Abstract
Pseudogenes populate the mammalian genome as remnants of artefactual incorporation of coding messenger RNAs into transposon pathways1. Here we show that a subset of pseudogenes generates endogenous small interfering RNAs (endo-siRNAs) in mouse oocytes. These endo-siRNAs are often processed from double-stranded RNAs formed by hybridization of spliced transcripts from protein-coding genes to antisense transcripts from homologous pseudogenes. An inverted repeat pseudogene can also generate abundant small RNAs directly. A second class of endo-siRNAs may enforce repression of mobile genetic elements, acting together with Piwi-interacting RNAs. Loss of Dicer, a protein integral to small RNA production, increases expression of endo-siRNA targets, demonstrating their regulatory activity. Our findings indicate a function for pseudogenes in regulating gene expression by means of the RNA interference pathway and may, in part, explain the evolutionary pressure to conserve argonaute-mediated catalysis in mammals.
Small-RNA-directed gene silencing pathways have been adapted to accept numerous inputs and to act on many types of downstream targets. In few places is this more apparent than in animal germ lines where two classes of small RNAs—microRNAs (miRNAs) and Piwi-interacting RNAs (piRNAs)—with distinct biogenesis mechanisms and biological functions have been reported. Although miRNAs, as a group, are ubiquitously expressed, piRNAs have thus far been found only in germ cells and in a few gonadal somatic cells types2. piRNAs repress the activity of mobile genetic elements, forming a small RNA-based, innate immune system with both genetically encoded and adaptive components2–9.
In mice, a homozygous mutation in any single Piwi family member causes male sterility accompanied by gonadal hypotrophy 5,10,11. In Mili and Miwi2 mutants, meiosis is not completed and germ cells are progressively lost5. This correlates with an activation of transposons, particularly the non-long terminal repeat (LTR) retrotransposon, L1 (refs 5, 12). DNA methylation of L1 elements is correspondingly lost. In contrast, females bearing homozygous mutations in individual Piwi genes are apparently normal and fertile 5,10,11. Because female germ cells must also control transposons, we sought to characterize their small RNA profiles to determine whether a piRNA system, similar to that operating in spermatocytes, also exists in oocytes.
Approximately 6,000 fully grown oocytes, arrested in prophase of meiosis I, were collected. Small RNA fractions from 19–24 nucleotides (lower fraction) and 24–30 nucleotides (upper fraction) were gel purified and used to prepare small RNA libraries. These were deeply sequenced3,4. A total of 1,037,355 sequences was obtained that could be mapped to the mouse genome (753,981 from the lower fraction and 283,374 from the upper fraction; Supplementary Table 1). In the lower fraction, 126,515 non-redundant sequences were obtained, falling into 24,271 non-overlapping clusters. In the upper fraction, 97,807 non-redundant sequences fell into 15,032 non-overlapping clusters.
An examination of the small RNAs in the upper fraction of the oocyte library revealed a piRNA population that resembled those found in early-stage spermatocytes3 (Fig. 1a, right). Roughly 62% of small RNAs correspond to annotated repeats (Supplementary Table 2), with 3% matching genic sequences and 3% matching unannotated, intergenic sites. The function of the latter species remains unknown. Roughly 30% of the library corresponded to presumed breakdown products of abundant, non-coding RNAs, such as ribosomal RNAs, transfer RNAs and small nucleolar RNAs.
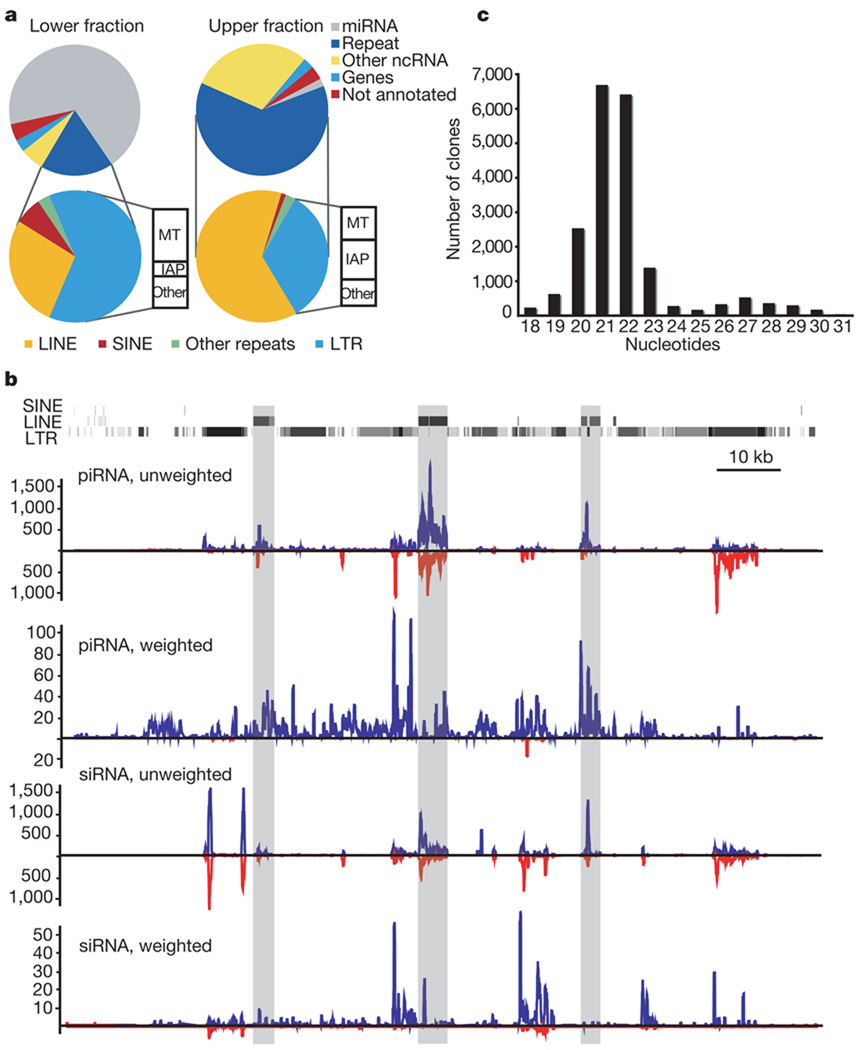
Figure 1. Both piRNA and siRNA systems control transposons in mouse oocytes.
a, Small RNA libraries from 19–24-nucleotide (lower fraction, left) and 24–30-nucleotide RNAs (upper fraction, right) were deeply sequenced. Reads were assigned an annotation as previously described12. The fraction of reads in each category is depicted. The repeat-annotated small RNAs were designated as LINE, SINE (short interspersed element), LTR and other, with the LTR category further divided between MT (mouse transcript), IAP and other. b, A representative piRNA locus on chromosome 10 is shown with the content of LINE, SINE and LTR fragments. Shading indicates the degree of match to the consensus element. Frequency plots for piRNAs and siRNAs from this locus are shown below. ‘Unweighted’ plots each match to the cluster; ‘weighted’ normalizes each match, dividing by its genomic frequency4. Blue and red lines indicate small RNAs mapping to the upper and lower genomic strand, respectively. c, RNAs from 19–30 nucleotides were deeply sequenced. Reads matching the piRNA cluster shown in b were used to construct a frequency plot by length.
As expected, oocyte piRNAs arise from discrete genomic loci in a strand-asymmetric fashion (Supplementary Table 3). A number of these loci share structural similarities to Drosophila piRNA loci, which act as master controllers of mobile elements3. One example (Fig. 1b) spans ~120 kb of chromosome 10 and contains an abundance of long interspersed elements (LINEs) and LTR elements. These have an orientation bias that results in the generation of predominantly antisense piRNAs (Fig. 1b, piRNA, weighted; see also Supplementary Fig. 1).
piRNAs have been proposed to act with transcripts from active transposons in a feed-forward amplification loop that confers signature features on a piRNA population that is mounting an ongoing transposon defence2–4,6,7. Primary piRNA-directed cleavage of transposon mRNAs creates the 5′ ends of secondary piRNAs4,6. This produces primary and secondary piRNA pairs that overlap by 10 nucleotides at their 5′ ends. The 5′ U bias of primary piRNAs thus leads to an enrichment of an A at position 10 of secondary piRNAs. These characteristics are prevalent in piRNA populations from mouse oocytes, particularly those that can be mapped to the L1 and intracisternal A particle (IAP) elements (Supplementary Fig. 2).
As expected, annotated miRNAs comprised the majority (69%) of 19–24-nucleotide RNAs (Fig. 1a, left; see also Supplementary Table 4). Among the highly abundant species are members of the let-7 family (let-7a/c/f), generally abundant miRNAs (miR-22, miR-16, miR-21, miR-26, miR-93 and miR-29a/b), and miRNAs abundant in ovary and placenta (miR-322, miR-503, miR-451). Finally, we detected miRNAs specifically expressed in male and female gonad (miR-103)13.
A substantial fraction of 19–24-nucleotide RNAs matched annotated transposons (Supplementary Table 2). Many that mapped uniquely to the genome could be traced to oocyte piRNA loci (Fig. 1b, siRNA, weighted). These species might represent piRNA degradation products, or oocyte piRNA clusters might generate both siRNAs and piRNAs.
Therefore, we independently mapped piRNAs and candidate siRNAs to consensus L1 and IAP sequences (Supplementary Fig. 3). Each gave characteristic profiles. Moreover, piRNAs and candidate siRNAs show distinctly different nucleotide biases, with piRNAs displaying their characteristic enrichment for a 5′ uridine residue and an A at position 10 (Supplementary Fig. 2). Candidate siRNAs lack a 10A bias and show enrichment for both A and U residues at their 5′ ends (Supplementary Fig. 2). Finally, we gel purified 19–30-nucleotide RNAs from mouse oocytes as a single fraction and deeply sequenced this population. A length distribution of small RNAs that match the piRNA cluster shown in Fig. 1b yields two distinct peaks (Fig. 1c). siRNAs 21–22 nucleotides in length apparently predominate over the piRNA population, which averages ~27 nucleotides. We conclude that transposon-rich loci in oocytes give rise to both siRNAs and piRNAs. Although siRNAs are apparently more abundant, piRNA cloning frequencies could be reduced by the 2′-O-methyl modification that occurs on their 3′ termini2. Our results raise the possibility that piRNA and siRNA systems may act redundantly to repress transposons in mouse oocytes, perhaps explaining the lack of substantial phenotypic consequences of individual Piwi mutations in females5,10,11.
Although many transposons were targeted by both piRNAs and siRNAs, some relied more heavily on a particular pathway. For example, MTB and MTC, both LTR retrotransposons, matched almost exclusively to siRNAs. Moreover, the most prominent cluster that produces MTB/MTC small RNAs contains an inverted repeat with strong potential to produce a Dicer substrate. Notably, this transposon class showed increased expression in Dicer-null oocytes, consistent with its being regulated predominantly if not exclusively by the siRNA system14.
Small RNA libraries often contain genic sequences. In other tissues, these correspond exclusively to sense sequences that probably represent contaminating degradation products. However, in oocytes, numerous sense and antisense siRNAs corresponding to protein-coding genes could be identified (Supplementary Table 5). As mammals lack any identifiable RNA-dependent RNA polymerase, this raised the question of how antisense siRNAs might be generated.
On the basis of polymorphisms, uniquely mapping sense siRNAs could often be assigned to the functional protein-coding copy of a gene, whereas antisense siRNAs mapped to a homologous pseudogene (Fig. 2a and Supplementary Table 5). Thus, oocyte endo-siRNAs might be processed from double-stranded (ds)RNAs that form by hybridization of transcripts derived from two unlinked loci. A similar process in which transcripts from active transposons hybridize to antisense transposon fragments transcribed from piRNA clusters could explain the genesis of transposon siRNAs.
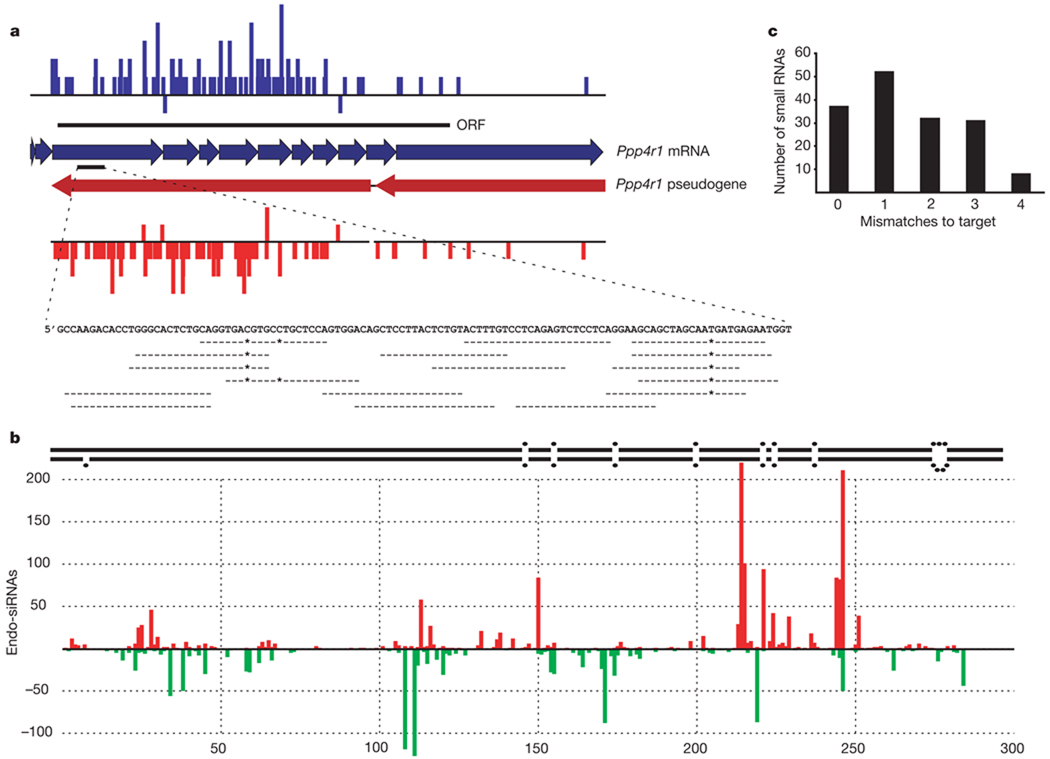
Figure 2. Gene–pseudogene interactions produce endogenous siRNAs.
a, Endo-siRNAs unambiguously mapped to the functional Ppp4r1 mRNA are plotted in blue, above the mRNA (individual exons indicated as thick arrows). The extent of the open reading frame (ORF) is indicated. Endo-siRNAs from the Ppp4r1 pseudogene are shown in red below the pseudogene. Arrows indicate two segments of Ppp4r1 homology. siRNAs plotted above each line are sense oriented, with respect to the functional mRNA, and those below are antisense. Shown below is an enlargement of one section of the mRNA with individual antisense siRNAs aligned. A dash indicates a match; an asterisk indicates a mismatch. The heights of the bars indicate the number of siRNAs starting at each position. b, Endogenous siRNAs homologous to Ran-GAP are shown below a schematic of the genomic inverted repeat structure from which they arise (the ~800-base loop is not depicted). Those shown above and below the x axis come from the upper or lower arm of the hairpin, respectively. No siRNAs were sequenced from the ~800-base loop that separates the two stem arms. c, siRNAs antisense to Ppp4r1 counted and plotted by the number of mismatches to the functional mRNA.
siRNAs from gene–pseudogene pairs arise exclusively from regions of complementarity between the partners. Because many sense-oriented siRNAs cross exon–exon junctions, we propose that mature, spliced mRNAs from genes interact with antisense pseudogene transcripts to form Dicer substrates (Supplementary Fig. 4). In one case (Fig. 2b), both sense and antisense siRNAs to the GTPase-activating protein for Ran (Ran-GAP) were produced from a pseudogene locus containing a ~300-base pair (bp) inverted repeat with an intervening ~800-base loop. siRNAs were derived only from the potentially double-stranded portion of this locus.
In some cases, Dicer processing of dsRNA substrates proceeds in an apparently processive fashion from a discrete initiation site, producing ‘phased’ small RNAs with an ~21-nucleotide periodicity15. Transposon-derived and genic siRNAs showed this property only very weakly (Supplementary Fig. 2). Notably, piRNAs also show a similar, very weak phasing signal, although with a period of ~27 nucleotides rather than ~21 nucleotides.
Pseudogenes have often diverged substantially from their functional ancestors. Thus, we wished to examine the possibility that pseudogene-derived antisense siRNAs could regulate corresponding protein-coding genes. We mapped antisense siRNAs to potentially relevant regulatory targets. Many small RNAs aligned to their targets either with no mismatches or with mismatches lying outside regions essential for slicer cleavage16,17 (Fig. 2a, c). Thus, antisense, pseudogene-derived siRNAs might be capable of regulating homologous protein-coding genes through a conventional RNA-interference mechanism.
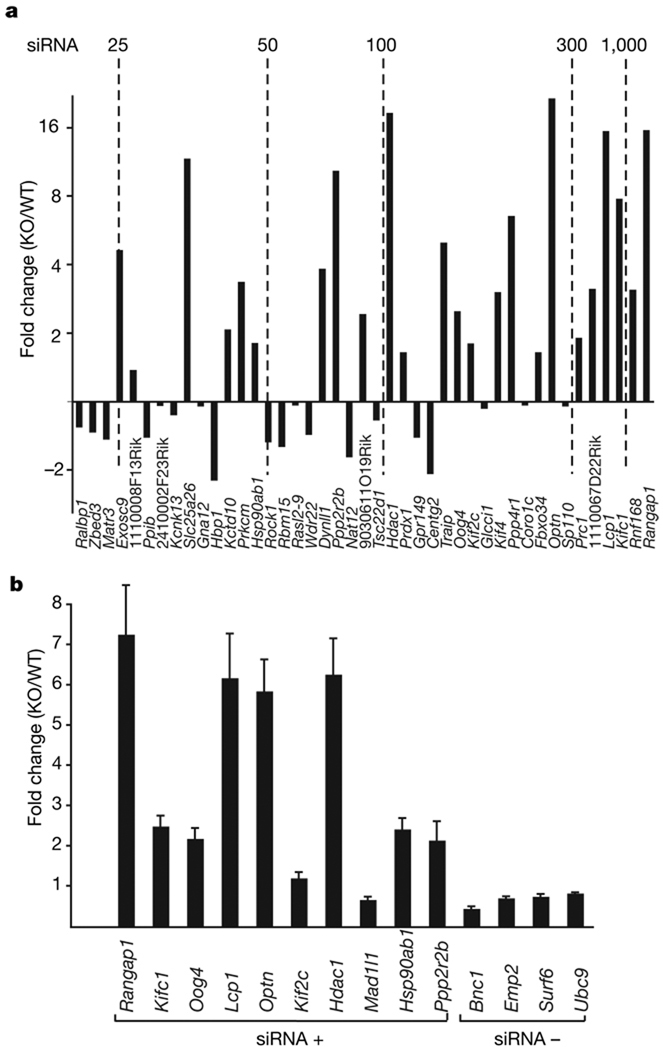
To test the regulatory potential of pseudogene-derived siRNAs, we assessed the effects of Dicer loss on their putative targets. We have previously shown that deletion of Dicer in growing oocytes causes the production of non-functional gametes with defects in spindle organization and chromosome segregation14,18. We compared the expression of candidate endo-siRNA targets in wild-type and Dicer-null cells14. Many genes with abundant, pseudogene-derived siRNAs showed significant increases in expression following Dicer loss (Fig. 3a). We verified candidates derived from the array data by semi-quantitative polymerase chain reaction with reverse transcription (qRT–PCR, Fig. 3b).
Figure 3. Endo-siRNAs have a role in gene regulation.
a, We had previously compared expression levels in wild-type and Dicer-null oocytes by microarray14. Genes with a large number of siRNAs were examined. For those with significant changes in expression (P < 0.1), the fold change in Dicer-null (KO) versus wild type (WT) was plotted. The graph was arranged according to the number of antisense siRNAs per gene in our data set, increasing from left to right (with benchmarks shown). The identity of the gene represented by each bar is given below. b, Fourteen genes (with and without siRNAs in our data set, indicated by siRNA+ and siRNA−, respectively), comprising a set in which some showed significant changes and some did not, were tested for changes in mRNA levels in Dicer-null versus wild-type oocytes by quantitative PCR. Gene names are indicated below each bar; error bars represent the mean ± s.e.m. (three replicates).
Collectively, our data indicate that in mammalian oocytes, protein-coding mRNAs interact with pseudogene transcripts to form dsRNAs that are processed into endo-siRNAs. Examination of Dicer knockouts indicates a function for endo-siRNAs in gene regulation. At present, we cannot distinguish whether these small RNAs direct target cleavage or whether the act of siRNA production per se, which consumes the coding mRNA, is sufficient for repression. However, the specific case of HDAC1 may point to a RNA-induced silencing complex (RISC)-based mechanism. Few uniquely mapping siRNAs are generated from the Hdac1 gene itself, suggesting that it is not used prominently as a Dicer substrate. Instead, most uniquely mapping sense and antisense siRNAs can be assigned to a series of Hdac1 pseudogenes. On the basis of its increased expression in Dicer-null oocytes, we propose that pseudogene-derived, antisense siRNAs direct RISC to cleave Hdac1 mRNAs.
The catalytic potential of at least one argonaute protein has been conserved through mammalian evolution from platypus to humans19,20. However, mammalian miRNAs, with one known exception, act through translational mechanisms without the need for mRNA cleavage21. The discovery of endogenous siRNAs in mammalian oocytes not only expands the realm of mammalian small RNA classes but also provides one possible explanation for the evolutionary pressure to conserve argonaute enzymatic activity.
Pseudogenes have long been considered to be non-functional artefacts of transposition pathways acting on protein-coding mRNAs. In a few cases, regulatory roles have been posited for pseudogenes, largely through antisense mechanisms22–24. Our findings, and those of the accompanying paper25, provide a role for a subset of mammalian pseudogenes in the production of functional siRNAs. The production of dsRNAs by interaction between sense and antisense transcripts from distinct loci has not been observed in other tissues and may require the unique environment of oocytes, which substantially lack a protein kinase R response (a dsRNA-induced general translational repression pathway) and are geared for mRNA stabilization and storage26–28. The fact that many targets of this pathway are related to microtubule dynamics (including microtubule-based processes, P = 0; kinesin complex, P = 0; motor activity, P < 1 × 10−254; spindle, P < 8 × 10−239; and microtubule-associated complex, P < 3 × 10−60; Supplementary Fig. 5) suggests that the regulatory circuits that we describe may have important biological roles, as the consequences of Dicer loss in growing oocytes is disruption of proper spindle formation and defects in chromosome segregation14,18.
METHODS SUMMARY
Mouse oocytes were collected from primed mice and used to prepare small RNA fractions. These were cloned and deeply sequenced as previously described2,4. Bioinformatic analysis of the sequences was performed as described in the Methods. For semi-quantitative RT–PCR, RNA was extracted from fully grown oocytes from Dicerflox/flox and Dicerflox/flox Zp3-cre mice. Quantitative PCR was performed using TaqMan probes.
Supplementary Material
Acknowledgements
We thank members of the Hannon laboratory for discussions. O.H.T. is a Bristol-Meyers Squibb fellow and A.G. is a Florence Gould Foundation Scholar of the Watson School of Biological Sciences. E.P.M. is supported by a fellowship from the Australian-American Association. This work was supported in part by grants from the NIH (R.M.S. and G.J.H.) and gifts from Kathryn W. Davis and the Stanley family (G.J.H. and E.H.). G.J.H. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Small RNA data sets can be accessed in GEO with the following accession numbers GSM261957, GSM261958 and GSM261959.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.D’Errico I, Gadaleta G, Saccone C. Pseudogenes in metazoa: origin and features. Brief. Funct. Genomics Proteomics. 2004;3:157–167. doi: 10.1093/bfgp/3.2.157. [DOI] [PubMed] [Google Scholar]
- 2.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 3.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 4.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 5.Carmell MA, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 59 end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 7.Houwing S, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 8.Saito K, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 10.Kuramochi-Miyagawa S, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 11.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 12.Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 13.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murchison EP, et al. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Martinez J, Tuschl T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr. Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Tang F, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshua-Tor L. The Argonautes. Cold Spring Harb. Symp. Quant. Biol. 2006;71:67–72. doi: 10.1101/sqb.2006.71.048. [DOI] [PubMed] [Google Scholar]
- 20.Murchison EP, et al. Conservation of small RNA pathways in platypus. Genome Res. doi: 10.1101/gr.073056.107. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Korneev SA, Park JH, O’Shea M. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J. Neurosci. 1999;19:7711–7720. doi: 10.1523/JNEUROSCI.19-18-07711.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weil D, Power MA, Webb GC, Li CL. Antisense transcription of a murine FGFR-3 psuedogene during fetal developement. Gene. 1997;187:115–122. doi: 10.1016/s0378-1119(96)00733-0. [DOI] [PubMed] [Google Scholar]
- 24.Zhou BS, Beidler DR, Cheng YC. Identification of antisense RNA transcripts from a human DNA topoisomerase I pseudogene. Cancer Res. 1992;52:4280–4285. [PubMed] [Google Scholar]
- 25.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. doi: 10.1038/nature06908. doi:10.1038/nature06908 (this issue) [DOI] [PubMed] [Google Scholar]
- 26.Stein P, Zeng F, Pan H, Schultz RM. Absence of non-specific effects of RNA interference triggered by long double-stranded RNA in mouse oocytes. Dev. Biol. 2005;286:464–471. doi: 10.1016/j.ydbio.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Bettegowda A, Smith GW. Mechanisms of maternal mRNA regulation: implications for mammalian early embryonic development. Front. Biosci. 2007;12:3713–3726. doi: 10.2741/2346. [DOI] [PubMed] [Google Scholar]
- 28.Stitzel ML, Seydoux G. Regulation of the oocyte-to-zygote transition. Science. 2007;316:407–408. doi: 10.1126/science.1138236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.