Abstract
Hereditary breast and ovarian cancer syndromes can be caused by loss-of-function germline mutations in one of two tumour-suppressor genes, BRCA1 and BRCA2 (ref. 1). Each gene product interacts with recombination/DNA repair proteins in pathways that participate in preserving intact chromosome structure. However, it is unclear to what extent such functions specifically suppress breast and ovarian cancer. Here we analyse what is known of BRCA gene function and highlight some unanswered questions in the field.
The amino acid sequences of BRCA1 and BRCA2 originally provided few clues to their specific functions (Fig. 1a). BRCA1 was found to localize to sub-nuclear foci during the S and G2 phases of the cell cycle. These foci also contain Rad51, a protein central to homologous recombination2. Endogenous BRCA1-Rad51 complexes were detected in cell extracts. BRCA2 also interacts and co-localizes with Rad51 and BRCA1 (refs 3 and 4). The three genes are co-expressed in development1.
Figure 1.
Structure and localization of BRCA proteins. a, Structure of the BRCA1 and BRCA2 polypeptides. BRCA1 shows RING domain, BRCT motifs, implicated in DNA damage response pathways. BRCA2 shows BRC repeats, involved in the Rad51 interaction. Some associated proteins are denoted in blue. b, Localization of BRCA1 during meiotic prophase I. Depicted is a single primary, human spermatocyte nucleus, stained with an antibody to the synaptonemal complex protein, SCP3 (white), and with BRCA1 antibody (red). BRCA1 localizes to the unsynapsed regions (axial elements) of developing synaptonemal complexes. Copyright held by Cell Press, reproduced with permission.
Human Rad51, like its homologues in other eukaryotes (RAD51) and prokaryotes (RecA), operates in the repair of double-strand DNA breaks (DSB) by binding single-stranded (ss) DNA to form a nucleoprotein filament that can invade a homologous duplex DNA molecule. Saccharomyces cerevisiae rad51 mutants are defective in double-strand break repair (DSBR) and meiosis, two processes dependent upon homologous recombination.
Rad51 also exhibits characteristic localization patterns during meiosis. The synaptonemal complex (SC) is a linear protein/DNA-bearing structure that assembles at the onset of meiotic prophase I, a tetraploid stage during which homologous chromosomes become paired before meiotic recombination. The SC forms by the apposition (‘synapsis’) of two homologous axial elements, each containing two identical sister chromatids and associated proteins. Rad51 binds axial elements. Strikingly, BRCA1 and BRCA2 act similarly2,4 (Fig. 1b), suggesting that BRCA proteins function in meiotic recombination, and hinting at a role in sister chromatid interactions.
Relocation of BRCA1 to sites of DNA synthesis
After hydroxyurea (HU)-mediated replication arrest or exposure of S-phase cells to ultraviolet (UV) light, BRCA1 rapidly relocalizes from its ‘native’ nuclear foci to sites of DNA synthesis and becomes hyperphosphorylated5. This suggests that BRCA1 is involved in the repair of abnormal DNA structures generated at sites of DNA synthesis after DNA damage. Among the proteins that interact and co-localize with BRCA1 in nuclear foci are BRCA2 and BARD1. Like BRCA1, BARD1 contains an amino-terminal RING domain and carboxy-terminal BRCT motifs6. BARD1, BRCA2 and Rad51 all accompany BRCA1 in its re-localization after DNA damage4,5. This implies that these multiprotein complexes are dedicated, at least in part, to repair of replication-associated DNA damage.
In yeast, cell cycle control responses to HU or to certain DNA damaging agents delivered in the S phase are controlled by S-phase checkpoint gene products, which include S. cerevisiae MEC1 and Schizosaccharomyces pombe rad3, two large nuclear kinases with C-terminal PI3 kinase-like domains. Atr is a mammalian homologue of these proteins and, like BRCA proteins, it also localizes to axial elements during meiotic prophase I7. Recently, Atr was found to colocalize with BRCA1 in somatic cells, both before and after replication arrest41. Atr, in part, controls BRCA1 phosphorylation following HU treatment41. BRCA1 is also phosphorylated following gamma irradiation5, under the control of a related kinase, Atm8.
Death by checkpoint
Mouse gene targeting experiments provided insights into the relationship between BRCA mutation and cancer (reviewed in refs 1 and 9). Nullizygous BRCA1 or BRCA2 embryos die around the time of gastrulation. Unexpectedly, these embryos reveal a proliferative defect and induction of the p53-dependent cell cycle inhibitor, p21 (ref. 10). It was argued that, if BRCA1 regulates DNA repair, its inactivation might lead to spontaneous abnormalities in DNA structure. If so, induction of p21 in BRCA1-/- embryos might reflect the activation of a DNA damage-dependent checkpoint2. The resulting cell cycle delay could have catastrophic effects on a gastrulating embryo, leading to ‘death by checkpoint’. Consistent with this hypothesis, p53 or p21 nullizygosity delays the death of BRCA1-/- or BRCA2-/- embryos (reviewed in refs 1 and 9).
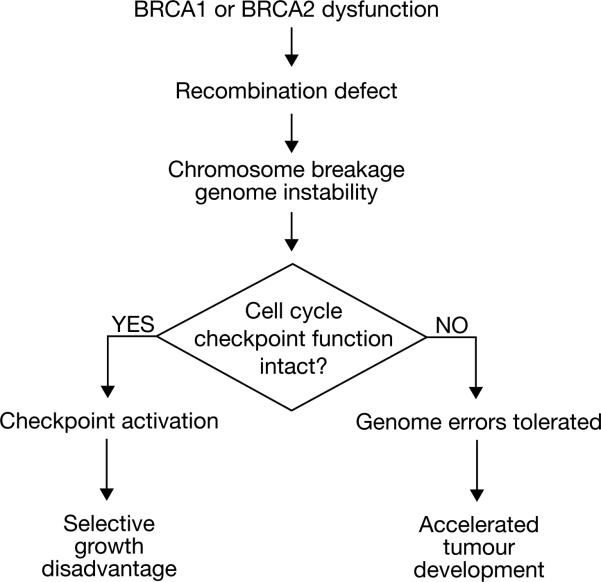
BRCA1 dysfunction might have similar effects in adult cells; it may precipitate spontaneous abnormalities of DNA structure and, thereby, provoke ‘death by checkpoint’. If, however, loss of BRCA1 function were to occur in a pre-malignant cell that had already suffered inactivation of key checkpoints, the aberrant DNA structures resulting from BRCA1 inactivation might be tolerated without cell cycle arrest2 (Fig. 2). This might promote neoplastic development. BRCA1 or BRCA2 inactivation, if it were to lead to cancer, would then not be the first ‘hit’ during tumorigenesis. Alternatively, BRCA gene inactivation in an otherwise wild-type cell would have to be closely followed by the inactivation of key checkpoints.
Figure 2.
Checkpoint inactivation and BRCA gene-mediated tumorigenesis. Inactivation of DNA damage-responsive cell cycle checkpoints may contribute to the cellular response to BRCA gene dysfunction. Loss of BRCA function and associated chromosome breakage in a cell trigger checkpoints that deselect it. If such a checkpoint has already been disabled (as in a pre-cancerous cell), the chromosome breakage syndrome may be tolerated and lead to accelerated neoplastic progression.
Support for the notion that BRCA genes suppress genome instability comes from several sources. Embryonic tissue lacking wt (wild-type) BRCA1 or BRCA2 reveals ionizing radiation (IR) hypersensitivity, consistent with a defect in DSBR3,9. BRCA1 or BRCA2 homozygous mutant mouse embryo fibroblasts (MEFs) undergo spontaneous chromosome breakage accompanied by checkpoint-mediated growth arrest9,11. Moreover, severe aneuploidy and centrosome amplification are observed in these cells1,9. The full identity of the checkpoints responsible for cell cycle arrest of BRCA1 or BRCA2 mutated cells is unclear. Among the proposed candidates is the spindle pole checkpoint1,9. Rad51-/- mouse embryos also suffer early death that is partially rescued by p53 germline inactivation, and cultured cell lines deprived of Rad51 undergo spontaneous chromosome breakage.
Interestingly, BRCA gene and checkpoint-mediated mechanisms may also suppress genome instability in breast tissue. Recently, breast-specific murine BRCA1 gene inactivation was achieved9. These mice developed late-onset breast cancer with frequent, tumour-associated p53 mutations. On a p53+/- background, earlier-onset and higher-frequency disease was detected. p53 hemizygosity was also found to promote breast tumour development in BRCA1+/- mice exposed to ionizing radiation1,9. Thus, p53-mediated checkpoint functions may contribute to proliferation control of BRCA1-deficient breast ductal cells. Loss of these p53 functions could increase tolerance of genome instability and promote tumorigenesis.
BRCA-mediated recombination and tumour suppression
Human genetics has defined numerous mutant BRCA1 and BRCA2 alleles defective in tumour suppression. This has provided a tool for testing the relationship between BRCA1 tumour suppressor function and its role in genome integrity maintenance. Recently, a breast cancer cell line, HCC1937, that lacks wtBRCA1 was found to be hypersensitive to IR and to exhibit delayed kinetics of DSBR. Stable expression of wtBRCA1 in these cells reversed IR sensitivity and restored efficient DSBR13. In contrast, several clinically important mutant alleles, each bearing a missense mutation in one of three distinct BRCA1 protein domains—the N-terminal RING domain, the C-terminal BRCT motifs, or a centrally located region—failed to rescue the phenotype. The DSBR function of BRCA1, therefore, arises from the participation of diverse structural elements of the protein. As each of these domains also serves as an interaction centre for associated proteins, a scaffolding role for BRCA1 in DSBR seems possible. Moreover, given the inactivation of this function by a diverse set of disease-producing, missense mutations, DSBR may be a key BRCA1 tumour-suppressor function.
At least two distinct processes, homologous recombination (HR) and non-homologous end joining (NHEJ), contribute to DSBR in mammalian cells. The interaction of BRCA1 with the NBS1–MRE11–Rad50 complex (reviewed in ref. 1) potentially implicates it in either of these processes. Current data suggest a specific contribution of BRCA genes to HR. For example, BRCA1 mutant mice reveal arrested spermatogenesis in meiotic prophase I, the stage at which BRCA1 is normally localized to the axial elements of developing synaptonemal complexes (reviewed in refs 1 and 9). A unique, viable BRCA1 homozygous mutant murine embryonic stem (ES) cell clone revealed reduced rates of HR or single-strand annealing in response to a site-specific DSB14. Gene targeting, an HR-dependent process, was enhanced by expression of wtBRCA1 in the same BRCA1-mutated ES cell clone15. By contrast, where measured, NHEJ appeared normal in BRCA1 or BRCA2 mutant cells11,14.
DNA polymerase stalling and S-phase checkpoint activation
If the BRCA genes act as tumour suppressors by supporting HR, which HR-dependent process is the key disease-relevant target? One candidate is sister chromatid recombination (SCR), a potentially error-free process which operates, in part, in the repair of recombinogenic DNA lesions generated during S phase16. SCR may be provoked by attempted replication across lesions that induce DNA polymerase stalling, such as those caused by UV irradiation or DNA adduction17. Indeed, work in prokaryotes indicates that replication arrest is a common event during DNA synthesis and that recombination functions are required for replication restart, genome integrity maintenance, and cell viability (reviewed in ref. 18).
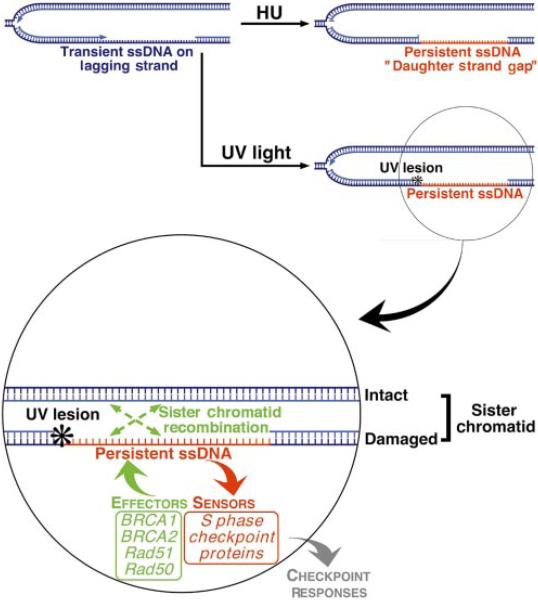
Hydroxyurea exposure may mimic such DNA polymerase-stalling events by causing genome-wide replication arrest. HU or UV treatment of S phase cells may each give rise to persistent ssDNA tracts—‘daughter strand gaps’ (DSG)—in close proximity to a replication fork19 (Fig. 3). We propose that such DSGs contribute to the rapid relocalization of BRCA1 and associated proteins to sites of replication following HU or UV treatment5. DSGs, or their derivatives, might activate S phase checkpoint signalling and also serve as substrates of recombinational responses, such as SCR (Fig. 3). Atr now appears to be a regulator of these events41. Interestingly, ATR-/-embryos suffer early embryonic lethality with spontaneous chromosome breakage—a phenotype reminiscent of BRCA-/- and Rad51-/- embryos20.
Figure 3.
Proposed role for the BRCA proteins in sister chromatid recombination. DNA polymerase stalling activates S-phase checkpoint signalling and recruits BRCA1, BRCA2, Rad51 and associated proteins to sites of arrested DNA synthesis. We propose that this recruitment arises from the existence of tracts of persistent ssDNA close to a replication fork. Such lesions, or their derivatives, may activate S-phase checkpoint signalling and act as a substrate for BRCA protein-mediated recombinational repair.
Whereas S-phase ‘checkpoint’ kinases signal to BRCA1, little is known of how these phosphorylations alter BRCA1 function. We do know that, unlike Atr, intact BRCA proteins are not required for S-phase checkpoint function in itself (ref. 11; R.S. and D.M.L., unpublished work). This implies that BRCA1 plays a checkpoint-associated not a checkpoint-intrinsic function, possibly a direct role in SCR (Fig. 3). This is supported by the frequent “chromatid-type” errors that arise in BRCA-mutant MEFs11,16.
Homozygous germline mutations in ATM produce genome instability and a cancer-prone syndrome in humans, and Atm participates in certain checkpoint responses to IR21. Indeed, Atm and a second checkpoint kinase, Chk2, signal to BRCA1 after IR8,22, but Atm does not appear to contribute to BRCA1 phosphorylation after HU exposure5,8. Moreover, unlike ATR-/- embryos, ATM-/- mice are viable. Taken together, the available data imply that the pathways that trigger BRCA1 phosphorylation by these two proteins and the biological outcomes of these sets of events are, at least in part, distinct. Perhaps Atr and Atm activation are triggered by distinct categories of DNA structure.
Other genome integrity functions of the BRCA complex
Defective transcription-coupled repair of oxidative DNA damage was demonstrated in BRCA1 and BRCA2 mutated cells23,42. Cells homozygous for a BRCA1 allele lacking exon 11 revealed anomalous G2/M checkpoint responses to gamma irradiation, implying a role for BRCA1 in this process9. Recently, a partial purification of BRCA1 revealed its association with mismatch repair proteins, the Bloom's syndrome helicase, and certain replication factors24. The functional significance of these particular associations is unclear. However, it is worth noting that the Bloom's Syndrome gene and certain mismatch repair genes have established tumour-suppressor function.
Unlike the homologues of the RAD52 epistasis group of S. cerevisiae, which are represented in all eukaryotic genomes, there are no clear homologues of BRCA genes in yeast, flies or worms. Therefore, the biochemical contributions of BRCA1 and BRCA2 to DNA repair may be as regulators of more conserved repair functions.
Transcriptional functions of BRCA1 and BRCA2
Certain fragments of each BRCA protein, when fused to a GAL4 DNA-binding domain, can transactivate a GAL4 reporter, except when they bear clinically relevant mutations25–27. This suggested that BRCA1 and/or BRCA2 function as transcriptional regulators of specific target genes, the identification of which might, in turn, shed light on the mechanism of BRCA tumour suppression. Interactions have been found between BRCA proteins and sequence-specific transcription factors such as c-myc (BRCA1) and p53 (BRCA1 and BRCA2), as well as with certain transcription regulation proteins that do not recognize a specific, canonical DNA sequence (reviewed in refs 1 and 28). The search for BRCA1 target genes has also pointed to candidates in the p53 pathway: p21 and GADD45 (reviewed in refs 1 and 28, and references therein). This connection suggests a different kind of link between BRCA1 function and genome integrity control in which its function is tied to the expression of genes that do the work of checkpoint control and/or DNA repair. BRCA1, when overproduced, also suppresses oestrogen receptor (ER) transactivation (cited in ref. 1). Notably, p53 mutation and ER negativity are frequent findings in BRCA1-linked tumours, so BRCA1 tumour-suppressor action appears not to be exclusively linked to p53- or ER-dependent functions.
Chromatin remodelling activity of BRCA multiprotein complexes
Chromatin remodelling functions have been attributed to both BRCA1 and BRCA21,28. A recent biochemical purification of BRCA1 suggests that it interacts with a SWI/SNF-containing complex29. The impact of chromatin structure and of its remodelling upon transcriptional events is widely accepted. Equally important is the relationship between chromatin structure and the repair of recombinogenic DNA lesions. For example, in S. cerevisiae, the SIR proteins function in transcriptional silencing of subtelomeric heterochromatin, but are rapidly released from such sites and recruited to the proximity of a DSB where their presence is required for efficient repair30. This is an attractive paradigm against which to consider the BRCA proteins. Perhaps a DSB triggers the release of BRCA proteins from their ‘civilian’ functions or loci and promotes their recruitment to the ‘locale’ of the break.
If this concept is correct, what defines the ‘locale’ of a DSB? Recent studies have shown that chromatin is actively modified around a DSB in mammalian cells. Specifically, the mammalian histone H2A subspecies, H2AX, is phosphorylated on a residue in its C-terminal tail within minutes of gamma irradiation. Indeed, phosphorylated H2AX was shown to appear around an experimentally induced ‘line’ of DSBs in interphase nuclei (cited in ref. 31). Conceivably, phosphorylation of H2AX participates in defining the ‘locale’ of a DSB. Some phosphorylated H2AX also exists in undamaged cells, and it forms nuclear foci that co-localize, in part, with the BRCA dots31. This suggests that the BRCA dots may contain a specialized form of chromatin structure.
Although there are few clues to the innate biochemical activities of the BRCA gene products, it is notable that the BRCA1 RING domain region, which interacts tightly, perhaps stoichiometrically, with the RING domain of BARD1, participates in an in vitro ubiquitin ligation reaction, like RING domains of some ubiquitin ligases32. Perhaps ubiquitination, sumoylation, or nedd 8 coupling of selected protein targets are mediated by BRCA1 and/or BARD1.
BRCA genes and breast or ovarian cancer predisposition
There is good reason to accept that defective maintenance of genomic integrity, as described in BRCA-deficient cells, is an accelerator of cancer progression. If so, how does such a generic defect translate into specifically increased breast or ovarian cancer risk?
The breast epithelium proliferates rapidly during puberty and under the influence of oestrogenic hormones. Unlike the cells of many rapidly proliferating epithelia, such as those of the intestine or of the uterine endometrium, progeny of this proliferative burst are retained within the breast epithelium. This is demonstrated by the finding that breast lobules are clonal33. If, before lobular development, a lobular precursor cell had sustained a cancer-predisposing mutation at a relevant locus (such as p53), the entire lobule would then carry that mutation. This, in turn, could amplify the risk of subsequent neoplastic progression. Some germline p53 mutations (such as in Li–Fraumeni syndrome) confer a particularly elevated risk of adult-onset breast cancer. Similarly, high-dose ionizing chest radiation during puberty, or even in early childhood (for example, atom-bomb exposure; mantle zone therapeutic radiation), specifically predisposes women to adult-onset breast cancer34,35.
These observations could be relevant to BRCA-linked disease, especially if the BRCA+/- genotype were haplo-insufficient in genome integrity maintenance function—an unanswered question at present. BRCA gene haplo-insufficiency could, in principle, increase the risk of additional cancer-promoting mutations occurring during breast development, including mutation of the BRCA gene, itself. Indeed, if puberty constitutes a limited ‘window’ during which a BRCA mutation carrier is at special risk of developing carcinogenic mutations, this might account for the relative absence of BRCA gene inactivation in sporadic breast or ovarian cancer. In sporadic disease, biallelic BRCA gene inactivation might occur too late to have an impact on disease risk36.
That the breast and ovary are oestrogen-responsive tissues could also be relevant to the tissue specificity of BRCA disease risk. Some oestrogen metabolites can adduct DNA, and so could act as tissue-specific carcinogens (so-called “remote carcinogenesis”)37. Conceivably, this effect might be exacerbated by BRCA mutation, if the relevant DNA repair pathways were dysfunctional.
BRCA proteins associate with chromosomal pairing events on the synaptanemal complex. In some model organisms, homologous chromosomal pairing is also known to occur in certain somatic cells where it can influence development by affecting transcriptional regulation (“transvection”) as well as imprinting38. Potentially analogous, homology-dependent transcriptional regulation may also operate in mammals39 and could represent a link between homologous pairing and tissue-specific gene expression. If homologous chromosomal pairing occurs in the breast or ovarian epithelium, are BRCA proteins involved and does such an involvement contribute to their organ-specific tumour suppression function?
Human cells express alternatively spliced BRCA1 transcripts, the biological significance of which is unclear40. Whether products of such transcripts contribute to the tissue specificity of BRCA tumour suppression bears future investigation.
Acknowledgements
We thank many colleagues for stimulating discussions and for sharing data before publication. In particular, J. Feunteun, R. Tibbetts, R. Abraham, W. Bonner, M. Gellert and P. Adams.
References
- 1.Welcsh PL, Owens KN, King MC. Insights into the functions of BRCA1 and BRCA2. Trends Genet. 2000;16:69–74. doi: 10.1016/s0168-9525(99)01930-7. [DOI] [PubMed] [Google Scholar]
- 2.Scully R, et al. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 3.Sharan SK, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, et al. Stable interaction between the products of the BRCA1 and BRCA2 tumoursuppressor genes in mitotic and meiotic cells. Mol. Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 5.Scully R, et al. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 6.Wu LC, et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nature Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 7.Keegan KS, et al. The Atr and Atm protein kinases associate with different sites along meiotically pairing chromosomes. Genes Dev. 1996;10:2423–2437. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- 8.Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 9.Deng CX, Scott F. Role of the tumor suppressor gene Brca1 in genetic stability and mammary gland tumor formation. Oncogene. 2000;19:1059–1064. doi: 10.1038/sj.onc.1203269. [DOI] [PubMed] [Google Scholar]
- 10.Hakem R, et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 11.Patel KJ, et al. Involvement of Brca2 in DNA repair. Mol. Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 12.Sonoda E, et al. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scully R, et al. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol. Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- 14.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol. Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 15.Snouwaert JN, et al. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a brca1 transgene. Oncogene. 1999;18:7900–7907. doi: 10.1038/sj.onc.1203334. [DOI] [PubMed] [Google Scholar]
- 16.Scully R, Puget N, Vlasakova K. DNA polymerase stalling, sister chromatid recombination and the BRCA genes. Oncogene. doi: 10.1038/sj.onc.1203971. in the press. [DOI] [PubMed] [Google Scholar]
- 17.Fornace AJ., Jr Recombination of parent and daughter strand DNA after UV-irradiation in mammalian cells. Nature. 1983;304:552–554. doi: 10.1038/304552a0. [DOI] [PubMed] [Google Scholar]
- 18.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 19.Cordeiro-Stone M, Makhov AM, Zaritskaya LS, Griffith JD. Analysis of DNA replication forks encountering a pyrimidine dimer in the template to the leading strand. J. Mol. Biol. 1999;289:1207–1218. doi: 10.1006/jmbi.1999.2847. [DOI] [PubMed] [Google Scholar]
- 20.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 21.Rotman G, Shiloh Y. ATM: a mediator of multiple responses to genotoxic stress. Oncogene. 1999;18:6135–6144. doi: 10.1038/sj.onc.1203124. [DOI] [PubMed] [Google Scholar]
- 22.Lee JS, Collins KM, Brown AL, Lee CH, Chung JH. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. 2000;404:201–204. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]
- 23.Gowen LC, Avrutskaya AV, Latour AM, Koller BH, Leadon SA. BRCA1 required for transcription-coupled repair of oxidative DNA damage. Science. 1998;281:1009–1012. doi: 10.1126/science.281.5379.1009. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, et al. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 25.Chapman MS, Verma IM. Transcriptional activation by BRCA1. Nature. 1996;382:678–679. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- 26.Monteiro AN, August A, Hanafusa H. Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc. Natl Acad. Sci. USA. 1996;93:13595–13599. doi: 10.1073/pnas.93.24.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milner J, Ponder B, Hughes-Davies L, Seltmann M, Kouzarides T. Transcriptional activation functions in BRCA2. Nature. 1997;386:772–773. doi: 10.1038/386772a0. [DOI] [PubMed] [Google Scholar]
- 28.Irminger-Finger I, Siegel BD, Leung WC. The functions of breast cancer susceptibility gene 1 (BRCA1) product and its associated proteins. Biol. Chem. 1999;380:117–128. doi: 10.1515/BC.1999.019. [DOI] [PubMed] [Google Scholar]
- 29.Bochar DA, et al. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- 30.Guarente L. Diverse and dynamic functions of the Sir silencing complex. Nature Genet. 1999;23:281–285. doi: 10.1038/15458. [DOI] [PubMed] [Google Scholar]
- 31.Paul TP, et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 32.Lorick KL, et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquiti-nation. Proc. Natl Acad. Sci. USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kordon EC, Smith GH. An entire functional mammary gland may comprise the progeny from a single cell. Development. 1998;125:1921–1930. doi: 10.1242/dev.125.10.1921. [DOI] [PubMed] [Google Scholar]
- 34.Tokunaga M, et al. Malignant breast tumors among atomic bomb survivors, Hiroshima and Nagasaki, 1950–74. J. Natl Cancer Inst. 1979;62:1347–1359. [PubMed] [Google Scholar]
- 35.Wolden SL, Lamborn KR, Cleary SF, Tate DJ, Donaldson SS. Second cancers following pediatric Hodgkin's disease. J. Clin. Oncol. 1998;16:536–544. doi: 10.1200/JCO.1998.16.2.536. [DOI] [PubMed] [Google Scholar]
- 36.Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 37.Fishman J, Osborne MP, Telang NT. The role of estrogen in mammary carcinogenesis. Ann. NY Acad. Sci. 1995;768:91–100. doi: 10.1111/j.1749-6632.1995.tb12113.x. [DOI] [PubMed] [Google Scholar]
- 38.Henikoff S. Nuclear organization and gene expression: homologous pairing and long-range interactions. Curr. Opin. Cell Biol. 1997;9:388–395. doi: 10.1016/s0955-0674(97)80012-9. [DOI] [PubMed] [Google Scholar]
- 39.Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 41.Tibbetts RS, et al. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. doi: 10.1101/gad.851000. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Page F, et al. BRCA1 and BRCA2 are necessary for the transcription-coupled repair of the oxidative 8-oxoguanine lesion in human cells. Cancer Res. 2000;60:5548–5552. [PubMed] [Google Scholar]