJ. Proc. Natl. Acad. Sci. U. S. A. 1991;88:7451–7455. doi: 10.1073/pnas.88.16.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Veldhuizen EJA, Waring AJ, Walther FJ, Batenburg JJ, van Golde LMG, Haagsman HP. Biophys. J. 2000;79:377–384. doi: 10.1016/S0006-3495(00)76299-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Waring AJ, Walther F, Gordon LM, Hernandez-Juviel J, Hong T, Sherman MA, Alonso C, Alig T, Brauner JW, Bacon D, Zasadzinski J. J. Pept. Res. 2005;66:364–374. doi: 10.1111/j.1399-3011.2005.00300.x. [DOI] [PubMed] [Google Scholar]
- (30).Dohm MT, Brown NJ, Seurynck-Servoss SL, Bernardino de la Serna J, Barron AE. Biochim. Biophys. Acta-Biomem. 2010 doi: 10.1016/j.bbamem.2010.04.012. in minor revision. [DOI] [PubMed] [Google Scholar]
- (31).Kirshenbaum K, Barron AE, Goldsmith RA, Armand P, Bradley EK, Truong KTV, Dill KA, Cohen FE, Zuckermann RN. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4303–4308. doi: 10.1073/pnas.95.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Appella DH, Christianson LA, Karle IL, Powell DR, Gellman SH. J. Am. Chem. Soc. 1996;118:13071–13072. [Google Scholar]
- (33).Cheng RP, Gellman SH, DeGrado WF. Chem. Rev. 2001;101:3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- (34).Hayen A, Schmitt MA, Ngassa F, Thomasson KA, Gellman SH. Angew. Chem. Int. Ed. 2004;43:505–510. doi: 10.1002/anie.200352125. [DOI] [PubMed] [Google Scholar]
- (35).Horne WS, Gellman SH. Acc. Chem. Res. 2008;41:1399–1408. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Zuckermann RN, Kerr JM, Kent SBH, Moos WH. J. Am. Chem. Soc. 1992;114:10646–10647. [Google Scholar]
- (37).Gelman MA, Lynn DM, Weisblum B, Gellman SH. Org. Lett. 2004;6:557–60. doi: 10.1021/ol036341+. [DOI] [PubMed] [Google Scholar]
- (38).Schmitt MA, Weisblum B, Gellman SH. J. Am. Chem. Soc. 2007;129:417–428. doi: 10.1021/ja0666553. [DOI] [PubMed] [Google Scholar]
- (39).Shai Y. Biochim. Biophys. Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- (40).Mowery BP, Lee SE, Kissounko DA, Epand RF, Epand RM, Weisblum B, Stahl SS, Gellman SH. J. Am. Chem. Soc. 2007;129:15474–15476. doi: 10.1021/ja077288d. [DOI] [PubMed] [Google Scholar]
- (41).Epand RF, Mowery BP, Lee SE, Stahl SS, Lehrer RI, Gellman SH. J. Mol. Biol. 2008;379:38–50. doi: 10.1016/j.jmb.2008.03.047. [DOI] [PubMed] [Google Scholar]
- (42).Mowery BP, Lindner AHW,B, Stahl SS, Gellman SH. J. Am. Chem. Soc. 2009;131:9735–9745. doi: 10.1021/ja901613g. [DOI] [PubMed] [Google Scholar]
- (43).Putz G, Goerke J, Taeusch HW, Clements JA. J. Appl. Physiol. 1994;76:1425–1431. doi: 10.1152/jappl.1994.76.4.1425. [DOI] [PubMed] [Google Scholar]
- (44).Seurynck SL, Brown NJ, Wu CW, Germino KW, Kohlmeir EK, Ingenito EP, Glucksberg MR, Barron AE, Johnson M. J. Appl. Physiol. 2005;99:624–633. doi: 10.1152/japplphysiol.00748.2004. [DOI] [PubMed] [Google Scholar]
- (45).Tanaka Y, Takei T, Aiba T, Masuda K, Kiuchi A, Fujiwara T. J. Lipid Res. 1986;27:475–485. [PubMed] [Google Scholar]
- (46).Zhang J, Kissounko DA, Lee SE, Gellman SH, Stahl SS. J. Am. Chem. Soc. 2009;131:1589–1597. doi: 10.1021/ja8069192. [DOI] [PubMed] [Google Scholar]
- (47).Ryan MA, Qi X, Serrano AG, Ikegami M, Perez-Gil J, Johansson J, Weaver TE. Biochemistry. 2005;44:861–872. doi: 10.1021/bi0485575. [DOI] [PubMed] [Google Scholar]
- (48).Serrano AG, Ryan MA, Weaver TE, Perez-Gil J. Biophys. J. 2006;90:238–249. doi: 10.1529/biophysj.105.073403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Wang YD, Rao KMK, Demchuk E. Biochemistry. 2003;42:4015–4027. doi: 10.1021/bi027344h. [DOI] [PubMed] [Google Scholar]
- (50).Brown NJ, Bernardino de la Serna J, Barron AE. Biophys. J. 2010 in revision. [Google Scholar]
- (51).Hashimoto K. Prog. Polym. Sci. 2000;25:1411–1462. [Google Scholar]
- (52).Cheng JJ, Deming TJ. J. Am. Chem. Soc. 2001;123:9457–9458. doi: 10.1021/ja0110022. [DOI] [PubMed] [Google Scholar]
- (53).Cochrane CG, Revak SD. Science. 1991;254:566–568. doi: 10.1126/science.1948032. [DOI] [PubMed] [Google Scholar]
- (54).Nadolski MJ, Linder ME. FEBS J. 2007;274:5202–5210. doi: 10.1111/j.1742-4658.2007.06056.x. [DOI] [PubMed] [Google Scholar]
- (55).Chu-Kung AF, Bozzelli KN, Lockwood NA, Haseman JR, Mayo KH, Tirrell M. Bioconj. Chem. 2004;15:530–535. doi: 10.1021/bc0341573. [DOI] [PubMed] [Google Scholar]
- (56).Bringezu F, Ding JQ, Brezesinski G, Zasadzinski JA. Langmuir. 2001;17:4641–4648. [Google Scholar]
- (57).Bernardino de la Serna J, Perez-Gil J, Simonsen AC, Bagatolli LA. J. Biol. Chem. 2004;279:40715–40722. doi: 10.1074/jbc.M404648200. [DOI] [PubMed] [Google Scholar]
- (58).Schurch S, Qanbar R, Bachofen H, Possmayer F. Biol. Neonate. 1995;67:61–76. doi: 10.1159/000244207. [DOI] [PubMed] [Google Scholar]
- (59).Veldhuizen EJA, Haagsman HP. Biochim. Biophys. Acta. 2000;1467:255–270. doi: 10.1016/s0005-2736(00)00256-x. [DOI] [PubMed] [Google Scholar]
- (60).Ryan MA, Akinbi HT, Serrano AG, Perez-Gil J, Wu HX, McCormack FX, Weaver TE. J. Immunol. 2006;176:416–425. doi: 10.4049/jimmunol.176.1.416. [DOI] [PubMed] [Google Scholar]
- (61).Andersson M, Curstedt T, Jornvall H, Johansson J. FEBS Lett. 1995;362:328–332. doi: 10.1016/0014-5793(95)00268-e. [DOI] [PubMed] [Google Scholar]
- (62).Merrifield RB. J. Am. Chem. Soc. 1963;85:2149–2154. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.