Abstract
Detection of antibodies to Kaposi’s sarcoma-associated herpesvirus (KSHV or Human Herpesvirus 8) is a topic of ongoing controversy. KSHV expresses multiple antigens and host responses are highly variable. We have previously described and algorithm for determining KSHV infection based on K8.1 ELISA and LANA Immunofluorescence assay (IFA). Here we describe the development of a recombinant ELISA for LANA and an improved testing strategy using ELISAs for LANA and K8.1. We assessed mammalian and baculovirus expression systems for the production of full length recombinant LANA. We evaluated the performance of LANA ELISAs using human serum samples from several sources including blood donors and clinical patients diagnosed with Kaposi’s sarcoma and compared them to LANA IFA. Both LANA ELISAs exhibited comparable sensitivity and specificity to LANA IFA but showed considerably greater reliability. The LANA ELISA can thus be used in conjunction with the previously described K8.1 ELISA to enable the highly sensitive and specific detection of antibodies to KSHV. Use of this testing strategy will provide a more accurate and reliable diagnostic assessment of KSHV status.
Keywords: KSHV serology, ELISA, LANA, K8.1
1. INTRODUCTION
Kaposi’s Sarcoma associated Herpesvirus (KSHV), also known as Human Herpesvirus 8 (HHV8), is the causative agent of Kaposi’s Sarcoma (KS) (Chang et al. 1994, Whitby et al. 1995). It is also associated with primary effusion lymphoma (PEL) (Cesarmen et al. 1995) and multicentric Castleman’s disease (Soulier et al. 1995).
KSHV encodes multiple antigenic proteins, some of which are expressed only during lytic replication (Chandran et al. 1998). The major antigenic proteins are the latently expressed latency associated nuclear antigen (LANA) encoded by ORF 73 (Kedes et al. 1996, 1997, Rainbow et al1997) and the lytically expressed K8.1 (Raab et al. 1998) and ORF 65 (Simpson et al. 1996). First generation serological assays were based on immunofluorescence of latently infected PEL cells (Gao et al. 1996, Kedes et al 1996, Simpson et al. 1996) or PEL cells induced by TPA treatment to produce lytic antigens (Lennette et al. 1996). Though a gold standard for the diagnosis of KSHV infection has never emerged, immunofluorescent assays have since been considered reference tests for the detection of antibodies to KSHV. However, IFAs have several major disadvantages: they are labor intensive, operator dependent, and cannot readily be automated for screening large numbers of samples. In addition, non-specific antibody reactions or high background may result in false positives or false negatives, respectively.
Enzyme linked immunosorbent assays (ELISA) for KSHV antibody detection have been developed using recombinant proteins or peptides (Engels et al. 2000b, Lam et al 2002, Laney et al. 2006, Nascimiento et al. 2007, Simpson et al. 1996). Humoral responses to KSHV antigens do not follow a predictable pattern and concordance between assays detecting antibodies to these antigens is moderate or poor (Biggar et al. 2003, Pellet et al. 2003, Rabkin et al. 1998). Detection of antibodies to a single KSHV antigen may therefore underestimate KSHV prevalence in cross sectional studies. We have previously reported the development of a robust recombinant protein ELISA to detect antibodies to K8.1 with excellent specificity and sensitivity (Engels et al. 2000a, 2000b). Here we describe a latent ELISA to detect antibodies to LANA which complements our lytic K8.1 ELISA to provide a more complete assessment of KSHV antibody status.
The 3.4 kb KSHV orf73 contains a series of nucleic acid repeats, covering nearly 1.8 kb in the central part of the gene, that encode short 5–7 amino acid sequences consisting of mostly aspartate, glutamate, proline, leucine, and glutamine residues (Kellam et al. 1999). Examination of Genbank ORF73 sequences from various KSHV strains shows dramatic variability in the number and makeup of repeats within the central gene region. Protein mapping studies indicate that antigenic regions are located throughout Orf73/LANA, including the repetitive region, and suggest that full length recombinant protein is necessary for optimal ELISA sensitivity. Assays based on LANA peptides or on bacterially-expressed full-length recombinant LANA demonstrated limited sensitivity compared to IFA (Olsen et al. 2000). In contrast, assays developed with baculovirus-expressed LANA produced in insect cells are highly sensitive and specific (Zhu et al. 1999). These reported sensitivity differences suggest that correct folding of LANA may be essential for optimal immunoreactivity.
We previously reported a testing algorithm for KSHV based on K8.1 ELISA and LANA IFA (Pellet et al. 2003). In this report, we describe the development of two ELISAs: one based on baculovirus-expressed full-length LANA grown in insect cells and the other based on full-length LANA produced in mammalian cells. We considered the possibility that the three-dimensional structure of the Orf73 protein produced in mammalian cells may more closely resemble that of the native protein in infected human cells. If this is the case, then mammalian cell-derived Orf73 may contain better preserved antigenic epitopes which may lead to enhanced performance in the KSHV LANA ELISA. To test this hypothesis, we produced full-length Orf73 in 293E mammalian cells and Sf9 insect cells, purified the antigens and compared the performance of the 2 different recombinant proteins in ELISAs. We then compared the two LANA ELISAs to the previous gold standard IFA. Additionally, we describe an algorithm for the use of the LANA ELISA with the previously described K8.1 ELISA for the sensitive and specific detection of antibodies to KSHV.
2. MATERIALS and METHODS
2.1 Production of Recombinant Orf73/LANA
The KSHV orf73 region was amplified by PCR from a classic KS lesion clone and subcloned by Gateway LR recombination into two vectors, after verification of the sequence. The x1-8-orf73 bacmid DNA was transfected into Sf9 insect cells using SuperFect reagent (Qiagen, Valencia, CA) according to the manufacturer’s protocol and the cells were grown at 27°C for four days in HyQ SFX-Insect serum-free medium (HyClone, Logan, UT). The culture supernatant containing the recombinant virus was collected and used to infect Sf9 cells (plated at 1×106 cells/ml) to create the amplified virus stock. This baculovirus stock was titrated by end-point dilution. One-liter production runs were performed using optimized conditions, the LANA producing cells were collected, washed with cold PBS (Biosource, Camarillo, CA), centrifuged and snap frozen using an alcohol-dry ice bath. Cell pellets were stored at −80°C until processed.
The pExp721-orf73 plasmid was transfected into 293E cells using SuperFect reagent according to the manufacturer’s instructions. Cells were grown at 37°C for 48 hours, collected, washed with cold PBS, and snap frozen using an alcohol-dry ice bath. Cell pellets were stored at −80°C until processed.
2.2 His6-Orf73 Protein Purification
Transfected 293E or infected Sf9 cells pellets were resuspended with 4 ml of extraction buffer [20 mM sodium phosphate buffer, pH 7.5, (Biosource, Camarillo, CA), 100 mM NaCl, 45 mM imidazole, 5% glycerol, 5 mM MgCl2, 1 mM β-mercaptoethanol, (Sigma-aldrich, St. Louis, MO), plus 1 tablet of complete–EDTA protease inhibitor (Roche Molecular, Pleasanton, CA) per 100 ml of lysate] per gram of cell pellet. This mixture was sonicated using a Digital Sonifier 450 (Branson, Danbury, CT) for 30 seconds and treated with 1 unit of benzonase (Novagen, San Diego, CA) per ml for 20 minutes on ice and clarified by ultracentrifugation (110,000 × g for 50 min under vacuum at 4°C) in an Ultima L90K centrifuge (Beckman Coulter, Fullerton, CA). The sample was filtered through a Puradisc FP30 0.8 µm filter (Whatman, Middlesex, UK) and applied at 0.5 ml/min onto a 5 ml HisTrap column (GE Healthcare, Chalfont St. Giles, UK) previously equilibrated with extraction buffer. The column was washed with extraction buffer at 2.5 ml/min to baseline and the sample was eluted in a 100 ml gradient of 45–400 mM imidazole in extraction buffer and collected in 2 ml fractions that were analyzed by SDS-PAGE as described below.
For his6-Orf73 expressed in 293E cells, positive fractions were pooled, dialyzed to phosphate-buffered saline (PBS) pH 7.5 (Biosource) and flash frozen in aliquots. For his6-Orf73 produced in insect cells, positive fractions were pooled, dialyzed to 20 mM sodium phosphate pH 7.5, 100 mM NaCl, 5 % glycerol, 1 mM DTT and applied to a 5 ml Q-sepharose column. The protein was finally eluted with a 0.1–1 M NaCl gradient over 20 column volumes, analyzed by SDS-PAGE, pooled, dialyzed to PBS pH 7.5 and flash frozen in aliquots
2.3 SDS-PAGE and immunoblot Analysis
All gel samples were prepared using lithium dodecyl sulfate (LDS) sample buffer (Invitrogen, Carlsbad, CA) and Tris [2-carboxyethyl] phosphine (TCEP) reducing agent (BioRad, Hercules, CA) according to the manufacturer’s protocols. Samples were heated at 70°C for 5 minutes before application to Criterion Tris-glycine 10–20% pre-cast SDS-PAGE gels (BioRad) and were electrophoresed at 200 V for 55 minutes at room temperature. Proteins were visualized using Coomassie Brilliant Blue staining (Sigma-aldrich, St. Louis, MO). For immunoblots, gels were transferred to a nitrocellulose membrane in 1× Towbin's buffer with 15% methanol using a Criterion transfer apparatus (BioRad), at 100 volts for 50 minutes. The membrane were incubated with an anti-penta-His antibody:HRP conjugate (Qiagen), 1:5,000 for 90 minutes at room temperature in 1× TTBS + 2% non-fat dry milk. Chemiluminescent detection was performed with West Pico reagent (Pierce).
2.4 Subjects and Samples
The initial tests to qualify the LANA ELISAs were done on a small panel (n = 10) of titrated samples from patients enrolled in clinical protocols by the HIV and AIDS Malignancy Branch of the NCI at NIH, Bethesda. All patients gave informed consent and the protocols were approved by the NCI Institutional Review Board. All patients were HIV positive including one individual diagnosed with active KS at the time of blood draw. We evaluated the performance of the LANA IFA and ELISAs by repeatedly testing these clinical samples, in duplicate, over several runs.
During assay development, a small set of samples from patients with KS, and a panel of 100 blood donor samples were run as ELISA positive or negative controls, respectively. The same KS sera and a small panel of donor sera with consistently low seroreactivity in previously developed KSHV IFA and ELISA assays were selected to be used as positive and negative controls, during assay validation and routine assay performance. These controls allow us to monitor parameter changes and thereby control plate to plate and experiment to experiment variation.
For the establishment of the cutoff, a training set of specimens was set up, including 82 samples from patients with KSHV infection form the NCI HIV and AIDS Malignancy Branch, and a set of 247 samples from blood donors. Patients had been determined to be KSHV infected if they had KSHV detected in PBMCs by a previously described real time PCR assay. Blood donors from a local donor program were assumed to be KSHV negative, even though the prevalence of KSHV in analogous low risk populations may vary between 5–10%, as estimated by previously developed assays (de Sanjose et al. 2002).
Subsequently, a validation set was tested. A set reproducing a low prevalence sample was constructed to evaluate assay performances in a simulated low risk population. We used a panel of 1000 blood donor plasma specimens from the National Heart, Lung and Blood Institute’s Retrovirus Epidemiology Donor Study (REDS) General Leukocyte and Plasma Repository (Zuck et al. 1995). These samples were randomly selected from donors who tested negative for all routine serological tests performed by the 5 US blood centers that supply the repository. REDS also provided samples of undiluted serum from 20 KS patients enrolled in ongoing studies at the Center for Disease Control (CDC) and 20 replicate specimens of diluted serum from a single KS patient. These 40 samples were distributed throughout the panel and all samples were tested blinded.
2.5 LANA and K8.1 ELISAs
The LANA ELISAs were developed based on similar previously reported assays (Engels et al. 2000a, 2000b). Full length Orf73 antigen was diluted 1:500 in PBS (Invitrogen, Carlsbad, CA) and 100 µl per well was used to coat 96-well non-sterile Dynex HBX4 microtiter plates (VWR, Bridgeport, NJ) at 4°C overnight. The wells were washed 3 times with Dupont ELISA wash (Perkin Elmer, Wellesley, MA) using a model EL×405 automated plate washer (Bio-Tek Instruments, Winooski, VT) and blocked using 280 µl assay buffer [(2.5% BSA (Sigma-aldrich), 2.5% normal goat serum (Equitech-Bio, Kerrville, TX), 0.005% Tween 20 (Sigma-aldrich) and 0.005% Triton-X 100 (Sigma-aldrich) in PBS] per well for 3 hours at 37°C. Plates containing assay buffer were stored at −80°C until needed. Protein coating concentrations for both the insect and mammalian based LANA and assay working conditions were optimized by testing a panel of 20 titrated serum samples.
Diluted test and control sera (100 µl at 1:100 in assay buffer) were pipetted into the wells and incubated at 37°C for 90 minutes. Positive and negative controls were run in triplicate on each plate. Unbound antibodies were removed by washing 5 times with New England Nuclear (NEN) ELISA plate wash (Perkin Elmer). Goat anti-human IgG(g) alkaline phosphatase (ReserveAP™, KPL, Gaithersburg, MD) was added to each well and incubated at 37°C for 30 minutes. After 5 washes, substrate 1 step p-nitrophenyl phosphate, disodium salt (Pierce, Rockport, IL) was added to detect antigen-antibody complexes. The wells were allowed to develop at room temperature in the dark for 30 minutes and the reaction was stopped with 3N NaOH. Absorbance was read at 405 nm using a model EL 312e automated plate reader (Bio-Tek Instruments). The K8.1 ELISA was performed as described previously (Engels et al. 2000b).
2.6 Assay Cut-off and Titration
For the LANA assay, a cut off equal was empirically set at the average of the negative controls plus 0.35 for each plate. Positive samples were titrated by 8 two-fold serial dilutions (1:100 to 1:12800) to determine LANA titer based on a standard titration curve.
For the K8.1 ELISA the cut off was set as the average of the negative controls plus 0.85 for each plate. Positive samples were titrated from 1:20 to 1:2560 as described above for the LANA ELISA.
2.7 Assay Quality Control
The high performance of the LANA and K8.1 assays was maintained by implementing stringent quality controls. First, for the assays to be considered valid, each negative control values need to have an OD between 0 and 0.2, and between 50% and 150% of the mean of negative controls, after subtracting the blank well’s OD value. Secondly, a twofold titration of the same positive sample was performed every six plates to control experiment to experiment and batch to batch variability.
2.8 LANA Immunofluorescence Assay (IFA)
IFA was performed using the KSHV latently infected PEL cell line BCP-1 as the positive control and Ramos cells as the negative control. BCP-1 and Ramos cells were combined at a 4:1 ratio in PBS and fixed with cold 4% paraformaldehyde (Sigma, St Louis, MO) for 10 minutes. After washing and resuspending in PBS, the cells were permeabilized with 0.2% Triton-X 100 (Sigma) for 10 minutes. The cells were washed and resuspended in PBS and spotted onto 5 mm diameter wells of a Teflon coated slide (12 wells/slide; Erie Scientific, Portsmouth, NH) and allowed to dry. Coated slides were stored at −20°C until needed.
Twenty microliters of test sera diluted 1:100 in PBS containing 3% fetal bovine serum (Sigma) was applied to each coated well and allowed to react for 30 minutes at room temperature in a dark humid chamber to prevent sample dehydration. The slides were washed 4 times with PBS/3% FBS before conjugating with Alexa Fluor 488 chicken anti-human IgG, H & L (Molecular Probes, Eugene, OR) for 30 minutes at room temperature in a dark humid environment. After a single wash with PBS/FBS and 3 subsequent washes with plain PBS, the slides were examined by ultraviolet microscopy using a Leitz Ortholux II microscope (Wetzlar, Germany). Fluorescently positive samples were titrated by 12 two-fold serial dilutions from 1:100 to 1:204,800.
2.9 Statistical Methods
Assay performances were examined performing a non parametric analysis of the receiver operator characteristic of the assays. Confidence intervals were calculated with the De Long method. Comparison of LANA ELISA and IFA performance was carried out using Hoyt’s and Winer’s estimates. All statistical analyses were performed using STATA v10.1 (StataCorp College Station Tx)
3. RESULTS
3.1 Evaluation of Baculovirus and Mammalian-produced orf73/LANA
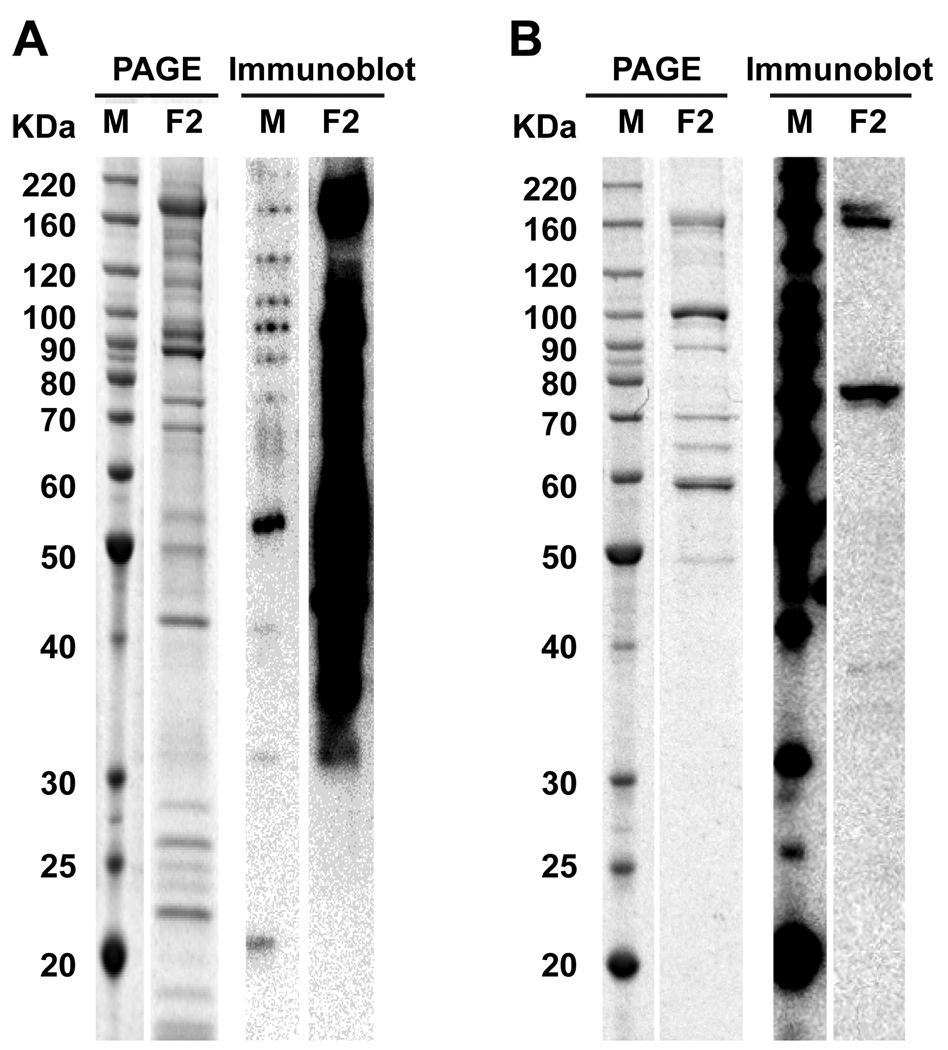
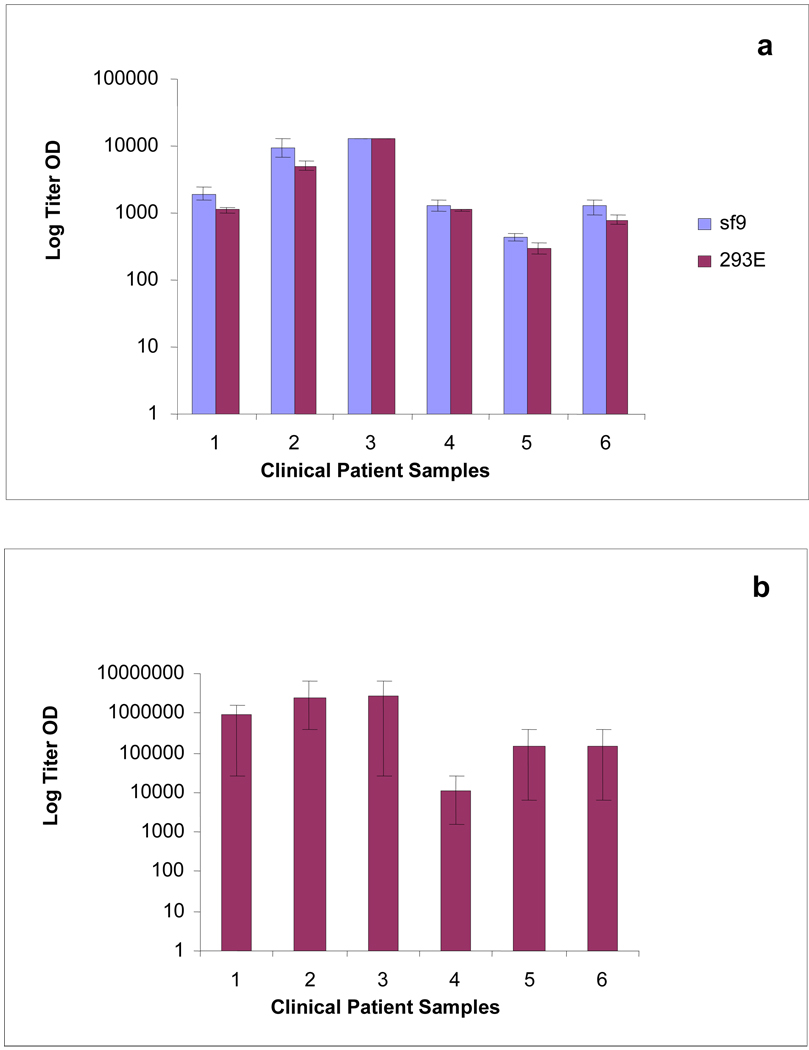
The performance of the LANA protein derived from mammalian and insect cells was assessed. SDS-PAGE and immunoblot analysis are shown in Figure 1. His6-Orf73 from Sf9 insect cells showed higher expression by SDS-PAGE, and a protein concentration of 0.025 µg/ml was sufficient for coating ELISA plates. In comparison, his6-Orf73 from 293E cells was considerably less abundant by SDS -PAGE and the protein concentration required for a satisfactory ELISA was more than twenty times higher (0.54 µg/ml). The panel of titrated serum samples from the NIH-NCI clinic (HIV and AIDS Malignancy branch) was repeatedly tested over several runs in ELISA using both the insect and mammalian cell derived LANA to ascertain which protein preparation was better suited for our assay (Figure 2). His6-Orf73 from Sf9 insect cells proved to produce consistently higher average optical density (OD) values for every patient sample than its mammalian counterpart, despite the lower concentration required. For this reason, in addition to the considerably lower production costs, all subsequent studies were performed using the baculovirus produced LANA.
Figure 1. SDS PAGE and immunoblot analysis of Insect or Mammalian derived KSHV LANA.
Five micrograms of insect derived protein (A), and 1.7 micrograms of mammalian derived product (B), fractions 1 and 2, were analyzed by SDS PAGE and immunoblot. For each of the two products, the most abundant elution fraction (F2) is showed.
Figure 2. Titration of a panel of clinical samples using KSHV LANA ELISA (Insect or Mammalian derived) or LANA IFA.
Average performance of the insect (Sf9) and mammalian cell (293E) derived LANA ELISAs (a) and the LANA IFA (b) on a panel of titrated serum samples from the NIH-NCI clinic tested repeatedly. Error bars show titer OD variation between experiments.
3.2 Comparison of ELISA and IFA Performance
To confirm the results generated by the Orf73/LANA ELISA and to determine comparative assay reliability, the NIH-NCI clinic panel was also tested using the LANA immunofluorescent assay (IFA). The IFA showed considerably greater variation in results between repeats than either the insect or mammalian LANA ELISAs (Figure 2). Statistical analyses of the LANA ELISA data showed it to be highly reproducible using either the insect or mammalian recombinant protein based assays, attaining near perfect reliability scores by Hoyt’s and Winer’s estimates (Table 1). In contrast, the LANA IFA showed less reliability and thus does not give as reproducible results.
Table 1. Comparison between LANA ELISA and IFA reliability.
Summary of reliability analyses for the LANA ELISA and LANA IFA calculated using Hoyt’s and Winer’s estimates.
| Hoyt estimate | Winer estimate | |
|---|---|---|
| Sf9 LANA ELISA | 0.985 | 0.980 |
| 293E LANA ELISA | 0.998 | 0.998 |
| LANA IFA | 0.406 | 0.243 |
3.3 Reproducibility of Orf73/LANA and K8.1 ELISAs
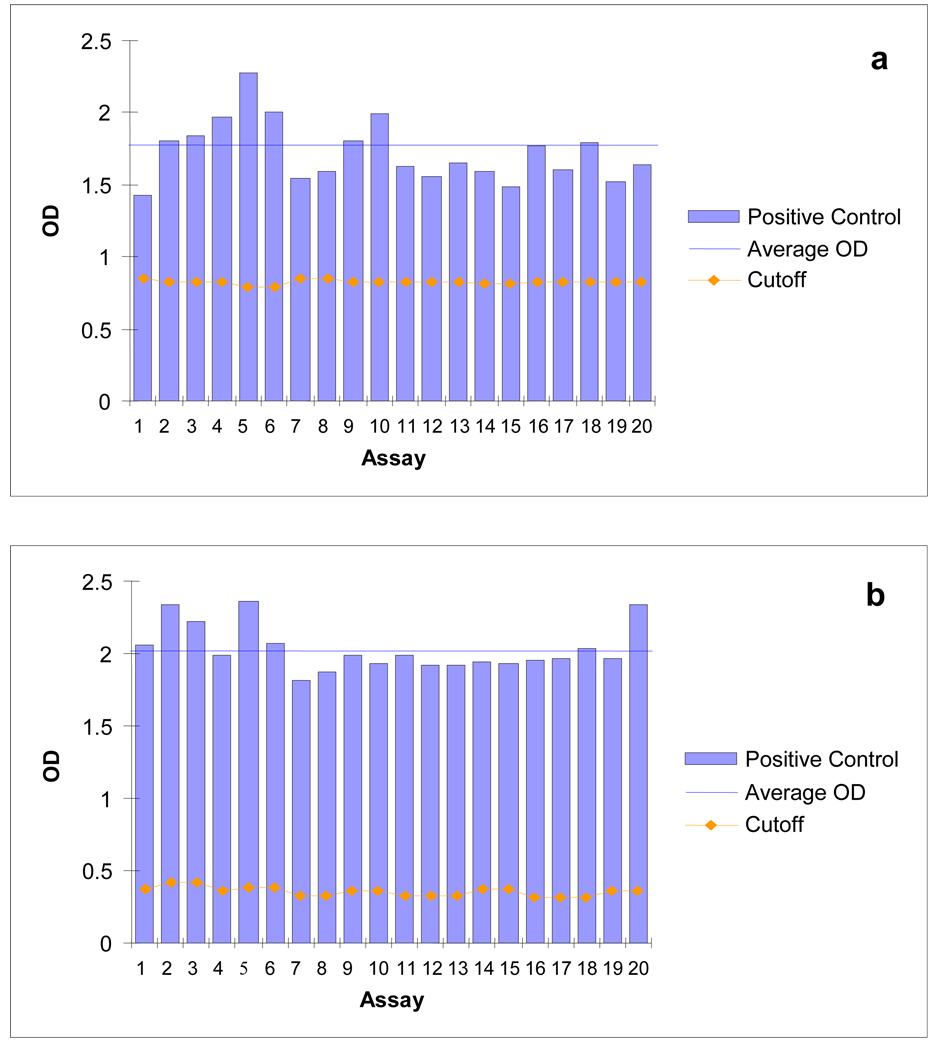
Consistency of performance for each KSHV ELISA was determined by comparing the OD values for 20 replicate specimens of diluted serum from a single KS patient distributed throughout the REDS panel as a positive control. OD values of each positive control aliquot showed only slight variability between plates for both the K8.1 and LANA ELISAs with the average positive control OD significantly higher than the cut off OD values (Figure 3).
Figure 3. Repeated testing of a diluted positive control sample using KSHV K8.1 and LANA ELISAs.
Performance of the K8.1 ELISA (a) and the LANA ELISA (b) on a diluted KS patient positive control sample repeatedly tested. The cutoff for each plate is shown as a gold line. The blue line indicates the average OD value of the positive control.
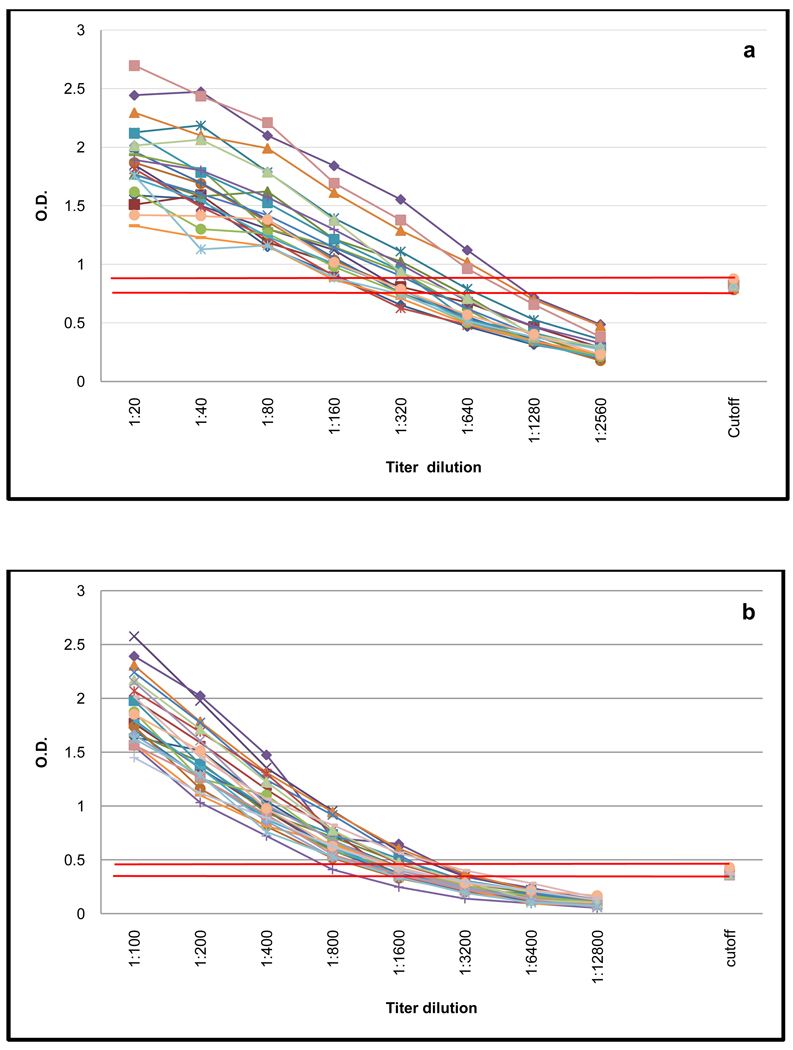
A positive control sample from an individual with KS was routinely titrated by 8 twofold serial dilutions on twenty individual ELISA plates. By monitoring the performance of these titrations we were able to verify plate to plate consistency throughout the batch of samples and confirm the reproducibility of each assay. The titration gradient is reasonably stable between plates for both assays with the cut off OD value consistently falling between the fifth and sixth titration point for both ELISAs (Figure 4).
Figure 4. Repeated titration of a positive control sample using KSHV K8.1 and KSHV LANA ELISA.
Performance of the K8.1 ELISA (a) the insect cell derived LANA ELISA (b) on a positive control titrated on 15 individual plates.
3.4 Performance of Orf73/LANA and K8.1 ELISAs
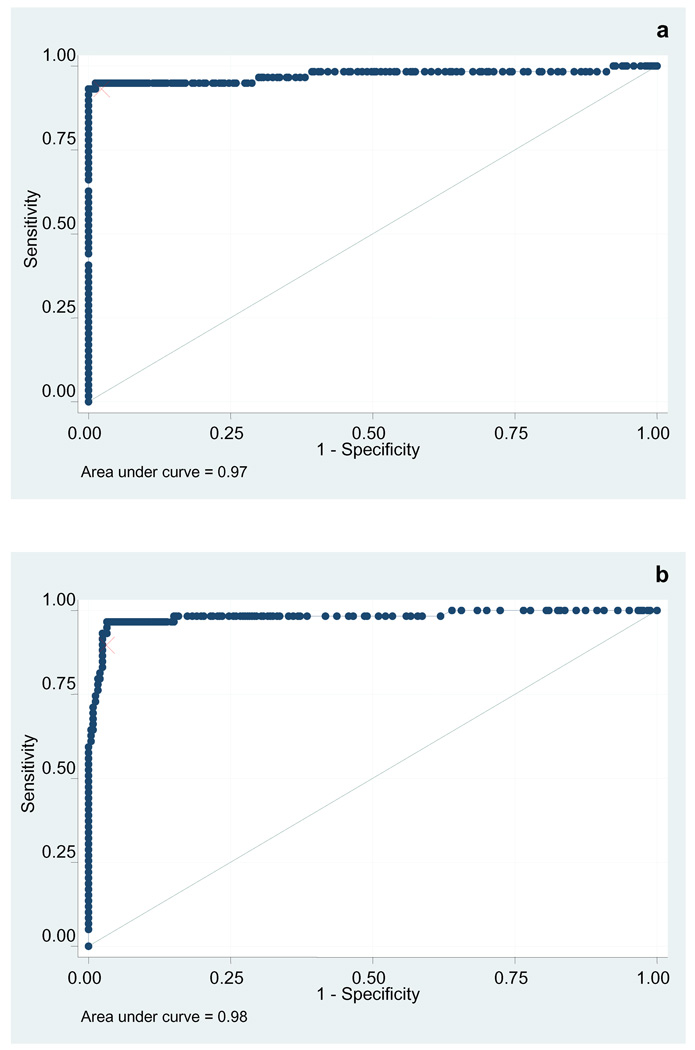
Receiver operating characteristic of both assays were first examined on the training set (Figure 5). The area under the curve was 0.98 (95% confidence interval 0.96 – 0.99) for the ORF 73 assay and 0.97 (95% confidence interval 0.95 – 0.99) for the ORFk8.1 assay. Analyzing individual cutoffs, it was found that for ORF73, a value corresponding to the mean of negative OD plus a constant of 0.35 afforded the highest accuracy (89.02% sensitivity and 97.57% specificity). For ORF K8.1, a second best classifying cutoff was selected, corresponding to the mean of negative OD plus 0.85 (98.78% sensitivity and 98.79% specificity).
Figure 5. ROC curve of the KSHV K8.1 and LANA ELISAs (training set).
Receiver operator characteristic of the KSHV k8.1 ELISA (a) and KSHV LANA ELISA (b) applied to the training set. The red mark indicates the optimal cutoff.
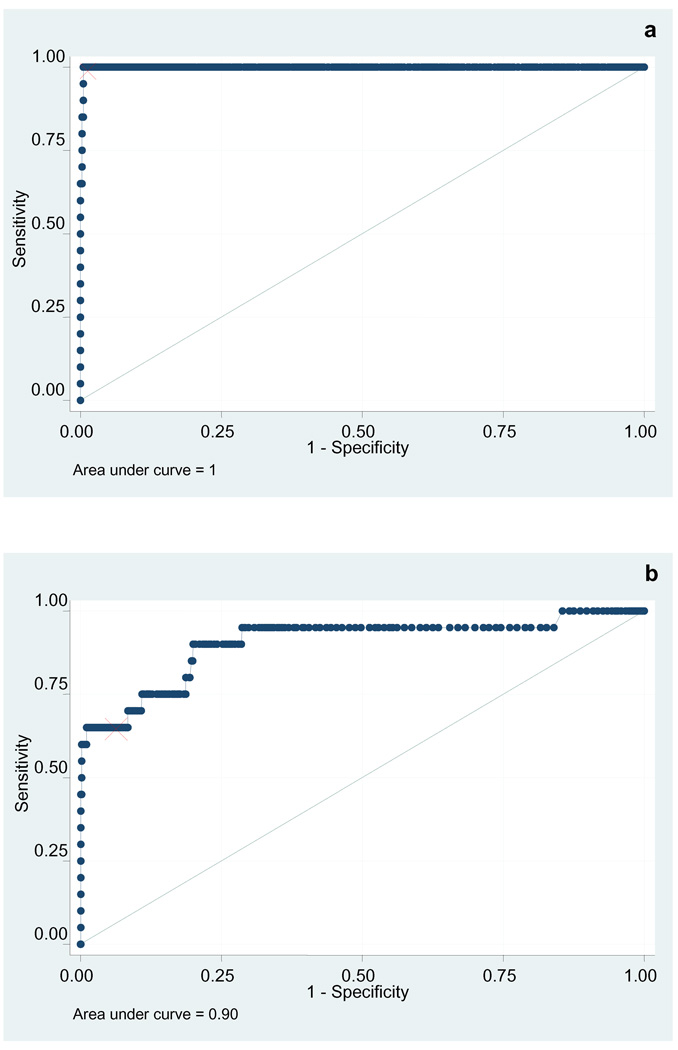
When the validation set was analyzed (Figure 6), the k8.1 assay retained an excellent characteristic (AUC 0.99, 95% confidence interval 0.99–1) while that of the ORF73 assay deteriorated ever so slightly (AUC 0.90, 95% confidence interval 0.88–0.92). Of note, while the K8.1 ELISA proved more sensitive in this validation set, the LANA ELISA displays much higher titers in all positive subjects. The reasons such differences in antigenic characteristic between the two proteins are yet to be elucidated.
Figure 6. ROC curve of the KSHV K8.1 and LANA ELISAs (validation set).
Receiver operator characteristic of the KSHV k8.1 ELISA (a) and KSHV LANA ELISA (b) applied to the validation set. The red mark indicates the optimal cutoff.
3.5 Analytical strategy for Determining KSHV Infection
By using the combined data from the K8.1 and LANA ELISAs, a test strategy was developed to better assess KSHV infection. Samples positive by both assays can be classified as KSHV positive with some confidence; likewise, samples negative by both assays can be classified as KSHV negative. Investigators may want to decide a priori how to interpret discordant results according to their particular research question and the population involved. The investigator may wish to take a broad approach and include them with the positives. In this validation set, when the two assays are performed in parallel and samples scoring positive in either assays are considered positive, the combined sensitivity of the two assays is 100%, while the specificity is 95.8%. Alternatively, a more focused approach may be more appropriate so that the discordant are counted as negative. In this example, testing would yield a sensitivity of 65% and a specificity of 99. 2%.
Applying this method to the 1000 US blood donor data from the REDS study yields an point estimate of the KSHV prevalence of 0.8% (exact 95% CI: 0.2–1.3%) using the most specific definition, and of 4.4% (exact 95% CI: 3.1–5.6%) using the more sensitive one. These values compare to those previously reported for other assays, 3.0 to 3.5% (Pellet et al. 2003).
4. DISCUSSION
Serological testing for KSHV antibodies has been controversial with different assays yielding very different prevalence estimates, especially in populations at low risk for KSHV infection (Pellet et al. 2003, Rabkin et al. 1997). We have previously described and utilized an algorithm to combine data from a K8.1 ELISA and an IFA on latently infected PEL cells (Pellet et al. 2003). However, we soon recognized the need for a protocol entirely based on ELISA, because of the limitations associated with IFAs. Indeed, immunofluorescent assays have seen recent developments; in particular, the use of insect cells expressing baculovirus-derived recombinant antigens (Minhas et al. 2008) has improved reliability. However, newer IFA retain some of the same disadvantages characterizing PEL based assays, that is, high labor intensiveness and operator dependence, and no immediate potential for scalability and automation.
Previously developed LANA ELISAs based on E.coli expressed protein had poor sensitivity compared to IFA (Engels et al. 2000b). We did evaluate the performances of peptide based assays for the detection of antibodies directed at ORF k8.1 and LANA, utilizing pools of 1 and 5 peptides per each ORF, respectively (data not shown). Such preliminary experiments indicated that all assays had a lower sensitivity compared with the corresponding protein based assays.
We here describe a newly developed ELISA utilizing full length recombinant insect or mammalian expressed LANA, which offers comparable or superior sensitivity to IFA, and the advantage of increased reproducibility as well as all other operational benefits that are typical of ELISAs. Moreover, we propose an approach for combining data from this assay and our K8.1 assay to better assess KSHV infection status in different settings.
Development of robust testing systems for KSHV has been hampered by a lack of “gold standard” for the diagnosis of KSHV. KS patients can be considered 100% infected by KSHV. However, the levels of antibody present in KS patient sera can be in the range of 1/5000 to 1/500,000 compared to typical titers in asymptomatic subjects of 1/50 to 1/500. For this reason, sensitivity estimates solely based on detection in KS patient sera can overestimate true sensitivity. Likewise, if subjects from low risk populations, such as US blood donors, are considered to be not infected, the specificity will likely be underestimated, since a small proportion of donors will in fact be infected with KSHV.
To overcome the first limitation, in our training set we included KSHV infected asymptomatic subjects in addition to KS patients, and in our validation set, we diluted sera from KS patients. Furthermore, our approach combines two different assays, utilizing one a latent and the other a lytic gene product, both of which are major antigens. It therefore affords a broader evaluation of antibody response across the spectrum of the natural history of the infection. Since KSHV has a large genome encoding for more than 80 proteins, we cannot assume that our combination of assays provides an exhaustive profile of each patient’s seroreactivity, and efforts are undergoing to further elucidate this issue. However it represent a major advancement in this direction, a particularly important one for cross-sectional studies, where a single time point is investigated.
Because of the different reactivity profiles of the two individual assays, our approach also allows tomaximize sensitivity, or specificity, depending on the particular hypothesis investigated.
Table 2. LANA/Orf73 and K8.1 ELISA data from 20 KS patient specimens.
Classification of 20 KS patient samples from the National Heart, Lung and Blood Institute’s Retrovirus Epidemiology Donor Study (REDS) General Leukocyte and Plasma Repository using the KSHV K8.1 ELISA and the insect-derived KSHV LANA ELISA.
| K8.1 | ||||
|---|---|---|---|---|
| + | − | total | ||
| LANA | + | 13 | 0 | 13 |
| − | 7 | 0 | 7 | |
| total | 20 | 0 | 20 | |
Table 3. LANA and K8.1 ELISA classification of 1000 blood donor plasma specimens.
Classification of 1000 blood donor specimens from the National Heart, Lung and Blood Institute’s Retrovirus Epidemiology Donor Study (REDS) General Leukocyte and Plasma Repository. using the KSHV K8.1 ELISA and the insect-derived KSHV LANA ELISA.
| K8.1 | ||||
|---|---|---|---|---|
| + | − | total | ||
| LANA | + | 8 | 24 | 32 |
| − | 12 | 956 | 968 | |
| total | 20 | 980 | 1000 | |
ACKNOWLEDGEMENTS
We thank Thomas Schulz of The University of Hanover for providing the KS lesion clone and Sabine Geisse of Novartis for the EBNA Origin Destination vectors. We also thank Deborah Todd of Westat, Inc. and Kathleen Wyvil and Karen Aleman of the National Cancer Institute’s HIV and AIDS Malignancy Branch. We are grateful to the National Heart, Lung and Blood Institute for establishing and providing the Human Herpes Virus 8 (HHV-8) Special Collection from the Institute’s Retrovirus Epidemiology Donor Study (REDS) General Leukocyte/Plasma Repository (GLPR), https://biolincc.nhlbi.nih.gov/studies/hhv8/?q=reds. This project has been funded in whole or in part with funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Biggar RJ, Engels EA, Whitby D, et al. Antibody reactivity to latent and lytic antigens to human herpesvirus-8 in longitudinally followed homosexual men. J. Infect. Dis. 2003;187:12–18. doi: 10.1086/345866. [DOI] [PubMed] [Google Scholar]
- Cesarman E, Chang Y, Moore PS, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS- related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- Chandran B, Smith MS, Koelle DM, et al. Reactivities of human sera with human herpesvirus-8-infected BCBL-1 cells and identification of HHV-8-specific proteins and glycoproteins and the encoding cDNAs. Virology. 1998;243:208–217. doi: 10.1006/viro.1998.9055. [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, F, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- de Sanjosé S, Marshall V, Solà J, et al. Prevalence of Kaposi's sarcoma-associated herpesvirus infection in sex workers and women from the general population in Spain. Int J Cancer. 2002;98(1):155–158. doi: 10.1002/ijc.10190. [DOI] [PubMed] [Google Scholar]
- Engels EA, Sinclair MD, Biggar RJ, et al. Latent class analysis of human herpesvirus 8 assay performance and infection prevalence in sub-saharan Africa and Malta. Int J Cancer. 2000;88:1003–1008. doi: 10.1002/1097-0215(20001215)88:6<1003::aid-ijc26>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Engels EA, Whitby D, Goebel PB, et al. Identifying human herpesvirus 8 infection: performance characteristics of serologic assays. J Acquir Immune Defic Syndr. 2000;23:346–354. doi: 10.1097/00126334-200004010-00011. [DOI] [PubMed] [Google Scholar]
- Gao SJ, Kingsley L, Li M, Zheng W, et al. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- Kedes DH, Lagunoff M, Renne R, et al. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J Clin Invest. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes DH, Operskalski E, Busch M, et al. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- Kellam P, Boshoff C, Whitby D, et al. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J Hum Virol. 1997;1:19–29. [PubMed] [Google Scholar]
- Kellam P, Bourboulia D, Dupin N, et al. Characterization of monoclonal antibodies raised against the latent nuclear antigen of human herpesvirus 8. J Virol. 1999;73:5149–5155. doi: 10.1128/jvi.73.6.5149-5155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LL, Pau CP, Dollard SC, et al. Highly sensitive assay for human herpesvirus 8 antibodies that uses a multiple antigenic peptide derived from open reading frame K8.1. J Clin Microbiol. 2002;40:325–329. doi: 10.1128/JCM.40.2.325-329.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney AS, Peters JS, Manzi SM, et al. Use of a multiantigen detection algorithm for diagnosis of Kaposi's sarcoma-associated herpesvirus infection. J Clin Microbiol. 2006;44:3734–3741. doi: 10.1128/JCM.00191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennette ET, Blackbourn DJ, Levy JA, et al. Antibodies to human herpesvirus type 8 in the general population and in Kaposi's sarcoma patients. Lancet. 1996;348:858–861. doi: 10.1016/S0140-6736(96)03240-0. [DOI] [PubMed] [Google Scholar]
- Minhas V, Crosby NL, Crabtree KL, et al. Development of an immunofluorescence assay using recombinant proteins expressed in insect cells to screen and confirm presence of human herpesvirus 8-specific antibodies. Clin.Vacc. Immunol. 2008;15(8):1259–1264. doi: 10.1128/CVI.00487-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento MC, de Souza VA, Sumita LM, et al. Comparative study of Kaposi's sarcoma-associated herpesvirus serological assays using clinically and serologically defined reference standards and latent class analysis. J Clin Microbiol. 2007;45:715–720. doi: 10.1128/JCM.01264-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SJ, Sarid R, Chang Y, Moore PS. Evaluation of the latency-associated nuclear antigen (ORF73) of Kaposi's sarcoma-associated herpesvirus by peptide mapping and bacterially expressed recombinant western blot assay. J Infect Dis. 2000;182:306–310. doi: 10.1086/315689. [DOI] [PubMed] [Google Scholar]
- Pellett PE, Wright DJ, Engels EA, et al. Multicenter comparison of serologic assays and estimation of human herpesvirus 8 seroprevalence among US blood donors. Transfusion. 2003;43:1260–1268. doi: 10.1046/j.1537-2995.2003.00490.x. [DOI] [PubMed] [Google Scholar]
- Raab MS, Albrecht JC, Birkmann A, et al. The immunogenic glycoprotein gp35-37 of human herpesvirus 8 is encoded by open reading frame K8.1. J Virol. 1998;72:6725–6731. doi: 10.1128/jvi.72.8.6725-6731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabkin CS, Schulz TF, Whitby D, et al. HHV-8 Interlaboratory Collaborative Group. Interassay correlation of human herpesvirus 8 serologic tests. J Infect Dis. 1998;178:304–309. doi: 10.1086/515649. [DOI] [PubMed] [Google Scholar]
- Rainbow L, Platt GM, Simpson GR, et al. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GR, Schulz TF, Whitby D, et al. Prevalence of Kaposi's sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet. 1996;348:1133–1138. doi: 10.1016/S0140-6736(96)07560-5. [DOI] [PubMed] [Google Scholar]
- Soulier J, Grollet L, Oksenhendler E, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- Whitby D, Howard MR, Tenant-Flowers M, et al. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- Zhu L, Puri V, Chandran B. Characterization of human herpesvirus-8 K8.1A/B glycoproteins by monoclonal antibodies. Virology. 1999;262:237–249. doi: 10.1006/viro.1999.9900. [DOI] [PubMed] [Google Scholar]
- Zuck TF, Thomson RA, Schreiber GB, et al. The Retrovirus Epidemiology Donor Study (REDS): rationale and methods. Transfusion. 1995;35:944–951. doi: 10.1046/j.1537-2995.1995.351196110900.x. [DOI] [PubMed] [Google Scholar]