Summary
The selective degradation of mRNAs by the nonsense-mediated decay pathway is a quality control process with important consequences for human disease. From initial studies using RNA hairpin-tagged mRNAs for purification of messenger ribonucleoproteins assembled on transcripts with HIV-1 3′ untranslated region (3′UTR) sequences, we uncover a two-step mechanism for Upf1-dependent degradation of mRNAs with long 3′UTRs. We demonstrate that Upf1 associates with mRNAs in a 3′UTR length-dependent manner and is highly enriched on transcripts containing 3′UTRs known to elicit NMD. Surprisingly, Upf1 recruitment and subsequent RNA decay can be antagonized by retroviral RNA elements that promote translational readthrough. By modulating the efficiency of translation termination, recognition of long 3′UTRs by Upf1 is uncoupled from the initiation of decay. We propose a model for 3′UTR length surveillance in which equilibrium binding of Upf1 to mRNAs precedes a kinetically distinct commitment to RNA decay.
Introduction
The nonsense-mediated decay (NMD) machinery executes important regulatory and quality control functions by targeting specific classes of messenger RNAs (mRNAs) for degradation (Chang et al., 2007). In addition to degrading transcripts containing premature termination codons (PTCs) resulting from mutation or rearrangement of genomic DNA or defects in mRNA biogenesis, the pathway is also responsible for regulating between 1–10% of all genes in diverse eukaryotes (He et al., 2003; Mendell et al., 2004; Rehwinkel et al., 2005; Wittmann et al., 2006; Weischenfeldt et al., 2008). Transcripts preferentially targeted by NMD include those with PTCs encoded by alternatively spliced exons, introns downstream of the termination codon (TC), long 3′ untranslated regions (3′UTRs), or upstream open reading frames (uORFs; reviewed in Nicholson et al., 2009; Rebbapragada and Lykke-Andersen, 2009). A characteristic common to many NMD decay substrates is an extended distance from the terminating ribosome to the mRNA 3′ end (i.e., 3′UTR length). Degradation of aberrant mRNAs by NMD can affect the progression of many human genetic disorders, an estimated one-third of which derive from PTCs (Kuzmiak and Maquat, 2006). In addition, shortening of 3′UTRs has been proposed to relax regulation of mRNA stability and translation, promoting cellular transformation (Sandberg et al., 2008; Wang et al., 2008; Mayr and Bartel, 2009). These findings underscore the importance of understanding the mechanisms by which 3′UTR length is sensed in the process of mRNA quality control.
The well-conserved superfamily I RNA helicase Upf1 is a crucial component of the core NMD machinery. Like other RNA helicases, Upf1 exhibits nonspecific but robust RNA binding activity modulated by ATP binding and hydrolysis (Weng et al., 1998; Bhattacharya et al., 2000). While the functional roles of Upf1’s ATPase and helicase activities are unclear, mutations that abolish its ATPase activity prevent NMD (Weng et al., 1996a, b; Sun et al., 1998). In addition, Upf1 participates in a network of interactions with additional factors proposed to mediate its association with mRNA targets and regulate a cycle of Upf1 phosphorylation and dephosphorylation required for establishment of translational repression and recruitment of RNA decay enzymes (reviewed in Nicholson et al., 2009; see below).
Within the context of a long 3′UTR, additional mRNA features and protein components of mRNPs can promote or inhibit decay. For example, the exon-junction complex (EJC), a multiprotein assembly deposited at exon-exon junctions in the process of splicing, acts through Upf1 to strongly activate decay (Le Hir et al., 2000; Kim et al., 2001; Le Hir et al., 2001; Lykke-Andersen et al., 2001). The competition between Upf1 and cytoplasmic poly(A)-binding protein 1 (PABPC1) for binding to the translation release factors eRF1 and eRF3 has been proposed to be a crucial factor in the decision to decay diverse transcripts (Ivanov et al., 2008; Singh et al., 2008). Upf1 binding to release factors at the terminating ribosome stimulates phosphorylation of Upf1 by the SMG-1 kinase, translational repression, and recruitment of decay factors (Kashima et al., 2006; Isken et al., 2008; Cho et al., 2009). Conversely, binding of PABPC1 to release factors is proposed to preserve transcript stability and translational competence. In support of this model, artificial tethering approaches and alterations in 3′UTR structure designed to mimic 3′UTR shortening by bringing PABPC1 in proximity to the termination codon can suppress Upf1-dependent decay (Amrani et al., 2004; Behm-Ansmant et al., 2007; Eberle et al., 2008; Ivanov et al., 2008; Silva et al., 2008).
Here, we use affinity purification of hairpin-tagged mRNAs to isolate and characterize endogenously assembled mRNP complexes. With this approach, we show that Upf1 assembles into mRNPs in a 3′UTR length-dependent manner. Upf1 co-purifies to some extent with all transcripts tested, but is highly enriched on mRNAs containing 3′UTRs derived from known NMD targets. The preferential association of Upf1 with mRNAs containing NMD-sensitive 3′UTRs is not affected by inhibition of translation and NMD. Together with our finding that the efficiency of Upf1 co-immunoprecipitation with 3′UTR-derived RNase H cleavage products correlates with fragment length, these observations suggest a direct role for Upf1 in 3′UTR length sensing. To further investigate the in vivo dynamics of 3′UTR length surveillance and decay, we use retroviral elements to induce translational readthrough of NMD-triggering termination codons. Surprisingly, periodic readthrough events can reduce steady-state Upf1 association with transcripts containing long 3′UTRs and robustly inhibit NMD. Moreover, we show that rare readthrough events permit steady-state Upf1 accumulation in mRNPs but prevent initiation of mRNA decay.
Our data inform a model in which equilibrium binding of Upf1 senses 3′UTR length and establishes an RNP state primed for decay. The identification of potential decay targets by Upf1 is coupled to a subsequent commitment to decay, the rate of which is dependent on other aspects of mRNP structure and composition. Furthermore, our data indicate that the decision to decay takes place over a kinetic interval corresponding to many translation termination events. This separation between 3′UTR length sensing and initiation of decay provides a mechanism to prevent aberrant degradation of normal RNAs and presents an opportunity for transcripts to evade cellular mRNA surveillance. Retroviruses may exploit this opportunity by inducing translational readthrough or frameshifting to periodically disrupt the recognition of viral mRNAs as potential decay substrates.
Results
RNA-based affinity purification identifies protein components of messenger RNPs
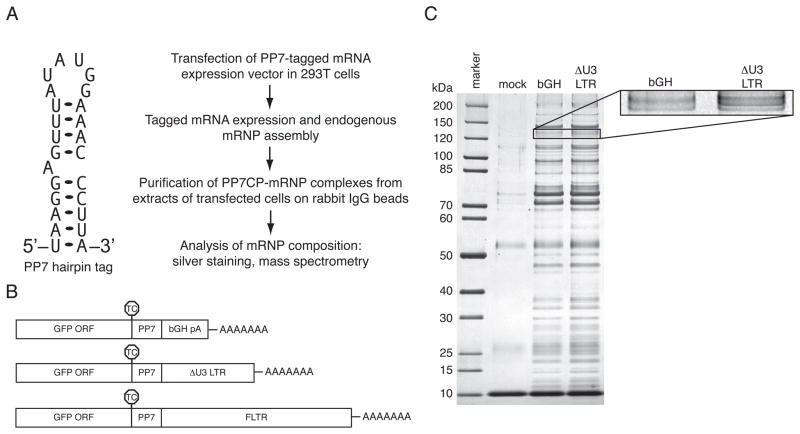
To better understand cellular mRNA biogenesis and decay, we have developed a generalizable technique for purification and characterization of endogenously assembled mRNP complexes. In this approach, we singly tag mRNAs with the naturally occurring Pseudomonas phage 7 coat protein (PP7CP) binding site, a 25 nucleotide (nt) stably folding hairpin (Figure 1A; Lim and Peabody, 2002). Tagged RNAs are transiently or stably expressed in appropriate mammalian cell lines, allowing progression through endogenous RNA processing pathways. RNPs assembled on the tagged RNAs are then purified from extracts using a version of the PP7CP tagged with tandem Staphylococcus aureus protein A domains. Previously, a similar method was used to isolate complexes associated with several noncoding RNAs (Hogg and Collins, 2007a, b). In the process of adapting this methodology to the purification of mRNPs, we found that the use of traditional agarose-based resins afforded inefficient purification of tagged mRNP complexes. In contrast, non-porous magnetic resins allowed purification of tagged mRNAs to near homogeneity following a single step of purification (Figures 1A and C, additional data not shown).
Figure 1. RNA hairpin-based affinity purification of PP7-tagged mRNAs.
A. Left, Predicted secondary structure of the PP7CP RNA binding site (Lim and Peabody, 2002). Right, scheme for purification and analysis of mRNPs containing specific mRNAs.
B. Tagged mRNAs used for RNA-based affinity purification. RNAs containing the GFP ORF, a single copy of the PP7 RNA hairpin, and either the bovine growth hormone polyadenylation element (bGH, top), a HIV 3′LTR variant containing a deletion in the U3 region (ΔU3 LTR, middle), or the full-length HIV 3′LTR (bottom). TC positions are indicated by octagons.
C. Purification of tagged mRNPs. Proteins co-purifying with bGH- or ΔU3 LTR-containing RNAs or present in mock purifications from extracts lacking tagged RNA were separated by SDS-PAGE and detected by silver staining. Inset: magnification of the band corresponding to Upf1 (see also Table S1).
Recent work in our laboratory has shown that HIV-1 3′LTR sequences play a crucial role in the regulation of viral mRNA biogenesis (Valente and Goff, 2006; Valente et al., 2009). To identify proteins specifically associated with HIV 3′LTR sequences, we constructed a series of PP7-tagged RNAs containing the GFP open reading frame and alternative 3′ UTRs (Figure 1B, see below). In our initial experiments, we used a version of the HIV 3′LTR containing a deletion in the U3 region (ΔU3 LTR). The bovine growth hormone polyadenylation (bGH pA) element of the pcDNA3.1 vector was used as a control, aiding discrimination of proteins specifically bound to HIV 3′LTR sequence-containing RNPs. Silver staining of complexes purified from whole cell extracts of transiently transfected 293T cells revealed that, as expected, each tagged RNA associates with a large number of proteins (Figure 1C, data not shown). Mock purifications from extracts lacking tagged RNAs exhibited very few contaminating proteins, indicating that the vast majority of the proteins visible by silver staining were isolated via their association with tagged mRNPs. Many components of the purified mRNPs are found in all complexes purified, including general translation factors, ribosomes, hnRNP proteins and other proteins that associate with common mRNA features (Figure 1C, data not shown).
Tandem mass spectrometry of gel slices excised from HIV ΔU3 LTR and bGH co-purifying material identified four peptides derived from the Upf1 protein in the ΔU3 LTR sample but none in the bGH control sample (Table S1). Immunoblotting of PP7-purified RNPs confirmed that Upf1 was enriched on transcripts containing HIV 3′LTR sequences, using immunoblotting for PABPC1 as a control for RNP recovery (Figure 2A). We detected Upf1 in association with RNAs containing the bGH pA element, but at much lower levels than those co-purifying with RNAs containing the ΔU3 LTR sequence. Still higher levels of Upf1 were isolated using a full-length LTR (FLTR) comprising intact U3, R, and U5 LTR segments from the pNL4.3 reference HIV genome (Figures 1B and 2A). In agreement with observations of human Upf1 co-sedimentation with bulk polysomes and co-immunoprecipitation with diverse mRNAs (Pal et al., 2001; Hosoda et al., 2005), we find that Upf1 associates to some degree with all RNAs tested (Figures 2A and 3A and data not shown). Importantly, we additionally observe substantial transcript-specific enrichment of Upf1 in mRNPs (see below).
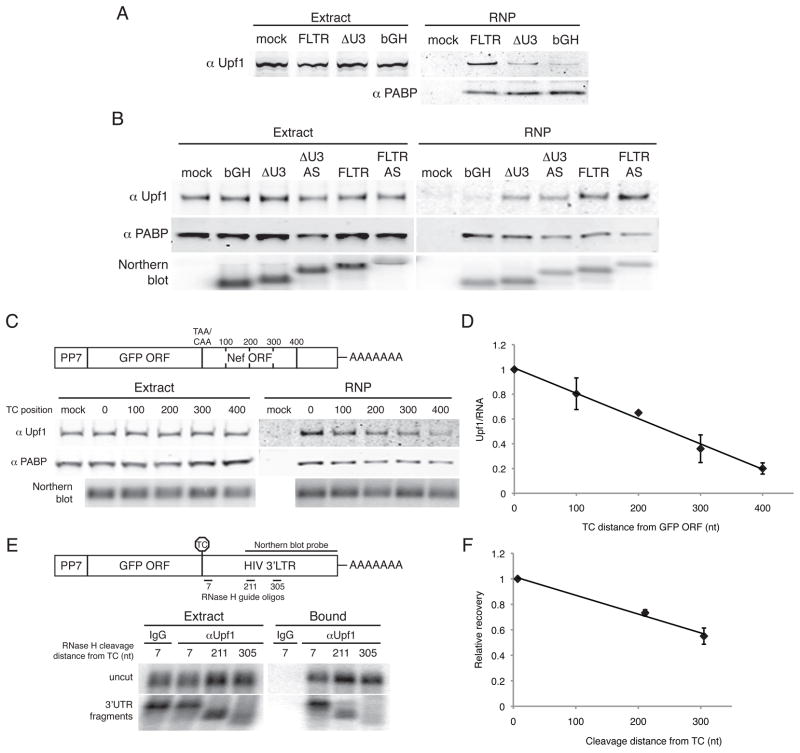
Figure 2. 3′UTR length-dependent interaction of Upf1 with mRNAs.
A. Enrichment of Upf1 on RNAs containing LTR sequence. Proteins in whole cell extracts of parental 293T cells (mock) or cells transiently transfected with the indicated PP7-tagged RNAs (Extract, left) or co-purifying with tagged RNAs (RNP, right) were detected by immunoblotting with antibodies against endogenous Upf1 (top) and PABPC1 (bottom).
B. Sequence-independent assembly of Upf1 in mRNPs. RNPs containing the bGH pA element or full-length or ΔU3 LTRs in the sense (FLTR and ΔU3 LTR) or antisense (ΔU3 AS and FLTR AS) orientations were subjected to purification and immunoblotting as in A. Bottom, small fractions of input extract and purified material were analyzed by Northern blotting to detect tagged RNAs.
C. Upf1 association depends on 3′UTR length. Top, constructs encoding RNAs in which the HIV 3′LTR was fused to the GFP ORF, placing the HIV nef ORF in frame. RNAs contained the standard GFP TC (0) or a CAA codon in place of the GFP TC in tandem with artificially introduced TCs at 100 nt intervals (100, 200, 300, and 400). Bottom, RNPs were purified and analyzed by immunoblotting and Northern blotting as in A.
D. Quantification of data in C. Upf1 signal was normalized to RNA signal from Northern blotting of a fraction of the purified material and arbitrarily set to 1 for the construct containing the standard GFP TC. Error bars denote +/− SEM (n = 2).
E. Co-immunoprecipitation of Upf1 with 3′UTR-derived RNase H cleavage products. RNase H-cleavage of PP7-GFP-FLTR mRNAs was directed using oligonucleotides hybridizing to FLTR sequences 7, 211, or 305 nt downstream of the GFP TC. Extracts were subjected to immunoprecipitation with an anti-Upf1 antibody or nonspecific IgG, and recovery of uncut mRNAs (top panel) and 3′UTR fragments (bottom panel) was monitored by Northern blotting using a probe against the indicated portion of the HIV 3′LTR sequence. See also Figure S1A.
F. Quantification of the data in E. Recovery of 3′UTR fragments was normalized to the abundance of the fragments in extracts, using the uncut mRNAs as internal controls. The recovery efficiency of the longest 3′UTR fragment (7) was arbitrarily set to 1. Error bars denote +/− SEM (n=2).
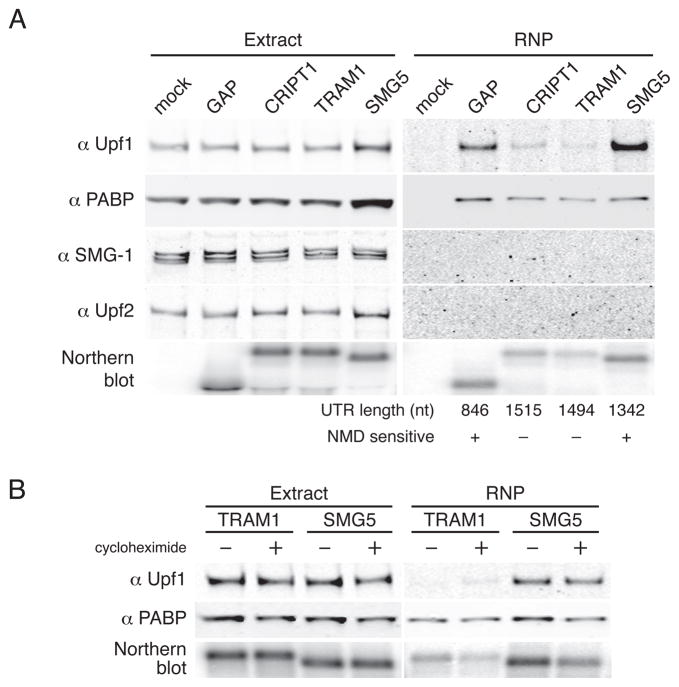
Figure 3. Upf1 preferentially associates with transcripts containing 3′UTRs known to trigger NMD.
A. PP7-tagged GFP mRNAs containing the indicated 3′UTRs were transiently expressed in 293T cells and subjected to affinity purification. Proteins present in whole cell extracts and purified RNPs were detected by immunoblotting with antibodies against endogenous Upf1, PABPC1, SMG-1, and Upf2. Bottom, RNA was isolated from small fractions of extracts and purified material and analyzed by Northern blotting. See also Figures S1A and S1B.
B. Upf1 recruitment is insensitive to cycloheximide treatment. 293T cells transiently transfected with PP7-tagged GFP mRNAs containing the TRAM1 or SMG5 3′UTRs were treated (+) or not treated (−) with cycloheximide for four hours prior to cell harvest and throughout extract preparation and affinity purification. Immunoblotting and Northern blotting were performed as in A. Inhibition of NMD by cycloheximide and persistence of Upf1 recruitment under conditions of translation inhibition by puromycin and 5′-proximal hairpins are illustrated in Figures S1C–F.
Upf1 associates with transcripts in a 3′UTR length-dependent manner
To address the specificity of Upf1 association with RNPs containing HIV 3′LTR sequences, we first created tagged RNA constructs in which the ΔU3 or full-length LTR elements were cloned in the antisense orientation, with the bGH pA element provided downstream to ensure proper 3′ end maturation. The requirement for an additional 3′ end-processing element caused the antisense 3′UTRs to be approximately 200 nt longer than their sense equivalents (Figure 2B, see Northern blot). As above, we observed increasing Upf1 co-purification with the bGH, ΔU3, and FLTR RNAs, respectively (Figure 2B). Surprisingly, the levels of Upf1 associated with the antisense LTR-containing RNAs were slightly higher than with the corresponding sense 3′UTRs. Thus, the observed recruitment of Upf1 to LTR-containing RNAs was not dependent on primary sequence or structural features. Instead, our data suggested that Upf1 accumulation in mRNPs might be dictated by 3′UTR length.
Current models suggest that 3′UTR length is a crucial determinant of NMD susceptibility (Mühlemann, 2008; Rebbapragada and Lykke-Andersen, 2009), but the mechanism by which 3′UTR length is sensed remains unclear. To test the hypothesis that Upf1 associates with transcripts in a 3′UTR length-dependent manner, we generated a series of 5′ PP7-tagged RNAs consisting of the GFP ORF fused to the HIV FLTR, such that the fragment of the HIV nef ORF contained in the LTR was in frame with the GFP ORF (Figure 2C). This series of constructs contains single termination codons at approximately 100 nucleotide intervals, starting with the original GFP termination codon and ending with the nef termination codon approximately 400 nt downstream. In this way, we varied 3′UTR length by making only one (ablation of the GFP TC) or two (ablation of the GFP TC combined with introduction of a new in-frame TC) point mutations to the RNA primary sequence. Using these constructs, we found that Upf1 co-purification with tagged mRNAs increased with 3′UTR length (Figure 2C). The relationship between Upf1 co-purification and 3′UTR length was strikingly linear, consistent with sequence non-specific recognition of long 3′UTRs by Upf1 (Figure 2D).
Our observations suggested that Upf1 might accomplish 3′UTR length sensing by associating with 3′UTRs. To better understand the basis for 3′UTR length-dependent accumulation of Upf1 in mRNPs, we used RNase H and a series of oligonucleotides directed against HIV 3′LTR sequence to site-specifically cleave 5′-tagged GFP-FLTR mRNAs at sites approximately 7, 211, and 305 nucleotides downstream of the GFP TC. Following RNase H digestion, we immunoprecipitated endogenous Upf1 and assayed mRNA recovery by Northern blotting using a probe against HIV 3′LTR sequence. The RNase H cleavage conditions were designed to leave a substantial fraction of the mRNAs intact, allowing the use of full-length mRNAs as recovery controls. FLTR-containing mRNAs were recovered with an antibody against Upf1 but not non-specific control goat IgG (Figures 2E and S1A). Consistent with our observations above, the efficiency of 3′UTR fragment co-immunoprecipitation increased with RNA length (Figures 2E and 2F). These data suggest that Upf1 association along the length of 3′UTRs accounts for the observed 3′UTR length-dependent accumulation in mRNPs.
Upf1 preferentially associates with transcripts containing NMD-sensitive 3′UTRs
Our observation that Upf1 association correlates with 3′UTR length mirrors prior findings that 3′UTR extension causes progressive transcript destabilization in mammalian cells (Bühler et al., 2006; Eberle et al., 2008; Singh et al., 2008). To assess the functional significance of the enrichment of Upf1 on specific mRNAs, we used a series of long 3′UTRs shown by Singh and colleagues (2008) to either promote or evade decay. As representative NMD-insensitive long 3′UTRs, we used the human CRIPT1 (1515 nt) and TRAM1 (1494 nt) 3′UTRs. To model targets of 3′UTR length-dependent NMD, we used the human SMG5 3′UTR (1342 nt) and an artificial 3′UTR comprising a portion of the GAPDH ORF and the GAPDH 3′UTR (GAP; 846 nt).
As above, we transiently transfected 293T cells with tagged RNA constructs containing model 3′UTRs and isolated mRNPs from whole cell extracts with PP7CP. Immunoblotting of purified RNPs revealed that Upf1 association strongly correlated with NMD sensitivity (Figure 3A). Very low levels of Upf1 co-purified with transcripts containing the NMD-insensitive CRIPT1 and TRAM1 3′UTRs. In contrast, transcripts containing the intronless GAP and SMG5 3′UTRs co-purified high levels of Upf1, with the SMG5 3′UTR-containing mRNAs showing the greatest Upf1 recruitment. Likewise, antibodies against Upf1 co-immunoprecipitated mRNAs containing the GAP and SMG5 3′UTRs at higher efficiencies than mRNAs containing the CRIPT and TRAM 3′UTRs (Figure S1A). We did not observe the NMD factors SMG-1 or Upf2 in PP7-purified mRNPs, despite robust detection of the proteins in whole cell extracts used for purification (Figure 3A). In similar experiments, comparable levels of Upf1 co-purified with mRNAs containing the intronless GAP 3′UTR and a version of the GAP 3′UTR containing the adenovirus major-late intron (GAP AdML; Figure S1B). This observation suggests that 3′UTR length is a more significant determinant of Upf1 association than the presence of a spliced intron downstream of the TC. Together, these findings indicate that the extent of Upf1 association with a transcript is diagnostic of its NMD susceptibility, consistent with previous experiments in yeast, C. elegans, and human cells (Johansson et al., 2007; Johns et al., 2007; Silva et al., 2008; Hwang et al., 2010). In addition, they raise the intriguing possibility that endogenous mRNAs with long 3′UTRs such the CRIPT1 and TRAM1 mRNAs evade NMD by preventing steady-state incorporation of Upf1 into mRNPs.
Preferential accumulation of Upf1 on NMD-sensitive transcripts is independent of ongoing NMD
We hypothesized that the enrichment of Upf1 on long 3′UTR-containing transcripts could reflect a direct role for the protein in 3′UTR-length sensing prior to the initiation of decay. To address this possibility, we tested the effect of suppressing NMD on Upf1 recruitment by treating cells with the translation elongation inhibitor cycloheximide (Figure 3B). Because initiation of NMD requires translation termination events, cycloheximide potently inhibits NMD (Figure S1C). Following cell growth and extract preparation in cycloheximide, we purified tagged RNAs containing the TRAM1 and SMG5 3′UTRs and analyzed Upf1 association by immunoblotting. Both in the presence and absence of cycloheximide, SMG5 3′UTR-containing RNAs exhibited enhanced co-purification of Upf1 relative to TRAM1 3′UTR-containing control RNAs (Figure 3B). Identical results were obtained using the CRIPT1 and GAP 3′UTRs (data not shown). Moreover, the same pattern of Upf1 accumulation in mRNPs was observed upon inhibition of translation elongation with puromycin or translation initiation with a cap-proximal stable hairpin (Figures S1D–F). These data demonstrate that Upf1 recruitment is independent of ongoing translation termination and NMD and is therefore well positioned to act as a key determinant of 3′UTR length sensing.
Translational readthrough events reduce Upf1 association with mRNAs containing long 3′UTRs
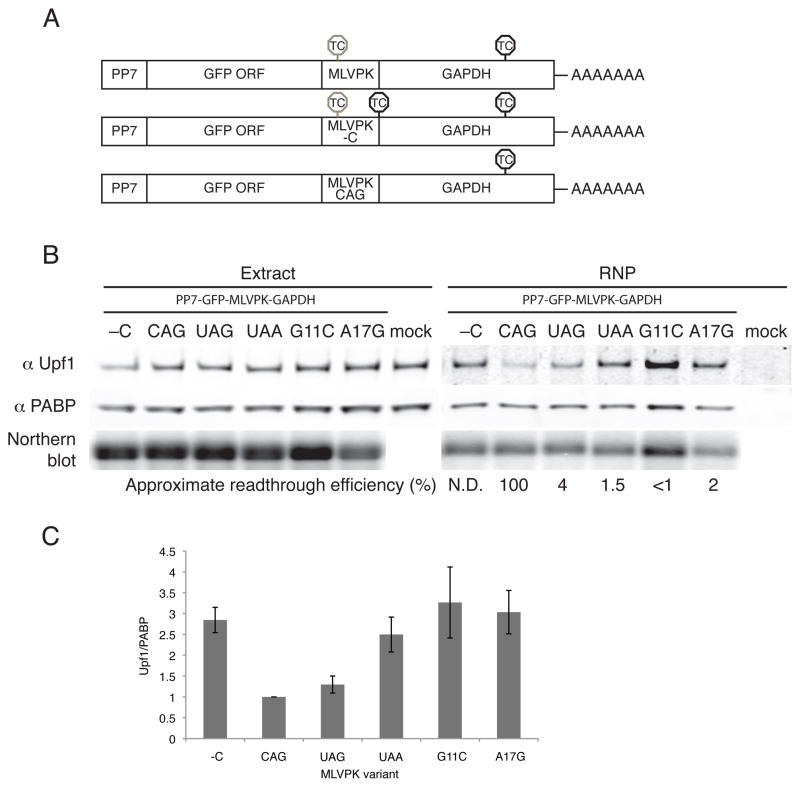
To probe the in vivo dynamics of 3′UTR length recognition by Upf1, we used retroviral elements to modulate the efficiency of translation termination upstream of an NMD-inducing 3′UTR. Retroviruses control the relative production of Gag and Gag-Pol precursor proteins using RNA motifs that induce regulated readthrough or -1 frameshifing to bypass the gag termination codon (Bolinger and Boris-Lawrie, 2009). The Moloney murine leukemia virus pseudoknot (MLVPK), a well characterized example of the former class, causes misincorporation of an amino acid at the gag termination codon with approximately 4% frequency (Figure S2A; Wills et al., 1991). Because ribosomes transiting through 3′UTRs are presumably capable of inducing dramatic remodeling of RNP structure, we reasoned that retroviral readthrough-promoting elements might disrupt the recognition of potential NMD substrates.
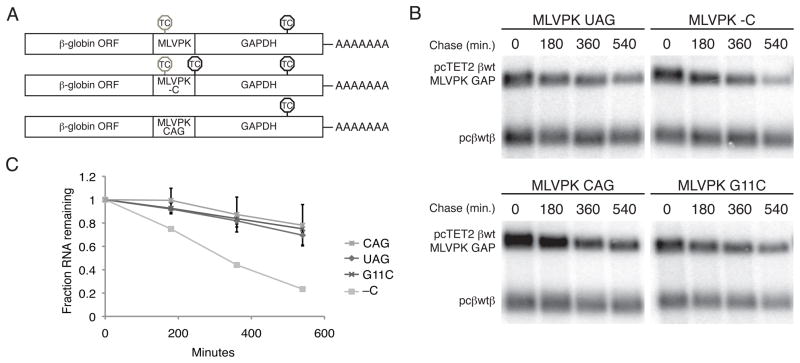
To determine the effects of translational readthrough on Upf1 accumulation and NMD, we inserted the readthrough-promoting MLVPK sequence in place of the standard GFP termination codon in PP7-tagged RNA constructs, upstream of the artificial GAP 3′UTR. This model NMD-triggering 3′UTR comprises 489 nt of the GAPDH open reading frame and 357 nt of the GAPDH 3′UTR. Readthrough events thus result in termination at the downstream GAPDH TC, a position that does not elicit NMD in reporter transcripts (Figure 4A, top; Singh et al., 2008; see below). As controls, we inserted an additional termination codon immediately downstream of the MLVPK to prevent readthrough into the GAPDH ORF (MLVPK -C, Figure 4A, middle) or mutated the upstream termination codon to CAG to allow constitutive translation of the GFP-GAPDH fusion protein (MLVPK CAG, Figure 4A, bottom). In addition, we used three MLVPK variants with reduced readthrough efficiency to assess the competiton between readthrough and Upf1 association: mutation of the wild type UAG termination codon to UAA (~1.5% readthrough; see Figure S2A for bicistronic dual-luciferase readthrough efficiency assays; Feng et al., 1990), G11C (numbering from termination codon; <1% readthrough; Felsenstein and Goff, 1992) and A17G (~2% readthrough; Wills et al., 1994)).
Figure 4. Effects of translational readthrough on Upf1 association.
A. PP7-tagged RNAs containing the GFP ORF in frame with a fragment of the GAPDH ORF were modified to contain the wild-type pseudoknot (top, MLVPK), the MLVPK with an additional in-frame TC downstream (middle, MLVPK –C), or an MLVPK sequence lacking a TC (bottom, MLVPK CAG). Positions of in-frame TCs are indicated. TCs subject to suppression by the MLVPK are indicated by gray octagons; normal TCs are indicated by black octagons.
B. Tagged mRNAs containing the indicated MLVPK variants were transiently expressed in 293T cells and used for RNA affinity purification. Proteins present in whole cell extracts and purified material were analyzed by immunoblotting for endogenous Upf1 and PABPC1, and tagged mRNAs were detected by Northern blotting. See also Figure S2 for determination of approximate readthrough efficiencies using a bicistronic luciferase assay, disruption of Upf1 binding to mRNAs containing the MMTV -1 frameshifting element, and the effects of puromycin on Upf1 recruitment to MLVPK-containing mRNAs.
C. Quantification of Upf1 co-purification normalized to PABPC1 co-purification, arbitrarily set to 1 for the CAG no-TC control. Error bars denote +/− SEM (n = 3).
As expected, immunoblotting of purified RNPs revealed that control mRNAs containing an additional termination codon downstream of the MLVPK efficiently recruited Upf1 (-C, Figure 4B). In contrast, mRNAs in which the wild-type MLVPK sequence (UAG termination codon) directed intermittent translation of the GAP ORF co-purified significantly reduced amounts of Upf1. In fact, mRNAs containing the wild-type MLVPK sequence co-purified levels of Upf1 similar to those associated with mRNAs lacking an upstream termination codon (CAG, Figures 4B and 4C). Interestingly, the extent of Upf1 association was dependent on the efficiency of readthrough, as the three MLVPK mutants exhibiting reduced readthrough activity co-purified high levels of Upf1, similar to the no-readthrough control (Figures 4B and 4C). As a control, we treated cells with puromycin prior to cell extract preparation to confirm that the modulation of Upf1 recruitment by MLVPK was dependent on translation. Indeed, mRNAs containing the wild-type MLVPK and the no-readthrough and constitutive-readthrough controls all co-purified similar levels of Upf1 under conditions of puromycin treatment (Figure S2C). In addition, the Mouse mammary tumor virus (MMTV) -1 frameshifting element also inhibited steady-state Upf1 association, demonstrating that the reduction in Upf1 recruitment was independent of the mechanism of termination codon evasion (Figure S2B). These data suggest that the periodic transit of ribosomes through the 3′UTR during readthrough events remodels the mRNP, displacing Upf1. Readthrough events caused by the wild-type MLVPK were sufficiently frequent to repress steady-state Upf1 accumulation, while less active MLVPK variants permitted recovery of Upf1 binding to mRNPs. Modulation of readthrough efficiency thus uncovers an equilibrium of Upf1 binding and displacement that marks long 3′UTRs of potential decay targets.
Rare readthrough events protect transcripts from decay independent of disruption of steady-state Upf1 association
The ability of readthrough-promoting elements to reduce steady-state Upf1 association with mRNPs raised the possibility that such elements could also stabilize targets of NMD in mammalian cells. To assess RNA decay, we introduced MLVPK variants into tetracycline (tet) regulated β-globin reporter RNAs containing the GAP artificial 3′UTR (Figure 5A, Singh et al., 2008). The indicated tet-regulated constructs were co-transfected with CMV promoter-driven β-globin control RNAs into Hela Tet-off cells. After an interval of transcription induction, we monitored the decay of tet-regulated RNAs over a 9-hour timecourse by Northern blotting (Figures 5B and 5C). As expected, mRNAs lacking an upstream TC (MLVPK CAG) were much more stable than RNAs containing the wild-type MLVPK sequence followed immediately by an additional termination codon (MLVPK –C). Remarkably, all of the MLVPK variants tested, including the minimally active G11C mutant, increased RNA stability to levels indistinguishable from control RNAs lacking an upstream TC (Figures 5B and 5C, additional data not shown). Using the MMTV -1 frameshift element to allow translation of the GAPDH ORF also rescued transcript stability, suggesting that readthrough events stabilize NMD targets independent of the mechanism of translational recoding (Figure S3).
Figure 5. Rare readthrough events stabilize targets of NMD.
A. Schematic of tet-regulated β-globin reporter mRNA constructs used in RNA decay assays. Constructs contained the β-globin ORF and the GAPDH ORF fragment in frame, with intervening MLVPK sequence to regulate readthrough. Positions of in-frame TCs are indicated. TCs subject to suppression by the MLVPK are indicated by gray octagons; normal TCs are indicated by black octagons.
B. Decay assays of reporter mRNAs containing MLVPK variants. Constructs encoding the tet-regulated transcripts described in A (pcTET2 βwt MLVPK GAP; top bands) were co-transfected with the constitutively expressed wild-type β-globin reporter (pcβwtβ; bottom bands) in HeLa Tet-off cells. RNA was harvested 30 minutes after transcription was halted by addition of doxycycline and at 3 hour intervals thereafter. See also Figure S3 for decay assays of mRNAs containing the MMTV -1FS element.
C. Quantification of decay assays. Levels of tet-regulated reporter mRNAs were normalized to levels of the wild-type β-globin transfection control. Error bars denote +/− SEM (n = 3).
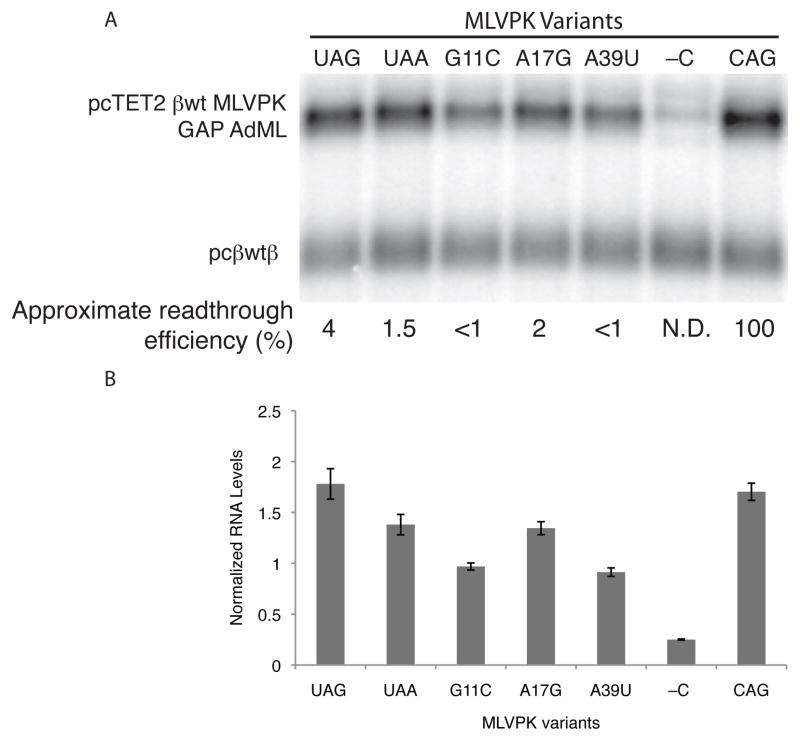
Impaired MLVPK variants allow steady-state Upf1 accumulation in mRNPs but robustly inhibit long 3′UTR-mediated mRNA decay. These findings imply that Upf1 recognition of long 3′UTRs is coupled to a kinetically deferred commitment to decay subject to inhibition by rare readthrough events. The presence of an EJC downstream of a TC substantially enhances mRNA decay, potentially by activating Upf1 recruited by long 3′UTRs. We therefore reasoned that the EJC might overcome the effects of readthrough by accelerating the initiation of decay. As a model for EJC-stimulated decay, we used the GAP AdML intron-containing 3′UTR previously shown to direct efficient degradation of reporter transcripts (Singh et al., 2008). Assays of GAP AdML 3′UTR-containing mRNA accumulation revealed that the ability of MLVPK variants to promote RNA stability correlated with readthrough efficiency (Figures 6A, 6B, and S4). The wild-type MLVPK sequence permitted mRNA accumulation to levels indistinguishable from those of mRNAs lacking an upstream TC (MLVPK CAG), while RNAs containing the mildly impaired UAA and A17G MLVPK variants accumulated to slightly lower levels. The more severe G11C and A39U mutations further reduced RNA accumulation but nevertheless allowed approximately four-fold higher RNA levels than the no-readthrough control (MLVPK -C). Decay assay timecourses confirmed that the MLVPK-containing RNAs were indeed stabilized relative to the no-readthrough control transcripts (Figure S4). These data show that even efficient EJC-stimulated NMD can be significantly impaired by instances of readthrough occurring at less than 1% of all possible termination events. In addition, the differential stability of transcripts undergoing readthrough of varying efficiency points to the existence of a rate-limiting step downstream of Upf1 association that can be accelerated by the EJC (see Discussion).
Figure 6. Readthrough inhibits EJC-stimulated NMD.
A. Northern blot of RNA accumulation in HeLa Tet-off cells co-transfected with constitutively expressed wild-type β-globin transcripts (pcβwtβ; bottom bands) and tet-regulated β-globin transcripts containing the GAP AdML 3′UTR and the indicated MLVPK variants (pcTET2 βwt MLVPK GAP AdML; top bands). See Figure S4 for decay assays.
B. Quantification of RNA accumulation assays. Levels of tet-regulated reporter RNAs were normalized to levels of the wild-type β-globin transfection control. Error bars denote +/− SEM (n = 3).
Discussion
The identification of proteins co-purifying with PP7-tagged RNAs by mass spectrometry allows unbiased analysis of the effects of RNA sequence, structure, and biogenesis pathway on mRNP composition. Using this system, we show that the extent of Upf1 association with specific transcripts strongly correlates with NMD susceptibility, as Upf1 is highly enriched on transcripts containing 3′UTRs derived from known NMD targets. Analysis of mRNPs containing HIV 3′LTR-derived and other model 3′UTR sequences revealed that Upf1 recruitment increases with 3′UTR length but is independent of ongoing translation and NMD. Moreover, we show that Upf1 co-immunoprecipitates 3′UTR-derived RNase H cleavage products in a fragment length-dependent manner, suggesting that Upf1 is associated along the length of 3′UTRs. The enrichment of Upf1 on long 3′UTR-containing transcripts may increase the probability that Upf1 will out-compete PABPC1 for release factor binding and trigger NMD, providing a potential mechanism for the correlation between 3′UTR length and transcript stability in human cells (Bühler et al., 2006; Eberle et al., 2008; Singh et al., 2008).
With the expectation that readthrough events would cause elongating ribosomes to periodically strip Upf1 and other proteins from the mRNA downstream of the bypassed termination codon, we modulated readthrough efficiency to probe the dynamics of Upf1 association. We show that the activity of wild-type MLVPK, which causes approximately 4% readthrough, suppresses steady-state Upf1 recruitment to the artificial GAP 3′UTR. Less frequent readthrough events, in contrast, allow complete recovery of Upf1 binding to mRNPs. Together, our observations suggest that Upf1 accumulation on transcripts is not simply dependent on the site of the vast majority of termination events but is instead determined by sequence-nonspecific association of Upf1 with 3′UTRs. Differential disruption of Upf1 binding by readthrough events of varying efficiency therefore reveals an equilibrium of Upf1 association that can serve as a mechanism for sensing 3′UTR length.
Equilibrium binding of Upf1 to mRNPs may be influenced by several factors, including Upf1 RNA binding affinity, interactions with additional mRNP components, and disruption by elongating ribosomes. The ATP binding and hydrolysis cycle of Upf1 modulates the protein’s sequence-nonspecific RNA binding activity, providing a potential mechanism to regulate Upf1 association with mRNPs (Weng et al., 1998; Bhattacharya et al., 2000). It is possible that Upf1 recruitment to long 3′UTRs is mediated by protein-protein interactions, but scrutiny of silver-stained gels of purified RNPs and immunoblotting for known NMD factors did not reveal additional proteins that showed patterns of co-purification similar to Upf1 (Figures 1C and 3A and data not shown). In addition, Upf1 association was maintained despite treatment with EDTA and inhibition of translation by multiple means, indicating that an interaction with intact ribosomes is not responsible for the observed accumulation of Upf1 in mRNPs (Figures 3B and S1C–G).
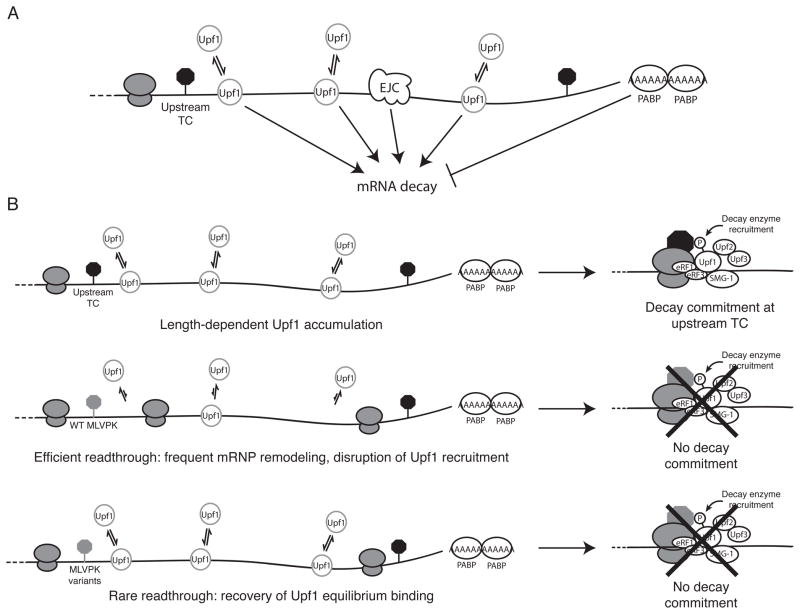
Based on our findings that rare readthrough events permit Upf1 3′UTR length-dependent accumulation in mRNPs but inhibit decay, we propose a two-step model in which Upf1 senses 3′UTR length to potentiate decay (Figure 7). In this model, length-dependent equilibrium binding of Upf1 marks 3′UTRs of potential decay substrates and increases the probability of Upf1 binding to release factors. Upf1 accumulation in mRNPs is necessary but not sufficient to initiate mRNA degradation and is instead followed by a kinetically distinct commitment to decay. The decision to decay is determined by the activity of Upf1 and additional mRNP factors, including the EJC and PABPC1. Here, we provide evidence that 3′ UTR recognition and decay commitment steps can be separated by modulating readthrough efficiency: frequent readthrough events disrupt 3′UTR length sensing by displacing Upf1 from 3′UTR sequence, and rare readthrough events allow Upf1 association but prevent one or more rate-limiting steps required for initiation of decay. Potential rate-limiting steps subject to disruption by infrequent translational readthrough include ATP binding and hydrolysis by Upf1, the formation of the Upf (Upf1-Upf2-Upf3b) or SURF (SMG-1-Upf1-eRF1-eRF3) complexes, Upf1 phosphorylation, and the recruitment and/or activity of the RNA degradation machinery. We find that the presence of a spliced intron downstream of the TC partially restores decay in the context of rare readthrough events, indicating that one key event may stimulated by the EJC, such as Upf1 ATPase activity or phosphorylation (Kashima et al., 2006; Wittmann et al., 2006; Chamieh et al., 2008).
Figure 7.
Model for 3′UTR length surveillance by Upf1. A. Equilibrium length-dependent binding of Upf1 marks long 3′UTRs as potential decay targets. Other aspects of RNP structure and composition such as the EJC and PABPC1 can stimulate or repress decay potentiated by Upf1 association. B. Inhibition of Upf1-dependent decay by translational readthrough. Top, 3′UTR length-dependent accumulation of Upf1 increases the likelihood of Upf1 binding to release factors and precedes the initiation of decay. Middle, readthrough induced by the wild-type MLVPK disrupts steady-state accumulation of Upf1 on mRNAs containing long 3′UTRs and inhibits decay. Bottom, MLVPK variants causing low levels of readthrough allow recovery of Upf1 equilibrium binding but inhibit a kinetically distinct decay commitment step. Candidate rate-limiting steps required for decay initiation are discussed in the main text.
An important consequence of the delay between length-dependent Upf1 accumulation in mRNPs and initiation of decay may be improved quality control fidelity. Based on a release factor-dependent nonsense error rate of 1 in 105 codons (Jorgensen et al., 1993), aberrant termination events are predicted to occur on approximately 50% of transcripts encoding 50 kDa proteins during the course of 100 translation events. Integrating the decision to decay over several termination events provides a mechanism to avoid degradation in response to translational errors while preserving the ability to recognize DNA or RNA-encoded PTCs. This mechanism may also prevent immediate decay upon binding of Upf1 to release factors at a normal TC, which may occur at a significant frequency as suggested by studies of Upf1 and PABP release factor association (Ivanov et al., 2008; Singh et al., 2008).
In mammals, NMD has been proposed to exclusively target newly exported mRNA during a pioneer round or rounds of translation that are biochemically distinguished by the presence of the nuclear cap binding proteins CBP80/20 in the mRNP (Maquat et al., 2010). Replacement of CBP80/20 with translation initiation factor eIF4E marks entry of an mRNP into bulk translation and is thought to confer resistance to NMD. We observe substantial stabilization of transcripts containing elements that direct readthrough at less than 1% efficiency, suggesting that the decision to decay spans an interval corresponding to many translation termination events. These findings mirror the effects of altering termination efficiency in yeast, in which NMD surveillance is conducted throughout the lifetime of a transcript (Zhang et al., 1997; Maderazo et al., 2003; Keeling et al., 2004). The ability of rare readthrough events to inhibit NMD in mammalian cells is supported by multiple reports of readthrough-promoting drugs or inefficient selenocysteine incorporation at UGA codons inducing accumulation of decay targets (Bedwell et al., 1997; Moriarty et al., 1998; Weiss and Sunde, 1998; Mehta et al., 2004; Allamand et al., 2008; Salvatori et al., 2009). Therefore, we hypothesize that mammalian NMD is able to degrade mRNAs that have proceeded beyond the pioneer round(s) of translation.
Experimental procedures
Constructs
For details of plasmid construction, see Extended Experimental Procedures.
Cell culture and extracts
293T cells were maintained, transfected, and used for cell extract preparation essentially as described (Hogg and Collins, 2007b). For details, see Extended Experimental Procedures.
RNA-based affinity purification of mRNPs and immunoprecipitation
All mRNP purification steps were performed at 4°C. Whole cell extracts prepared in 20 mM HEPES pH 7.6, 150 mM NaCl, 2 mM MgCl2, 10% glycerol, 1 mM DTT, and protease inhibitors (HLB150) were supplemented with 0.1% NP-40 and approximately 1 ug/mL ZZ-tev-PP7CP expressed and purified in E. coli as described (Hogg and Collins, 2007b). After rotating for 60 minutes, 4.5 mg of M-270 expoxy Dynabeads (Invitrogen) conjugated with rabbit IgG (Oeffinger et al., 2007) were added per mL of extract, followed by an additional 60-minute incubation. Beads were collected using a magnetic particle concentrator (Invitrogen) and extensively washed in HLB150 + 0.1% NP-40. RNP proteins were eluted from beads using LDS buffer (Invitrogen) for immunoblotting or 1:50 diluted RNase A/T1 mix (Ambion) in 100 mM ammonium bicarbonate for silver staining and mass spectrometry. For details of mass spectrometry and RNase H-cleavage and immunoprecipitation experiments, see Extended Experimental Procedures.
RNA Decay and Accumulation Assays
RNA decay assays were performed as described, with modifications (Singh et al., 2008). For details, see Extended Experimental Procedures.
Detection of RNA and protein
Northern blots were imaged on a Typhoon Trio and quantified using ImageQuant software (G.E.). Immunoblots were imaged and quantified on the Odyssey Infrared Imaging System (LI-COR). For information on probes and antibodies used, see Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Jens Lykke-Andersen and Brian Houck-Loomis for generously providing reagents and Kathleen Collins, Lisa Postow, and Jason Rodriguez for critical reading of the manuscript. Mass spectrometry was performed by Mary Ann Gawinowicz in the Columbia University Medical Center protein core facility. J.R.H. is supported by NRSA postdoctoral fellowship 1F32GM087737. S.P.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allamand V, Bidou L, Arakawa M, Floquet C, Shiozuka M, Paturneau-Jouas M, Gartioux C, Butler-Browne GS, Mouly V, Rousset JP, et al. Drug-induced readthrough of premature stop codons leads to the stabilization of laminin α2 chain mRNA in CMD myotubes. J Gene Med. 2008;10:217–224. doi: 10.1002/jgm.1140. [DOI] [PubMed] [Google Scholar]
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- Bedwell DM, Kaenjak A, Benos DJ, Bebok Z, Bubien JK, Hong J, Tousson A, Clancy JP, Sorscher EJ. Suppression of a CFTR premature stop mutation in a bronchial epithelial cell line. Nat Med. 1997;3:1280–1284. doi: 10.1038/nm1197-1280. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26:1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–1235. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolinger C, Boris-Lawrie K. Mechanisms employed by retroviruses to exploit host factors for translational control of a complicated proteome. Retrovirology. 2009;6:8. doi: 10.1186/1742-4690-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler M, Steiner S, Mohn F, Paillusson A, Mühlemann O. EJC-independent degradation of nonsense immunoglobulin-μmRNA depends on 3′ UTR length. Nat Struct Mol Biol. 2006;13:462–464. doi: 10.1038/nsmb1081. [DOI] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Cho H, Kim KM, Kim YK. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell. 2009;33:75–86. doi: 10.1016/j.molcel.2008.11.022. [DOI] [PubMed] [Google Scholar]
- Eberle AB, Stalder L, Mathys H, Orozco RZ, Mühlemann O. Posttranscriptional Gene Regulation by Spatial Rearrangement of the 3′ Untranslated Region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein KM, Goff SP. Mutational analysis of the gag-pol junction of Moloney murine leukemia virus: requirements for expression of the gag-pol fusion protein. J Virol. 1992;66:6601–6608. doi: 10.1128/jvi.66.11.6601-6608.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YX, Copeland TD, Oroszlan S, Rein A, Levin JG. Identification of amino acids inserted during suppression of UAA and UGA termination codons at the gag-pol junction of Moloney murine leukemia virus. Proc Natl Acad Sci USA. 1990;87:8860–8863. doi: 10.1073/pnas.87.22.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- Hogg JR, Collins K. Human Y5 RNA specializes a Ro ribonucleoprotein for 5S ribosomal RNA quality control. Genes Dev. 2007a;21:3067–3072. doi: 10.1101/gad.1603907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA. 2007b;13:868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda N, Kim YK, Lejeune F, Maquat LE. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell. 2010;39:396–409. doi: 10.1016/j.molcel.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JWB, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJO, He F, Spatrick P, Li C, Jacobson A. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc Natl Acad Sci USA. 2007;104:20872–20877. doi: 10.1073/pnas.0709257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns L, Grimson A, Kuchma SL, Newman CL, Anderson P. Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons. Mol Cell Biol. 2007;27:5630–5638. doi: 10.1128/MCB.00410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen F, Adamski FM, Tate WP, Kurland CG. Release factor-dependent false stops are infrequent in Escherichia coli. J Mol Biol. 1993;230:41–50. doi: 10.1006/jmbi.1993.1124. [DOI] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling KM, Lanier J, Du M, Salas-Marco J, Gao L, Kaenjak-Angeletti A, Bedwell DM. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA. 2004;10:691–703. doi: 10.1261/rna.5147804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science. 2001;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- Kuzmiak HA, Maquat LE. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med. 2006;12:306–316. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim F, Peabody DS. RNA recognition site of PP7 coat protein. Nucleic Acids Res. 2002;30:4138–4144. doi: 10.1093/nar/gkf552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- Maderazo AB, Belk JP, He F, Jacobson A. Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution. Mol Cell Biol. 2003;23:842–851. doi: 10.1128/MCB.23.3.842-851.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread Shortening of 3′UTRs by Alternative Cleavage and Polyadenylation Activates Oncogenes in Cancer Cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Rebsch CM, Kinzy SA, Fletcher JE, Copeland PR. Efficiency of mammalian selenocysteine incorporation. J Biol Chem. 2004;279:37852–37859. doi: 10.1074/jbc.M404639200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Moriarty PM, Reddy CC, Maquat LE. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol. 1998;18:2932–2939. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlemann O. Recognition of nonsense mRNA: towards a unified model. Biochem Soc Trans. 2008;36:497–501. doi: 10.1042/BST0360497. [DOI] [PubMed] [Google Scholar]
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci. 2009;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Meth. 2007;4:951–956. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- Pal M, Ishigaki Y, Nagy E, Maquat LE. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA. 2001;7:5–15. doi: 10.1017/s1355838201000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr Opin Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatori F, Breveglieri G, Zuccato C, Finotti A, Bianchi N, Borgatti M, Feriotto G, Destro F, Canella A, Brognara E, et al. Production of β-globin and adult hemoglobin following G418 treatment of erythroid precursor cells from homozygous β039 thalassemia patients. Am J Hematol. 2009;84:720–728. doi: 10.1002/ajh.21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA. 2008;14:563–576. doi: 10.1261/rna.815108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke-Andersen J. A Competition between Stimulators and Antagonists of Upf Complex Recruitment Governs Human Nonsense-Mediated mRNA Decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Perlick HA, Dietz HC, Maquat LE. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci USA. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente ST, Gilmartin GM, Mott C, Falkard B, Goff SP. Inhibition of HIV-1 replication by eIF3f. Proc Natl Acad Sci USA. 2009;106:4071–4078. doi: 10.1073/pnas.0900557106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente ST, Goff SP. Inhibition of HIV-1 gene expression by a fragment of hnRNP U. Mol Cell. 2006;23:597–605. doi: 10.1016/j.molcel.2006.07.021. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Mönch K, Thoren LA, Nielsen FC, Jacobsen SEW, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss SL, Sunde RA. Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA. 1998;4:816–827. doi: 10.1017/s1355838298971990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996a;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996b;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA. 1998;4:205–214. [PMC free article] [PubMed] [Google Scholar]
- Wills NM, Gesteland RF, Atkins JF. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc Natl Acad Sci USA. 1991;88:6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills NM, Gesteland RF, Atkins JF. Pseudoknot-dependent read-through of retroviral gag termination codons: importance of sequences in the spacer and loop 2. EMBO J. 1994;13:4137–4144. doi: 10.1002/j.1460-2075.1994.tb06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann J, Hol EM, Jäck HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Welch EM, Hogan K, Brown AH, Peltz SW, Jacobson A. Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA. 1997;3:234–244. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.