Abstract
We report high-resolution crystal structures of six new α/β-peptide foldamers that have a regular α-residue/α-residue/β-residue (ααβ) backbone repeat pattern. All of these foldamers were crystallized from aqueous solution, and all display four-helix bundle quaternary structure in the crystalline state. These oligomers are based on the well-studied 33-residue α-peptide GCN4-pLI, which is an engineered derivative of the dimerization domain of GCN4, a yeast transcription factor. GCN4-pLI forms a stable tetramer in solution and crystallizes as a four-helix bundle (Harbury et al. Science 1993, 262, 1401–1407). Previously we described a foldamer (designated 1 here) that was generated from GCN4-pLI by replacing every third α-amino acid residue with the homologous β3-amino acid residue; this ααβ oligomer retains the side chain sequence of the original α-peptide, but the backbone contains 11 additional CH2 units, which are evenly distributed (Horne et al. Proc. Natl. Acad. Sci. USA 2008, 105, 9151–9156). Despite the expanded backbone, 1 was found to retain the ability to form a tetrameric quaternary structure in which the individual molecules adopt an α-helix-like conformation. Here we compare nine analogues of 1 that have the same ααβ backbone but in which one or more of the flexible β3-amino acid residues is/are replaced with an analogous cyclic β-residue. The motivation for β3→cyclic replacements is to enhance conformational stability; however, a crystal structure of the one previously reported example (designated 2 here) revealed a “stammer” distortion of the helix-bundle architecture relative to 1. The results reported here suggest that the stammer is a peculiarity of 2, because all six of the new α/β-peptides display undistorted four-helix bundle quaternary structures. More broadly, our results indicate that β3→cyclic replacements are generally well-accommodated in helix-bundle quaternary structure, but that such replacements can be destabilizing in certain instances.
Keywords: α/β-peptides, foldamers, helix-bundle, quaternary structure, α-helix mimics
Introduction
The information embedded in a polypeptide primary sequence underlies the complex folding behavior that is a prerequiste for most protein function.1 Understanding how a one-dimensional pattern of α-amino acid side chains encodes a specific three-dimensional conformation remains an important goal in chemistry and biology.2 Efforts to probe the sequence-structure relationship in polypeptides have typically relied on mutagenesis, that is, evaluation of the impact on folding behavior of modifying one side chain or multiple side chains. A complementary approach is to modify the polypeptide backbone while retaining the original side chain sequence. This experimental strategy can elucidate the contribution of the backbone to protein folding behavior. The absence of protein-like molecules with altered backbones in biology makes it tempting to assume that protein conformational preferences are highly dependent on the poly-α-amino acid backbone; examination of peptide and protein analogues with altered backbones is necessary to test this assumption.
Backbone alteration is more challenging to implement than is side chain alteration, since the biosynthetic machinery is largely intolerant of building blocks other than α-amino acids.3 Chemical synthesis can be used to generate small to medium-length peptidic oligomers with altered backbones; such oligomers can be employed for folding studies or used as precursors for longer protein analogues (obtained via segment condensation). These approaches have been used, for example, to prepare protein analogues in which α-amino acid residues are replaced by α-hydroxy acid residues bearing the same side chain. This modification replaces a backbone amide group with an ester, which enables one to examine the influence of backbone H-bonds on protein folding behavior.4–12 Replacement of a backbone amide group with an alkene isostere13–23 or with a triazole heterocycle24,25 has also been explored, as has the substitution of an olefin-based surrogate for an intramolecular i,i+4 C=O···N–H hydrogen bond between backbone amide groups in an α-helical peptide.26 Another line of research has involved replacement of a specific element of protein secondary structure with an unnatural subunit. This approach has been used to evaluate the impact of reverse turn replacement (two to four residues) on enzymatic function25,27 or on the thermodynamics and kinetics of protein folding.28,29 Beck-Sickinger et al. recently replaced the C-terminal α-helical segment of interleukin-8 with a helical segment composed entirely of β-amino acid residues; the resulting chimeric molecule retained weak signaling activity.30 However, an important control experiment showed that replacing the native C-terminal α-helical segment of IL-8 with a completely different α-helical segment caused little change in signaling relative to IL-8 itself, which indicates that the role of the C-terminal segment in IL-8-receptor interactions does not depend upon a specific side chain sequence.
The precedents summarized above involve backbone modifications at a single site or region within a polypeptide. Other efforts have focused on folded oligomers with entirely unnatural backbones,31–35 including those with the ability to adopt protein-like tertiary and quaternary structures.36–43 We have undertaken complementary studies that focus on the impact of β-amino acid substitutions introduced systematically throughout a natural polypeptide sequence.44 β-Amino acids are attractive in this regard because it is straightforward synthetically to replace an α-amino acid residue with a homologous β3-amino acid residue that retains the original side chain. Each modification of this type adds a CH2 unit into the backbone, relative to the pure α-residue prototype. Our efforts in this arena45 began with the dimerization domain of the yeast transcription factor GCN4. This 33-residue peptide (designated GCN4-p146) forms a parallel coiled-coil dimer, in which each component adopts an α-helical conformation, and the two helices supercoil slightly around one another. Dimerization is driven largely by burial of hydrophobic surface area on nonpolar side chains, which are intimately interdigitated at the helix-helix interface. The side chains that form the core of the dimer occur at positions a and d of the heptad repeat pattern (abcdefg) that is found in the GCN4-p1 sequence. We found that systematic α→β3 replacements at b and f positions in the GCN4 sequence produced an α/β-peptide that displays a modified self-recognition propensity relative to that of the α-peptide prototype: the α/β-peptide crystallizes as a three-helix bundle rather than a two-helix bundle.45 Each subunit in this quaternary structure displays an α-helix-like conformation, even though there is one extra CH2 unit per helical turn. The hydrophobic core of the three-helix bundle is formed by side chains contributed by α-residues at the a and d positions; the β-residues at b and f positions form a stripe that runs along the outward-facing side of each helix.
We extended this “sequence-based” design approach to GCN4-pLI,47 an engineered mutant of GCN4-p1 (Figure 1A). Systematic α→β3 replacements at b and f positions of GCN4-pLI led to an α/β-peptide that crystallizes as a parallel four-helix bundle that is very similar to the quaternary structure of GCN4-pLI itself (Figure 1B,C).45 Subsequent studies revealed that the GCN4-pLI sequence tolerates a variety of α→β3 replacement patterns without losing the ability to crystallize as a parallel four-helix bundle similar to that of parent α-peptide, which suggests that the self-recognizing capability encoded in the side chain sequence tolerates significant variations in backbone structure.48 This tolerance is particularly noteworthy for GCN4-pLI homologues with ααβ3 or αααβ3 backbone patterns. In these cases, a few side chains from β3-residues contribute to the hydrophobic core of the quaternary structure, as revealed by crystallographic analysis. This backbone modification approach has limits, however. Replacement of every other residue in GCN4-pLI with the homologous β3-residue leads to an oligomer that does not fold or self-associate.48 Replacement of all a and d residues with the homologous β3-residues generates an α/β-peptide that displays altered self-association behavior: this molecule forms an antiparallel four-helix bundle.49
Figure 1.
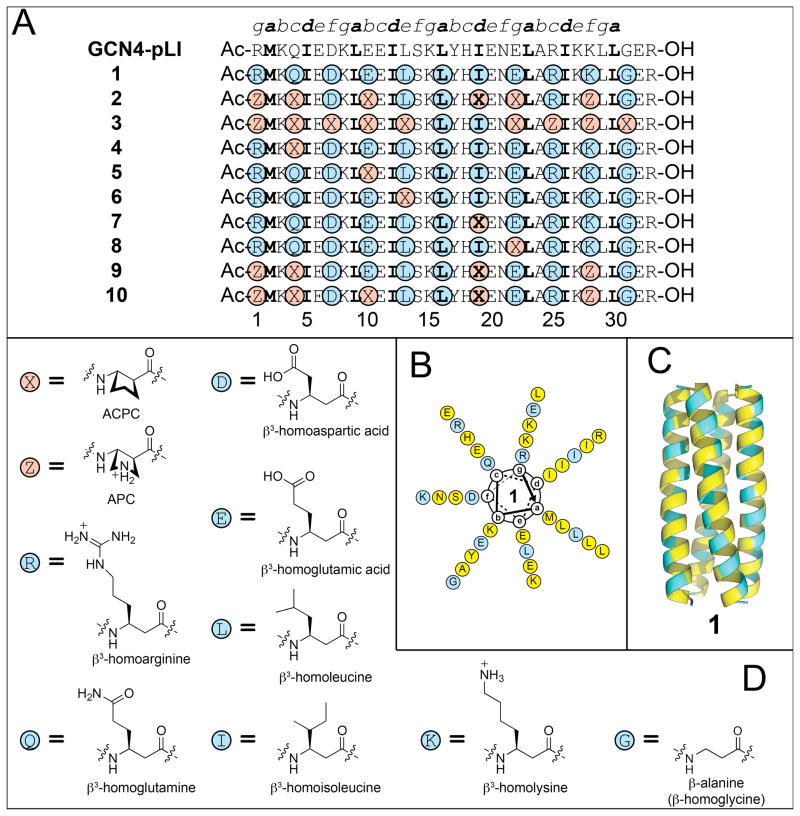
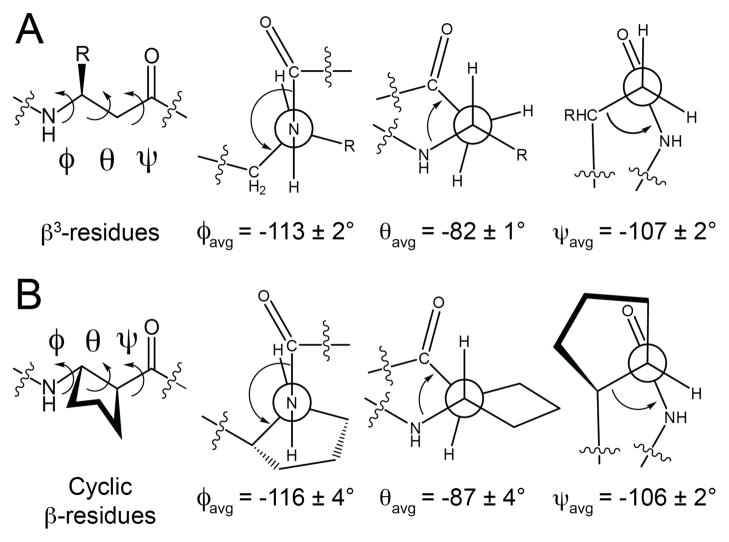
(A) Amino acid sequences of α/β-peptides 1–10 and α-peptide GCN4-pLI (refs. 47 and 48). α-Amino acids are abbreviated according to the standard one-letter code. β3-Amino acids are highlighted with blue circles and are abbreviated with the one letter code of the corresponding homologous α-amino acid. Cyclic β-amino acids are highlighted with red circles and are abbreviated as follows: X = ACPC, Z = APC. Hydrophobic a and d position residues are highlighted with boldface type. (B) Helical wheel diagram of α/β-peptide 1 viewed from the N-terminus (see ref. 48). α-Amino acids are highlighted with yellow circles and are abbreviated as described above. β-Amino acids are highlighted with blue circles and are abbreviated as described above. (D) Ribbon diagram of the tetrameric helix bundle formed by α/β-peptide 1 (PDB: 3C3G). (D) Structures of ACPC, APC, and the β 3-amino acids used in α/β-peptides 1–10.
If very similar helical conformations are adopted by an α-peptide and an α/β-peptide homologue containing α→β3 replacements, then it seems likely that the α/β-peptide conformation will be less stable because each α→β3 replacement adds a flexible bond to the backbone, and helix formation requires that each flexible backbone bond be torsionally constrained. Use of cyclic β-amino acid residues32,50 in place of β3-residues offers the prospect of enhancing α/β-peptide helix stability if the covalent ring promotes Cα -Cβ torsion angles that are consistent with the helix. Previous work suggested that the five-membered ring constraint of trans-2-aminocyclopentanecarboxylic acid (ACPC) provides the local folding propensity necessary for an α-helix-like conformation,51–54 and we have conducted preliminary efforts to stabilize four-helix bundle quaternary structures of GCN4-pLI-derived α/β-peptides via β3→cyclic residue replacements.48 This approach involves unavoidable deviation from the side chain sequence of the α-peptide prototype, but one can try to maintain the general character of the replaced side chain. For example, ACPC is a logical replacement for β3-residues with nonpolar side chains, while the analogous pyrrolidine-based residue (APC) is a logical replacement for β3-residues with basic side chains (Figure 1B). These considerations led us to compare two α/β-peptides based on GCN4-pLI, 1 and 2, which have the same ααβ backbone pattern. α/β-Peptide 1 contains only β3-residues, while in 2 six of the eleven β-positions are occupied by cyclic residues (ACPC or APC).48 Analytical ultracentrifugation (AU) indicates that both 1 and 2 form tetrameric assemblies in aqueous solution. Thermal disruption studies, monitored by circular dichroism (CD), suggest that the tetramer of 2 is more stable than the tetramer of 1. If helix formation occurs concomitantly with tetramerization, then this trend is consistent with our expectation that β3→cyclic replacements should enhance the conformational stability of the α/β-peptide helix. The crystal structure of 1 shows a four-helix bundle that is quite similar to the quaternary structure observed for the α-peptide prototype, GCN4-pLI. The crystal structure of 2, however, reveals an unexpected deviation at the quaternary structure level: the helices display a discontinuity in the heptad repeat, a feature that has been referred to as a “stammer” in the context of purely α-peptide helical assemblies.55 (The stammer discontinuity observed in crystal structure of 2 is described in detail in the Supporting Information.48)
The studies reported here were intended to reveal whether α/β-peptides containing cyclically constrained β-residues are prone to stammer-type deviations from classical helix-bundle association patterns, as observed for 2. We have recently shown that α/β-peptides can functionally mimic α-helical subunits of natural proteins that participate in specific protein-protein interactions.56–59 Use of α/β-peptides to inhibit disease-associated protein-protein interactions that are based on helix recognition could ultimately lead to novel therapeutic agents, and it seems reasonable to expect that cyclically constrained β-amino acid residues would be useful design tools. If, however, the stammer discontinuity observed in 2 turned out to be a common consequence of β3→cyclic replacements, then cyclic β-amino acid residues might have only limited utility for development of α/β-peptides intended to inhibit protein-protein associations based on α-helix recognition. Several considerations suggested that α/β-peptides 1 and 2 provide a good basis for exploring the relationship between β-residue identity and helix-bundle-forming propensities of α/β-peptides: (1) the backbone common to 1 and 2 is rich in β-residues, (2) the β-residues occupy a variety of positions in the heptad repeat, (3) the available structural and biophysical data for 1 and 2 provide benchmarks for comparisons, and (4) precedent with α-peptides and α/β-peptides in this family suggests that new analogues would have a strong tendency to form high-quality crystals.
Below we describe eight new α/β-peptides related to 1 and 2 (Figure 1). These new 33-mers (3–10) have been characterized in solution via CD and AU. High-resolution crystal structures have been obtained for six of the new α/β-peptides. Each of the new crystal structures shows a parallel four-helix bundle quaternary structure similar to that observed previously for 1 and for parent α-peptide GCN4-pLI. None of these new structures contains a helical stammer of the type observed in the crystal structure of 2. Data obtained in solution indicate that β3→cyclic replacements are well-tolerated at many positions within the sequence, although replacement at one position is quite destabilizing, and some replacements appear to cause changes in the preferred self-association stoichiometry. The new information strengthens our understanding of helical folding among α/β-peptide foldamers and enhances the prospects for rational design of α/β-peptides that inhibit protein-protein associations involving α-helix recognition.
RESULTS AND DISCUSSION
Design of new α/β-peptide sequences based on 1 and 2
An interest in determining whether α/β-peptides that contain cyclically constrained β-residues are prone to stammer-type deviations from classical helix-bundle association patterns (as was observed for 2) motivated us to prepare 3–10 (Figure 1A). In α/β-peptide 3, only two of the β3-residues from 1 have been retained, β3-hLeu16 (a-position) and β3-hIle19 (d-position); these are the only two β3-residues that occupy core positions in the heptad repeat of 1. All of the nine remaining β-positions are occupied by cyclic residues. In contrast, α/β-peptides 4–8 each contain only one β3→cyclic replacement. The replacement sites were selected to sample different types of positions in the heptad repeat, including a core position (7, d replacement; β3-hIle19→ACPC), flanking positions (6 and 8, e and g replacements, respectively; β3-hLeu13→ACPC and β3-hGlu22→ACPC) and solvent-exposed positions (4 and 5, c and b replacements, respectively; β3-hGln4→ACPC and β3-hGlu10→ACPC). Our final α/β-peptide designs contained multiple β3→cyclic replacements and were derived from 2. α/β-Peptide 9 contains four of the β3→cyclic replacements found in 2 (at residues 1, 4, 19 and 28); 9 differs from 2 in that β3-hGlu has been retained at positions 10 and 22, while ACPC occurs at these positions in 2. In contrast, α/β-Peptide 10 has ACPC at position 10 and β3-hGlu at position 22, thus differing from 2 only at position 22. The identity of residue 22 is of particular interest because the crystal structure of 2 suggests that ACPC22 plays a central role in the stammer.
Crystallographic analysis of α/β-peptide helix bundles
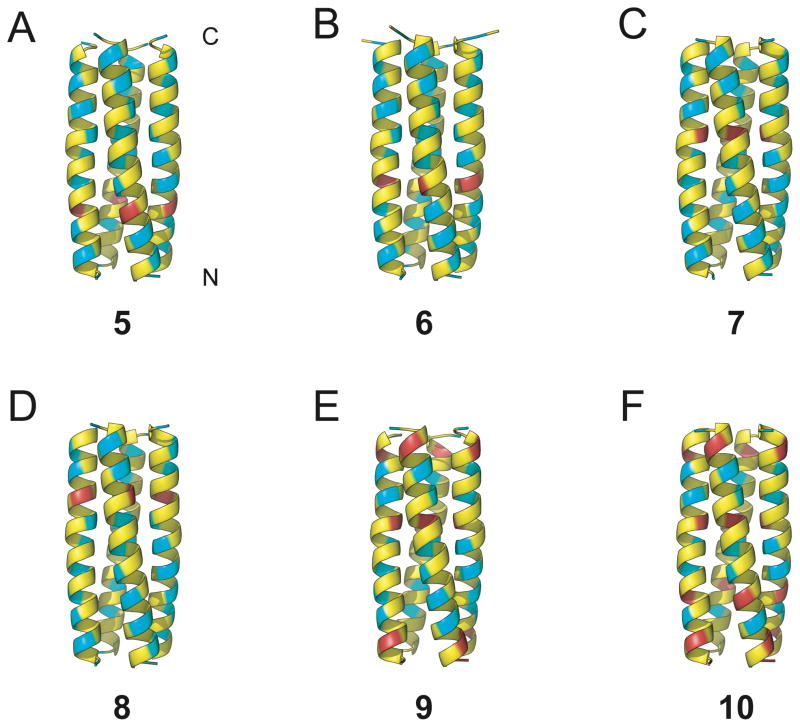
Crystals of α/β-peptides 5–10 were grown by hanging-drop vapor diffusion, and x-ray diffraction data were used to solve crystal structures to 2.1 Å (5), 2.0 Å (6), 2.0 Å (7), 2.0 Å (8), 2.8 Å (9) and 2.0 Å (10) resolution (attempts to obtain diffraction quality crystals from 3 and 4 were unsuccessful). Ribbon diagrams generated from these crystal structures are shown in Figure 2. Each of these six α/β-peptides forms a parallel tetrameric helix-bundle; all crystallize in the same space group. In the crystal structures of α/β-peptides 6–10 the asymmetric unit contains a single helix, and the four helix-bundle is generated by the crystallographic four-fold axis of symmetry. In α/β-peptide 5, the asymmetric unit is composed of two crystallographically distinct (but nearly identical) helices, each of which generates a distinct (but nearly identical) four-helix bundle via a four-fold axis of symmetry. In each structure, individual helices display intramolecular i,i+4 C=O···N–H hydrogen bonds between backbone amide groups, which is the same H-bonding pattern present in α-helical α-peptides.
Figure 2.
Ribbon diagrams from x-ray crystal structures of α/β-peptides (A) 5 (PDB: 3HET), (B) 6 (PDB: 3HEU), (C) 7 (PDB: 3HEV), (D) 8 (PDB: 3HEW), (E) 9 (PDB: 3HEX), (F) 10 (PDB: HEY), showing the helix-bundle tetramer quaternary structures formed in each case. α-Amino acid residues are colored yellow, β3-amino acid residues are colored blue, and cyclic β-amino acid residues are colored red. Unlike 2, α/β-peptides 5–10 have a continuous heptad repeat pattern, without a helical stammer.
All of the α/β-peptides display a continuous hydrophobic heptad repeat pattern from the N-terminus to the C-terminus, without the helical stammer that was observed for α/β-peptide 2. The absence of the helical stammer is particularly striking for α/β-peptide 10, which differs from 2 by only one residue (β3-hGlu instead of ACPC at position 22; see Figure 1A). This result suggests ACPC22 is a key structural determinant of the helical stammer in 2. However, the lack of a stammer discontinuity in the crystal structure of α/β-peptide 8 (in which ACPC at position 22 is the only cyclic residue) suggests that ACPC22 alone is not sufficient to induce the helical stammer.
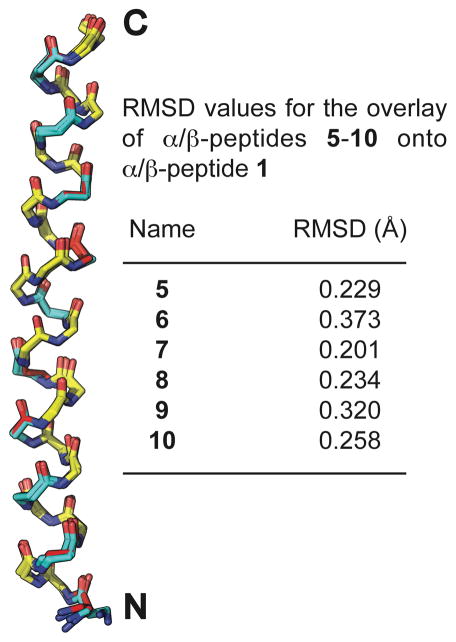
Figure 3 shows the alignment of individual helices from the crystal structures of α/β-peptides 5–10 with a single helix from the crystal structure of α/β-peptide 1, which contains no cyclic β-residues (we included only one of the two crystallographically distinct but nearly identical helices from the structure of α/β-peptide 5 in the overlay analysis). These overlays were generated by aligning each backbone α-carbon (Cα) from residues 1–30 (including α- and β-amino acids) of α/β-peptides 5–10 with Cα from the corresponding residue of 1. The root-mean-square deviation (rmsd) for each alignment is shown. The small size of these rmsd values highlights the similarity of the backbone conformations displayed by α/β-peptides 5–10 relative to the backbone conformation of 1. This similarity shows that the cyclic β-residues in 5–10 can readily adopt the backbone torsion angles preferred by flexible β3-residues in helix-bundle-forming α/β-peptides.
Figure 3.
Overlay of the peptide backbones from α/β-peptides 5–10 onto the peptide backbone of α/β-peptide 1. These overlays were generated by aligning the backbone α-carbons from residues 1–30 (including α- and β-amino acids) of α/β-peptides 5–10 with the backbone α-carbons from the corresponding residues in α/β-peptide 1 (we included only one of the two crystallographically distinct but nearly identical helices from the structure of α/β-peptide 5 in the overlay analysis). α-Amino acid residues are colored yellow, β3-amino acid residues are colored blue, and cyclic β-amino acid residues are colored red.
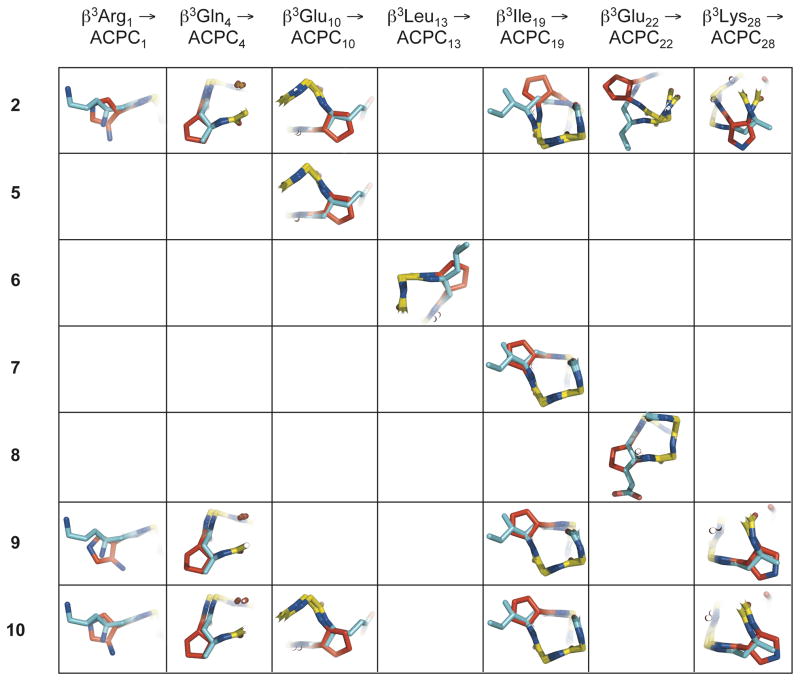
The ability of β-residues with a five-membered ring constraint to match the conformations displayed by flexible β-residues in an α-helix-like folding pattern is evident from the close overlay of individual ACPC and APC residues in α/β-peptides 5–10 with the corresponding β3-residues in 1 (Figure 4). Overlay of cyclic β-residues from 2 with corresponding β3-residues from 1 are included in this analysis for comparison. These overlays were generated by aligning backbone atoms (amino N, Cα, carbonyl C, carbonyl O, and for β-residues, Cβ) from residues 1–31 of α/β-peptides 2 and 5–10 with backbone atoms from corresponding residues of α/β-peptide 1 (only one of the two crystallographically distinct but nearly identical helix bundles from the structure of α/β-peptide 5 was included in the overlay analysis). Except for the N-terminal APC in 9 and 10, the backbone Cα and Cβ and the first carbon of the side chain (Cγ) of each cyclic β-residue in 5–10 align closely with Cα, Cβ and Cγ of the corresponding β3-residue in 1. In contrast, there is a substantial deviation between the positions of the cyclic β-residues at positions 19, 22 and 28 of 2 relative to the analogous β3-residues of 1 (upper right in Figure 4), as a result of the helical stammer discontinuity in 2. The high degree of positional similarity between the cyclic β-residues of 5–10 and the corresponding β3-residues of 1 suggests that the helical stammer observed in the crystal structure of α/β-peptide 2 is the exception rather than the rule, and that usually cyclic β-residues are structurally conservative replacements for β3-residues.
Figure 4.
Overlay of individual cyclic β-residues in α/β-peptides 2, and 5–10 with corresponding acyclic β3-residues in α/β-peptide 1, using the crystal structures of each peptide. These overlays were generated by aligning backbone atoms (amino N, Cβ, Cα, carbonyl C, carbonyl O, and for β-residues, Cβ) from residues 1–31 of α/β-peptides 2 and 5–10 with backbone atoms from the corresponding residues of α/β-peptide 1 (only one the two crystallographically distinct but nearly identical helix bundles from the structure of α/β-peptide 5 was included in the overlay analysis). Except for the ACPC and APC substitutions at β3Ile19, β3Glu22, and β3Lys28 in 2 (upper right), cyclic β-residues generally overlay well with their β3-residue counterparts, with backbone Cα and Cβ and side-chain Cγ atoms from each residue type located in close proximity. The close alignment of these structures suggests that cyclic β-residues are generally structurally conservative replacements for acyclic β3-residues in α/β-peptides. α-Amino acid residues are colored yellow, β3-amino acid residues are colored blue, and cyclic β-amino acid residues are colored red.
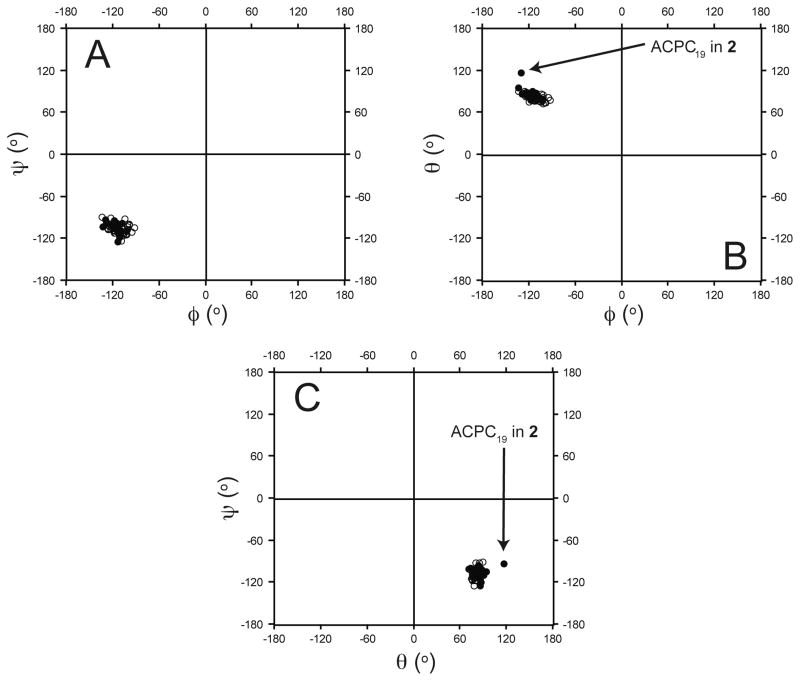
A comparison of the backbone dihedral angles of cyclic β-residues with those of analogous β3-residues in the crystal structures of α/β-peptides 1, 2 and 5–10 provides additional support for the conclusion that cyclic β-residues are structurally conservative replacements for β3-residues in this helical context. The average φ, θ and ψ torsion angles for the β3-residues in 1, 2 and 5–10 agree closely with the average φ, θ and ψ torsion angles for the cyclic β-residues (Figure 5; for this analysis, we used only one of the two crystallographically distinct, but nearly identical helices in the crystal structure of 5). The Ramachandran-type60–62 plots shown in Figure 6 (φ vs. ψ, φ vs. θ, and θ vs. ψ) highlight this similarity; β3-residues cluster tightly with cyclic β-residues in each plot with one exception, ACPC19 in 2, which is the residue that occupies an x-layer in the hydrophobic core one helical turn prior to the helical stammer.63 It is noteworthy that the backbone torsional preferences for cyclic residues reflected in Figure 6 are similar to those observed for ACPC residues in a large number of short α/β-peptides, with 1:1, 2:1 or 1:2 α:β repeat patterns, that crystallize without displaying helical bundle quaternary structure.64,65 The tight clustering of β3-residues with cyclic β-residues in Figure 6 suggests that cyclic β-residues can effectively imitate the local conformations of β3-residues in α/β-peptides that adopt an α-helix-like shape without necessarily causing the kinds of structural rearrangements that were observed in 2.
Figure 5.
Average backbone dihedral angles φ, θ, and ψ for (A) β3-residues and (B) cyclic β-residues in the crystal structures of α/β-peptides 1, 2, and 5–10 (only one the two crystallographically distinct but nearly identical helix bundles from the structure of α/β-peptide 5 was included in this analysis).
Figure 6.
Ramachandran plots of the dihedral angles adopted by β-residues (between residues 2–29) from α/β-peptides 1, 2, and 5–10 (only one the two crystallographically distinct but nearly identical helix bundles from the structure of α/β-peptide 5 was included in this analysis). (A) (φ, ψ), (B) (φ, θ), and (C) (θ, ψ) dihedral angles of β3- (open circles) and cyclic β-residues (filled circles). β3- and cyclic β-residues cluster in each of the (φ, ψ), (φ, θ), and (θ, ψ) plots with one exception (ACPC19 in 2; this is the residue that adopts an x-position in the hydrophobic core one helical turn prior to the stammer in 2). Overall, these data suggest that cyclic residues can readily adopt the conformations preferred by β3-residues in α/β-peptide helix bundles.
Characterization of 3–10 in aqueous solution
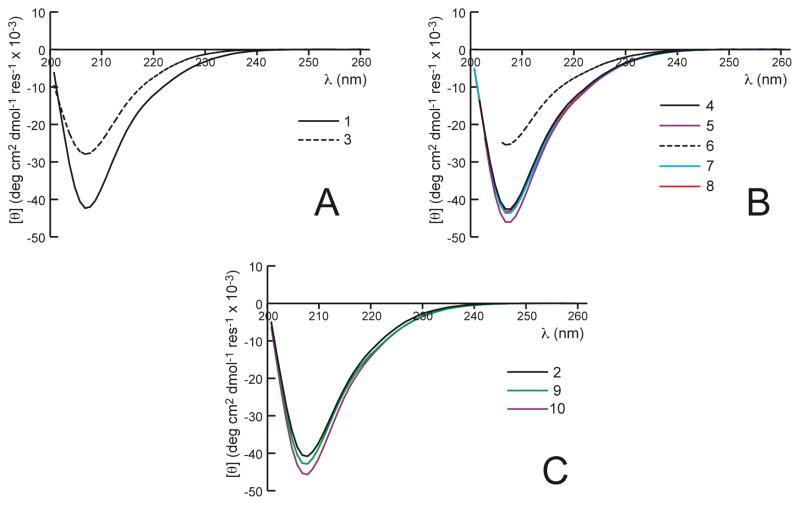
We used circular dichroism (CD) spectroscopy and sedimentation equilibrium analytical ultracentrifugation (AU) to investigate the effect of β3→cyclic replacements on the ability of α/β-peptides 3–10 to form helix-bundle quaternary structure in solution. Previous work has established that formation of an α-helix-like conformation by α/β-peptides is signaled by a strong CD minimum at ~206 nm; α/β-peptides that do not fold display little CD in this region.45,53,66 Among α/β-peptides such as 1 and 2, helical folding appears to occur concomitantly with helix bundle formation, as is true of comparable α-peptides (such as GCN4-p1 and its derivatives).46,67 Therefore, the intensity of the helix-characteristic CD signal at ~206 nm provides an indirect measure of quaternary structure formation. We expected that the nine β3→cyclic replacements used to generate 3 would provide increased conformational stability relative to 1, which has no cyclic β-residues. To our surprise, CD results suggest that 3 is significantly less folded than 1. Figure 7A shows that 100 μM 1 displays a much stronger minimum at 206 nm than does 100 μM 3, which implies that helix-bundle formation is less favorable for 3 than for 1, despite the presence of nine cyclic β-residues in 3. This result suggests that some cyclic β-residue replacements do not promote helix bundle formation in this α/β-peptide family, despite the general structural compatibility of β3- and cyclic β-residues demonstrated in the crystal structures of 5–10.
Figure 7.
CD spectra for (A) α/β-peptides 1 and 3, (B) 4–8, and (C) 2, 9, and 10 at 100 μM peptide in 10 mM NaOAc (pH 4.6) at 25 °C. In general, CD spectra of α/β-peptides with cyclic residues have the same shape as the CD spectrum of 1, with a global minimum near 206 nm that is close in magnitude to the global minimum in the CD spectrum of 1. α/β-Peptides 3 and 6 are exceptions, with minima near 206 nm that are significantly weaker than the minimum in the CD spectrum of 1, suggesting significantly diminished helicity in 3 and 6 relative to 1.
As described above, α/β-peptides 4–8 each contain only one β3→cyclic replacement. Comparison of CD spectra (Figure 7B) suggest that most of these single β3→cyclic replacements exert little impact on coupled folding and assembly relative to 1. The lone exception is α/β-peptide 6 (e, β3-hLeu13→ACPC), for which diminished intensity at 206 nm indicates diminished folding. The similarity between the CD signatures for 3 and 6 suggests that the β3→cyclic replacement at residue 13 is at least partially responsible for the diminished folding/assembly of 3 relative to other members of this α/β-peptide family. CD data suggest that 9 and 10 display high levels of folding and assembly (Figure 7C).
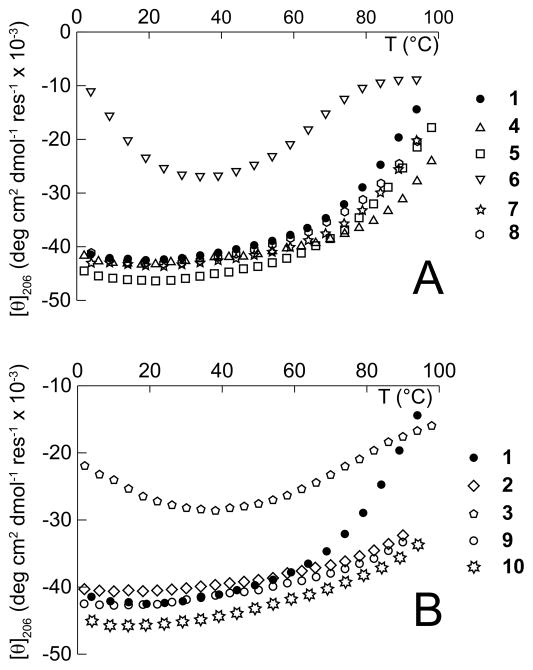
Variable temperature CD experiments (Figure 8) support the conclusions drawn from analysis of the CD spectra of α/β-peptides 3–10 at 25 °C, although the high stability of the structures adopted by α/β-peptides 1, 2, 4, 5 and 7–10 (incomplete transitions) and the non-monotonic behavior observed for α/β-peptides 3 and 6 make quantitative thermodynamic interpretation of the data impossible. Qualitatively, CD intensity for 4, 5, 7 and 8 (which each contain one cyclic β-residue) decreases by a smaller amount with increasing temperature than does the CD intensity for 1 (which has no cyclic β-residues, see Figure 8A), suggesting that the assemblies formed by 4, 5, 7 and 8 are likely to be a little more stable than the assembly formed by 1, and that replacing β3-residues with cyclic β-residues can stabilize helical secondary structure and helix-bundle quaternary structure. As noted previously, this stabilizing effect appears to be context dependent; α/β-peptide 6, which has a β3hLeu→ACPC mutation at position 13, appears to form an assembly that is significantly less stable than that of 1, which has no cyclic residues.
Figure 8.
Variable temperature CD (monitored at 206 nm) for α/β-peptides 1–10 (100 μM) in 10 mM NaOAc (pH 4.6). (A) α/β-Peptides 4–8 each contain one cyclic β-residue, and (B) α/β-peptides 2, 3, 9, and 10 each have multiple cyclic β-residues, whereas α/β-peptide 1 has no cyclic β-residues.
Increasing the number of cyclic β-residue substitutions appears to increase helix-bundle stability in three of four cases: CD intensity for 2, 9 and 10 (which each contain multiple cyclic β-residues) does not change much with increasing temperature (Figure 8B), suggesting that these α/β-peptides form more stable assemblies than do 4, 5, 7 or 8 (which each contain only one cyclic β-residue). The exception is 3, which has the β3hLeu13→ACPC mutation that is also present in 6 and forms a similarly unstable assembly. Thus, of the cyclic β3→cyclic replacements we have investigated, it seems that only one (β3-hLeu13→ACPC) causes destabilization of helical folding and assembly. We do not presently understand why the β3→cyclic replacement at position 13 is so destabilizing to 3 and 6 in solution, especially given the high structural similarity of 6 to 1, 5 and 7–10 in the crystalline state (Figures 2–4).63
Association stoichiometry in solution
Sedimentation equilibrium data were used to evaluate the stoichiometry of α/β-peptide self-association for 3–10 (Table 1). Previously we reported that 1 sediments as a tetramer when analyzed at 200 μM in 10 mM aqueous NaOAc, pH 4.6 containing 150 mM NaCl.48 α/β-Peptide 3 was evaluated at 100 μM in buffer lacking NaCl. Under these conditions, the AU data for 3 were best fit to a two-state model involving monomeric and trimeric states in a ~1:2 ratio. These results cannot be directly compared to those for 1 because of the difference in conditions, but we note that a lower association propensity of 3 relative to 1 would be consistent with the difference in CD signatures of these two α/β-peptides (Figure 7A).
Table 1.
Self-Association Behavior of α/β-peptides 1–10a as determined by Sedimentation Equilibrium Experiments
| Peptide | Best-fit Model for the Observed Equilibrium Sedimentation Behaviorb |
|---|---|
| 1 | 4 |
| 2 | 4 |
| 3 | (1,3)c |
| 4 | (1,4,5) |
| 5 | 4 |
| 6 | (1,4,5) |
| 7 | 4 |
| 8 | 4 |
| 9 | (1,5) |
| 10 | 4 |
Except as noted, sedimentation equilibrium data are for 200 μM peptide solutions in 10 mM aqueous sodium acetate, pH 4.6 + 150 mM NaCl, at 25 °C. Data for 1 and 2 are from reference 48 and are included for comparison.
See Supporting Information for details. A single number indicates that the best-fit model was for a single species of the following size: 1=monomer, 2=dimer, 3=trimer, etc. Multipe numbers in parentheses indicate that the best-fit model is for multiple species in equilibrium: (1,4,5) indicates a monomer-tetramer-pentamer equilibrium.
Data for 3 were collected at 100 μM peptide and in 10 mM sodium acetate, pH 4.6 (no NaCl).
Among the α/β-peptides containing single β3→cyclic replacements, 5, 7 and 8 displayed sedimentation equilibrium data consistent with the presence of a single tetrameric species. In contrast, the data for 6 were best fit with a three-state model involving monomer, tetramer and pentamer. The lower self-assembly propensity of 6 relative to 1, 5, 7 and 8 is consistent with the weaker CD intensity for 6 relative to the other four α/β-peptides. AU data for 4 were best fit with a monomer-tetramer-pentamer model, with roughly half the material in the tetrameric state and roughly a quarter in the monomeric state, which is surprising because the CD signature of 4 is similar to those of other α/β-peptides that appear to be entirely or largely tetrameric. The contrast between AU data for 4 and AU data for 1, 5, 7 and 8 is puzzling since the lone β3→cyclic replacement in 4 occurs at a c-position in the heptad repeat, which is not expected to participate in inter-helical packing.
Sedimentation equilibrium data for 9, which contains four β3→cyclic replacements relative to 1, are best fit with a two state model involving a monomer and a pentamer, with the latter dominant (>80%). In contrast, data for 10, which has one additional β3→cyclic replacement relative to 9, are best fit by a single-species model involving a tetramer. It is unclear why changing β3-hGlu10 in 9 to ACPC in 10 would exert such a profound effect on self-assembly behavior. This effect may be related to a change in the nature of the side chain at residue 10 (acidic in 9 to hydrophobic in 10). However, changing β3-hGlu10 in 1 to ACPC10 in 5 does not influence self-assembly behavior, which suggests that a subtle interplay among multiple sites of β3→cyclic replacement can affect the preferred association state of α/β-peptide helix bundles in solution.
The consistency of the tetramer stoichiometry observed for the helix-bundle forms of α/β-peptide 1, 2 and 5–10 in the crystalline state stands in contrast to the variability in self-association suggested by AU data. Most of these molecules display a strong preference for tetramer formation under the AU conditions, but 4, 6 and 9 deviate somewhat from this trend. Both 4 and 6 may form a small amount of pentamer under AU conditions. Given the complexity of three-state fitting to the experimental data, which was necessary for 4 and 6, perhaps the more conservative conclusion is simply that these α/β-peptides have some tendency to adopt solution stoichiometries other than tetramer, although the tetramer appears to be preferred. For 9, however, the AU data seem to suggest a preference for pentamer in solution, even though 9 crystallizes as a tetramer. Since all six of the new structures feature a similar crystal lattice, the behavior of 9 may suggest that lattice packing forces exert a significant effect on the quaternary structure observed crystallographically in this family of α/β-peptides.
Conclusions
We have provided a large amount of new high-resolution structural data for α/β-peptides in α-helix-like conformations. The results indicate that a five-membered ring constraint, such as that found in ACPC or heterocyclic analogue APC, supports the β-residue backbone torsion angles required for an α-helix-like secondary structure. These data provide a foundation for the design of foldamers that mimic α-helical segments of natural proteins.
If ACPC and APC are predisposed to adopt the local conformation required for α-helix mimicry, as is suggested by our structural data for α/β-peptide helix bundles and by earlier crystallographic evidence obtained with short, helical α/β-peptides,64,65 then replacement of flexible β3-residues by cyclic analogues should stabilize helical secondary structure and any helix-bundle quaternary structure that requires helical folding. We examined thermal disruption of helix-bundle quaternary structure formed by the α/β-peptides 1–10, but quantitative stability differences were impossible to ascertain, in part because even the most flexible molecule, 1, forms an assembly that is not fully disrupted at the maximum attainable temperature. Nevertheless, qualitative comparisons indicate that α/β-peptides 4–8 (which each have one cyclic β-residue) form assemblies that are at least as stable as that formed by 1 (which has no cyclic β-residues), and α/β-peptides 2, 9, and 10 form assemblies that are significantly more stable than the assemblies formed by α/β-peptides 1 and 4–8 (Figure 8). Previous work with short, non-associating α/β-peptides has indicated qualitatively that β3→cyclic replacements enhance helical folding.52 Recent results with a hybrid tertiary structure in which an α/β-peptide helix packs against an α-helix have shown that β3→cyclic replacements at solvent-exposed sites provide a ~0.4 kcal/mol structural stabilization (ΔG).54
The present results show that β3→cyclic replacement is not automatically stabilizing: α/β-peptide 6 contains a cyclic residue at just one position (ACPC13), but this cyclic residue causes a substantial destabilization of helix-bundle quaternary structure relative to 1, which contains no cyclic residues. The difference in quaternary structural stability between 1 and 6 presumably arises because steric repulsions experienced by the non-backbone atoms of ACPC13 in the folded/assembled state can overwhelm conformational benefits of limiting backbone flexibility. Repulsive interactions involving ACPC13 in 6 are not immediately apparent upon comparing the structure of 6 with that of 1,63 though such interactions could be important, especially if the solution and crystal structures of 6 differ significantly. This type of interplay between backbone preorganization and unfavorable interactions of non-backbone atoms may explain why mutating cyclic β-residues to β3 analogues had little effect on the binding of helical α/β-peptides to a complementary cleft on the protein Bcl-xL.68
The observation that some of the α/β-peptides (3, 4, 6 and 9) diverge from simple tetrameric stoichiometry in solution suggests that β3→cyclic replacements can exert variable effects on quaternary structure propensity, depending on context. Because of current limitations in the available pool of β-amino acid building blocks with a five-membered ring constraint, β3→cyclic replacements can alter side-chain encoded information, and such alterations may give rise to the apparent changes in self-association behavior that we have observed in some cases.
It is currently difficult to predict whether β3→cyclic replacement at any given position in a helical α/β-peptide will result in changes to helix-bundle self-association behavior (observed for α/β-peptides 3, 4, 6 and 9), or decreases in helix-bundle stability (observed for α/β-peptides 3 and 6). However, more than half of the α/β-peptides investigated here form assemblies that are at least as stable as that of the parent α/β-peptide 1 and are faithful to the tetrameric association state specified by the parent sequence. The β3→cyclic replacement design principles described here and elsewhere48,54 may not enhance α/β-peptide helix-bundle formation in every case, but they remain useful design tools for generating helical α/β-peptides that maintain the desired helicity and self-association behavior while benefitting from the structural preorganization imparted by the cyclic β-residues. Indeed, we have recently shown that β3→cyclic replacements can substantially improve the affinity of gp41-mimetic α/β-peptides for a complementary protein surface, which leads to improved efficacy for inhibiting HIV infection relative to α/β-peptides that lack cyclic β-residues.59 The results reported here should facilitate future efforts to use cyclically constrained β-amino acid residues to optimize α/β-peptides that block biomedically important protein-protein interactions.
Supplementary Material
Acknowledgments
We thank Prof. James Keck for providing access to x-ray instrumentation and PepTech for providing protected β3-amino acid derivatives at a discount. This work was supported by National Institutes of Health (NIH) Grant GM61238. J.L.P. was supported in part by a Chemistry-Biology Interface Training Grant (T32 GM008505), and W.S.H. was supported in part by an NIH postdoctoral fellowship (CA119875). CD and AU data were obtained at the University of Wisconsin Biophysics Instrumentation Facility, which was established with support from the University of Wisconsin, National Science Foundation Grant BIR-9512577 and NIH Grant S10 RR13790. The microwave reactor was purchased with support from the University of Wisconsin, Madison Nanoscale Science and Engineering Center (DMR-0832760).
Footnotes
Supporting Information Available. Supplementary AU data and experimental methods. This material is available free of charge via the Internet at http://pubs.acs.org. Coordinates for α/β-peptides 5–10 have been deposited in the Protein Data Bank (PDB: 3HET, 3HEU, 3HEV, 3HEW, 3HEX, 3HEY, respectively).
References
- 1.Anfinsen CB. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Schueler-Furman O, Wang C, Bradley P, Misura K, Baker D. Science. 2005;310:638–642. doi: 10.1126/science.1112160. [DOI] [PubMed] [Google Scholar]
- 3.Hartman MCT, Josephson K, Lin C-W, Szostak JW. PLoS ONE. 2007;2:e972. doi: 10.1371/journal.pone.0000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powers ET, Deechongkit S, Kelly JW. Adv Protein Chem. 2005;72:39–78. doi: 10.1016/S0065-3233(05)72002-7. [DOI] [PubMed] [Google Scholar]
- 5.Lu W, Qasim MA, Laskowski M, Kent SBH. Biochemistry. 1997;36:673–679. doi: 10.1021/bi9625612. [DOI] [PubMed] [Google Scholar]
- 6.Koh JT, Cornish VW, Schultz PG. Biochemistry. 1997;36:11314–11322. doi: 10.1021/bi9707685. [DOI] [PubMed] [Google Scholar]
- 7.Karle IL, Das C, Balaram P. Biopolymers. 2001;59:276–289. doi: 10.1002/1097-0282(20011005)59:4<276::AID-BIP1024>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 8.Beligere GS, Dawson PE. J Am Chem Soc. 2000;122:12079–12082. [Google Scholar]
- 9.Gordon DJ, Meredith SC. Biochemistry. 2003;42:475–485. doi: 10.1021/bi0259857. [DOI] [PubMed] [Google Scholar]
- 10.Silinski P, Fitzgerald MC. Biochemistry. 2003;42:6620–6630. doi: 10.1021/bi020700b. [DOI] [PubMed] [Google Scholar]
- 11.Deechongkit S, Nguyen H, Powers ET, Dawson PE, Gruebele M, Kelly JW. Nature. 2004;430:101–105. doi: 10.1038/nature02611. [DOI] [PubMed] [Google Scholar]
- 12.Gao J, Bosco DA, Powers ET, Kelly JW. Nat Struct Mol Biol. 2009;16:684–690. doi: 10.1038/nsmb.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gante J. Angew Chem Int Ed. 1994;33:1699–1720. [Google Scholar]
- 14.Wipf P, Xiao J, Stephenson CRJ. CHIMIA Int J Chem. 2009;63:764–775. doi: 10.2533/chimia.2009.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hann MM, Sammes PG, Kennewell PD, Taylor JB. Chem Commun. 1980:234–235. [Google Scholar]
- 16.Johnson RL. J Med Chem. 1984;27:1351–1354. doi: 10.1021/jm00376a023. [DOI] [PubMed] [Google Scholar]
- 17.Wipf P, Fritch PC. J Org Chem. 1994;59:4875–4886. [Google Scholar]
- 18.Gardner RR, Liang G-B, Gellman SH. J Am Chem Soc. 1995;117:3280–3281. [Google Scholar]
- 19.Gardner RR, Liang G-B, Gellman SH. J Am Chem Soc. 1999;121:1806–1816. [Google Scholar]
- 20.Jenkins CL, Vasbinder MM, Miller SJ, Raines RT. Org Lett. 2005;7:2619–2622. doi: 10.1021/ol050780m. [DOI] [PubMed] [Google Scholar]
- 21.Fu Y, Bieschke J, Kelly JW. J Am Chem Soc. 2005;127:15366–15367. doi: 10.1021/ja0551382. [DOI] [PubMed] [Google Scholar]
- 22.Dai N, Wang XJ, Etzkorn FA. J Am Chem Soc. 2008;130:5396–5397. doi: 10.1021/ja711021m. [DOI] [PubMed] [Google Scholar]
- 23.Oishi S, Kamitani H, Kodera Y, Watanabe K, Kobayashi K, Narumi T, Tomita K, Ohno H, Naito T, Kodama E, Matsuoka M, Fujii N. Org Biomol Chem. 2009;7:2872–2877. doi: 10.1039/b907983a. [DOI] [PubMed] [Google Scholar]
- 24.Horne WS, Yadav MK, Stout CD, Ghadiri MR. J Am Chem Soc. 2004;126:15366–15367. doi: 10.1021/ja0450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tam A, Arnold U, Soellner MB, Raines RT. J Am Chem Soc. 2007;129:12670–12671. doi: 10.1021/ja075865s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patgiri A, Jochim AL, Arora PS. Accounts of Chemical Research. 2008;41:1289–1300. doi: 10.1021/ar700264k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold U, Hinderaker MP, Nilsson BL, Huck BR, Gellman SH, Raines RT. J Am Chem Soc. 2002;124:8522–8523. doi: 10.1021/ja026114n. [DOI] [PubMed] [Google Scholar]
- 28.Liu F, Du D, Fuller AA, Davoren JE, Wipf P, Kelly JW, Gruebele M. Proc Natl Acad Sci USA. 2008;105:2369–2374. doi: 10.1073/pnas.0711908105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuller AA, Du D, Liu F, Davoren JE, Bhabha G, Kroon G, Case DA, Dyson HJ, Powers ET, Wipf P, Gruebele M, Kelly JW. Proc Natl Acad Sci USA. 2009;106:11067–11072. doi: 10.1073/pnas.0813012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.David R, Günther R, Baumann L, Lühmann T, Seebach D, Hofmann H-J, Beck-Sickinger AG. J Am Chem Soc. 2008;130:15311–15317. doi: 10.1021/ja802453x. [DOI] [PubMed] [Google Scholar]
- 31.Hecht S, Huc I, editors. Foldamers: Structure Properties, and Applications. Wiley-VCH; Weinheim: 2007. [Google Scholar]
- 32.Gellman SH. Acc Chem Res. 1998;31:173–180. [Google Scholar]
- 33.Kirshenbaum K, Zuckermann RN, Dill KA. Curr Opin Struct Biol. 1999;9:530–535. doi: 10.1016/S0959-440X(99)80075-X. [DOI] [PubMed] [Google Scholar]
- 34.Hill DJ, Mio MJ, Prince RB, Hughes TS, Moore JS. Chem Rev. 2001;101:3893–4012. doi: 10.1021/cr990120t. [DOI] [PubMed] [Google Scholar]
- 35.Czyzewski AM, Barron AE. AIChE Journal. 2008;54:2–8. [Google Scholar]
- 36.Raguse TL, Lai JR, LePlae PR, Gellman SH. Org Lett. 2001;3:3963–3966. doi: 10.1021/ol016868r. [DOI] [PubMed] [Google Scholar]
- 37.Cheng RP, DeGrado WF. J Am Chem Soc. 2002;124:11564–11565. doi: 10.1021/ja020728a. [DOI] [PubMed] [Google Scholar]
- 38.Daniels DS, Petersson EJ, Qiu JX, Schepartz A. J Am Chem Soc. 2007;129:1532–1533. doi: 10.1021/ja068678n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersson EJ, Schepartz A. J Am Chem Soc. 2008;130:821–823. doi: 10.1021/ja077245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pomerantz WC, Grygiel TLR, Lai JR, Gellman SH. Org Lett. 2008;10:1799–1802. doi: 10.1021/ol800622e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burkoth TS, Beausoleil E, Kaur S, Tang D, Cohen FE, Zuckermann RN. Chem Biol. 2002;9:647–654. doi: 10.1016/s1074-5521(02)00140-0. [DOI] [PubMed] [Google Scholar]
- 42.Lee B-C, Zuckermann RN, Dill KA. J Am Chem Soc. 2005;127:10999–11009. doi: 10.1021/ja0514904. [DOI] [PubMed] [Google Scholar]
- 43.Delsuc N, Léger JM, Massip S, Huc I. Angew Chem Int Ed. 2007;46:214–217. doi: 10.1002/anie.200603390. [DOI] [PubMed] [Google Scholar]
- 44.Horne WS, Gellman SH. Acc Chem Res. 2008;41:1399–1408. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horne WS, Price JL, Keck JL, Gellman SH. J Am Chem Soc. 2007;129:4178–4180. doi: 10.1021/ja070396f. [DOI] [PubMed] [Google Scholar]
- 46.O’Shea EK, Klemm JD, Kim PS, Alber T. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 47.Harbury PB, Zhang T, Kim PS, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 48.Horne WS, Price JL, Gellman SH. Proc Natl Acad Sci USA. 2008;105:9151–9156. doi: 10.1073/pnas.0801135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giuliano MW, Horne WS, Gellman SH. J Am Chem Soc. 2009;131:9860–9861. doi: 10.1021/ja8099294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng RP, Gellman SH, DeGrado WF. Chem Rev. 2001;101:3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 51.Hayen A, Schmitt MA, Ngassa FN, Thomasson KA, Gellman SH. Angew Chem Int Ed. 2004;43:505–510. doi: 10.1002/anie.200352125. [DOI] [PubMed] [Google Scholar]
- 52.Schmitt MA, Choi SH, Guzei IA, Gellman SH. J Am Chem Soc. 2005;127:13130–13131. doi: 10.1021/ja0536163. [DOI] [PubMed] [Google Scholar]
- 53.Price JL, Horne WS, Gellman SH. J Am Chem Soc. 2007;129:6376–6377. doi: 10.1021/ja071203r. [DOI] [PubMed] [Google Scholar]
- 54.Price JL, Hadley EB, Steinkruger JD, Gellman SH. Angew Chem Int Ed. 2010;49:368–371. doi: 10.1002/anie.200904714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown JH, Cohen C, Parry DAD. Prot Struct Funct Gen. 1996;26:134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 56.Sadowsky JD, Schmitt MA, Lee H-S, Umezawa N, Wang S, Tomita Y, Gellman SH. J Am Chem Soc. 2005;127:11966–11968. doi: 10.1021/ja053678t. [DOI] [PubMed] [Google Scholar]
- 57.Horne WS, Boersma MD, Windsor MA, Gellman SH. Angew Chem Int Ed. 2008;47:2853–2856. doi: 10.1002/anie.200705315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee EF, Sadowsky JD, Smith BJ, Czabotar PE, Peterson-Kaufman KJ, Colman PM, Gellman SH, Fairlie WD. Angew Chem Int Ed. 2009;48:4318–4322. doi: 10.1002/anie.200805761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horne WS, Johnson LM, Ketas TJ, Klasse PJ, Lu M, Moore JP, Gellman SH. Proc Natl Acad Sci USA. 2009;106:14751–14756. doi: 10.1073/pnas.0902663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kleywegt GJ, Jones TA. Structure. 1996;4:1395–1400. doi: 10.1016/s0969-2126(96)00147-5. [DOI] [PubMed] [Google Scholar]
- 61.Ramakrishnan C, Ramachandran GN. Biophys J. 1965;5 doi: 10.1016/S0006-3495(65)86759-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramachandran GN, Sasisekharan V. Adv Protein Chem. 1968;23:283–437. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 63.See supporting information for details.
- 64.Choi SH, Guzei IA, Spencer LC, Gellman SH. J Am Chem Soc. 2008;130:6544–6550. doi: 10.1021/ja800355p. [DOI] [PubMed] [Google Scholar]
- 65.Choi SH, Guzei IA, Spencer LC, Gellman SH. J Am Chem Soc. 2009;131:2917–2924. doi: 10.1021/ja808168y. [DOI] [PubMed] [Google Scholar]
- 66.Schmitt MA, Weisblum B, Gellman SH. J Am Chem Soc. 2007;129:417–428. doi: 10.1021/ja0666553. [DOI] [PubMed] [Google Scholar]
- 67.O’Shea EK, Lumb KJ, Kim PS. Curr Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 68.Sadowsky JD, Fairlie WD, Hadley EB, Lee H-S, Umezawa N, Nikolovska-Coleska Z, Wang S, Huang DCS, Tomita Y, Gellman SH. J Am Chem Soc. 2007;129:139–154. doi: 10.1021/ja0662523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.