Abstract
The development of organisms requires concerted changes in gene activity. The free radical theory of development proposes that oxygen serves as a morphogen to educe development by influencing the production of metabolic oxidants such as free radicals and reactive oxygen species. One of the central tenets of this theory is that these metabolic oxidants influence development by altering the antioxidant capacity of cells by changing their production of glutathione (GSH). Here we extend upon these principles by linking GSH production and oxygen sensing in the control of gene expression to establish the epigenotype of cells during development. We prescribe this novel role to GSH and oxygen during development because these metabolites influence the activity of enzymes responsible for initiating and perpetuating epigenetic control of gene expression. Increased GSH production influences epigenetic processes including DNA and histone methylation by limiting the availability of S-adenosylmethionine, the cofactor utilized during epigenetic control of gene expression by DNA and histone methyltransferases. Moreover, the recent discovery of histone demethylases that require oxygen as a cofactor directly links epigenetic processes to oxygen gradients during development.
Keywords: Epigenetics, oxygen, S-adenosylmethionine, DNA methylation, histone, glutathione, Jumanji, HIF
Introduction
Development is process that requires the formation of complex tissues and organ systems. The primary force behind these processes in development is changes in gene activity. Gene activity during development can be dictated through morphogens. However, at this time the mechanisms driving differential gene activity remain unknown. Much speculation has been regarding the role of free radicals during development. The balance between the production and removal of oxidants are known to change during the development of organisms. Rajindar Sohal and others first hypothesized a role of oxygen free radicals in differentiation in 1985 and later in 1986 [1-3]. Three years after this Allen and Balin expanded upon this hypothesis to put forth the free radical theory of development [4]. The central tenets of this theory are that changes in oxygen gradients and an alteration in the balance between the formation and removal metabolically generated oxidants can elicit developmental events. Sohal, and later Allen and Balin, suggested that both oxygen gradients and an altered redox state in cells caused by free radicals can educe development by changing gene expression. When the free radical theory of development was first put down our understanding of the complex processes regulating gene expression were first emerging. Because every cell contains the same genetic information, changes in gene expression are controlled by processes functioning independently of alterations in DNA sequence. One such manner by which gene expression can be dynamically controlled independent of changes in DNA sequence is by epigenetic processes.
In the past decade the role of epigenetic processes in governing gene expression during development and carcinogenesis has received much attention. Epigenetic control of gene expression is largely facilitated by DNA methylation and the post-translation modification of histones. Both DNA methylation and histone modifications are known to play a role in controlling gene expression during development. Global changes in DNA and histone modifications take place during the development of organisms [5]. However, the basic mechanisms eliciting changes in epigenetic control of gene expression during development are largely unknown. We speculate that dynamic changes in epigenetic processes are related to oxygen gradients and the redox status observed during development. The purpose of this article is to reexamine the free radical theory of development and its relation to epigenetic control of gene expression. We will begin by covering the basic principles of the free radical theory of development. We will then introduce the fundamentals of epigenetic processes and how they regulate gene expression during development. Our primary focus will be to discuss how redox status might influence development by affecting the production of S-adenosylmethionine (SAM), the cofactor for DNA and histone methylation reactions. The emerging influence of oxygen gradients on epigenetic processes and their potential role in regulating gene expression will also be discussed.
The Free Radical Theory of Development
The free radical theory of development centers on the role reactive oxygen species play to bring forth cellular differentiation. Free radicals bring about differentiation by progressively changing the intracellular environment of cells towards an oxidizing state. This topic has already been exhaustively reviewed [1-4], therefore we will summarize the main points that relate to our epigenetic perspective. We will separate our discussion into two main parts. First we will focus on changes in ROS production and their role in altering antioxidant enzyme levels during development; and second we will discuss redox alterations during development by changes in glutathione (GSH) synthesis.
Increasing oxygen concentration greatly influences the production of reactive oxygen species (ROS) like superoxide (O2•−) and hydrogen peroxide (H2O2). Oxygen tension across developing tissues is proportional to their distance from a source of oxygen (i.e. a vessel). This change in oxygen concentration alters the metabolic scope of cells by allowing them to utilize oxidative phosphorylation to produce ATP, rather than relying on glycolysis alone. Allen and Balin described this as a “metabolic gradient” that influences the development of tissues [4]. Likewise, Sohal et al. postulated that increasing oxygen drives development by increasing the production of oxidative species like O2•− and H2O2 in cells [2]. To counter increased production of ROS during development cells simultaneously increase their level of antioxidant defenses. These antioxidant defenses exist in two forms: antioxidant enzymes with catalytic activity, and small molecular weight antioxidants that eliminate ROS in a stoichiometric manner. The expression of antioxidant enzymes such as superoxide dismutases (SODs) changes during development [6, 7]. In mammals the most dramatic change in SOD activity is seen following birth. This is likely due to exposure of fetal tissues to atmospheric oxygen after parturition. The expression and activity of manganese superoxide dismutase (MnSOD) increases in many tissues and continues to do so for the first few weeks of life [6, 7]. Furthermore, sod2−/− knockout mice apparently develop normally in utero [8]. It is only after experiencing life in oxygen that sod2−/− pups succumb to oxygen toxicity, most likely by leaving O2•− unchecked. It is even more interesting to look at the sod2+/− animals. These pups develop to adulthood normally without any apparent developmental defects even though they only 50% of the MnSOD activity of wild type littermates [8, 9].
Alterations in cellular redox during development can principally be attributed to changes in GSH production. As a general rule poorly differentiated cells have high levels of GSH, while well differentiated cells have a lower concentration of GSH [10, 11]. This dynamic change in GSH production can be attributed to alterations in the level of expression of enzymes that are producing glutathione [12]. Central to the free radical theory of development is that a change in redox potential from a reducing to an oxidizing intracellular environment must occur as cells differentiate. Maintaining a reducing environment by forcing organisms to overproduce GSH blocks this oxidative switch from occurring and impedes their development. Over expression of the catalytic subunit of γ-glutamylcysteine ligase (GCLc), the rate limiting enzyme in glutathione synthesis, in Drosophila dramatically increases their glutathione production and delays aging. However, further increasing GSH production by simultaneously over expressing GCLc and its modifier subunit GCLm inhibits Drosophila metamorphosis [13]. This block in Drosophila development can be attributed to the increased level of GSH that maintains larvae in a reducing environment. If maintaining an overly reducing environment inhibits development, does a prooxidant environment elicit or accelerate differentiation? This type of prooxidizing environment can be achieved in by interrupting GSH synthesis, or by exposing organisms a prooxidant such as superoxide. Blocking glutathione synthesis in the slime mold Physarum polycephalum with the GCLc inhibitor buthionine sulfoximine (BSO) decreases their production of GSH and accelerates their rate of differentiation [2]. Furthermore Physarum differentiation can also be also be educed with the superoxide generator paraquat [2]. Taken together these studies demonstrate a relationship between the redox state and differentiation in lower organisms. In mammals a burst of GSH synthesis and redox changes occur during two distinct points in development, gametogenesis, and after fertilization. During gametogenesis the GSH content of spermatogonia and oogonia increases dramatically as they mature [14-16]. Increased GSH synthesis occurs again between fertilization and blastogenesis. During these early stages of development it GSH is most likely required to protect the developing embryo from oxidative stress. Most studies have focused on comparing GSH in specific tissues between fetal, neonatal, and adult stages of development. These studies reveal a general trend between age and GSH content, with fetal tissues being the most reducing and adult tissues the most oxidizing [11].
The means by which free radicals are exerting an influence on development is by changing gene activity. A widely held opinion is that increased ROS production and redox changes can lead to genetic damage and altered gene expression during carcinogenesis [17, 18]. While this might well be the case in cancer, proper development is insistent upon mutations being kept to a minimum. In the original renderings of the free radical theory of development it was speculated that ROS generation by oxygen influences the redox state and affects gene expression by altering chromatin configuration [2, 4]. This effect of oxygen and ROS on chromatin structure would be referred to today as an epigenetic process. We speculate that oxygen and redox conditions influence the availability of cofactors required by enzymes that initiate and perpetuate epigenetic events. By influencing the activity of these enzymes epigenetic events can be dynamically changed during development and thus alter gene activity. The remainder of our discussion will focus on the influence of free radicals and oxygen on epigenetic processes in development.
Epigenetics: The science of development
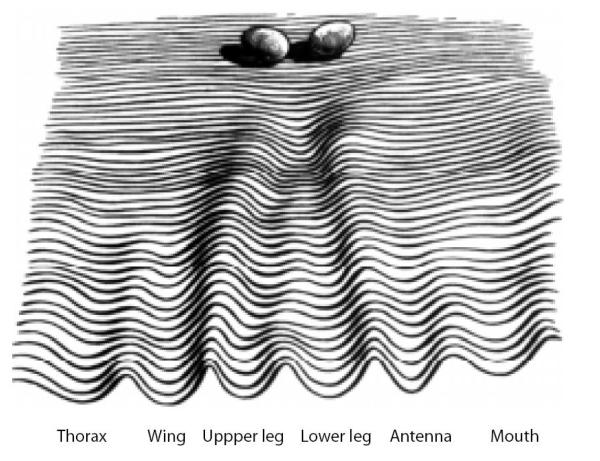
Conrad Waddington first coined the term epigenetics in 1938 where he defined it as “the science concerned with the causal analysis of development” [19]. At that time there was no evidence to support a genetic component of development as we understand it. Waddington depicted the concept of epigenetics during development as a fertilized egg rolling down a theoretical “epigenetic landscape” [20]. Each egg in Waddington’s illustration represents a developing cell. As each egg rolls down the epigenetic landscape it enters a series of canals in a process Waddington referred to as “canalization”, which decides the developmental fate of cells (Fig. 1). A contemporary understanding of epigenetics as it relates to gene expression during development has been put forth by Robin Holliday where he broadly describes epigenetics as the “unfolding of the genetic program for development” [21]. Under this contemporary view, epigenetics can loosely be defined as changes in gene expression independent of changes in DNA sequence. While these definitions describe the role of epigenetics during development they lack a mechanistic component. Recently a unified definition of epigenetics has been proposed by Adrian Bird where he describes it as “the structural adaptation of chromosomal regions so as to register, signal or perpetuate altered activity states” [22]. It is by altering chromosomal regions that epigenetics affects gene activity during development. Chromosomal changes that modify gene expression are primarily facilitated by two processes, methylation of CpG di-nucleotides, and the post-translational modification of histone tails. Both of these processes dynamically control gene expression in higher eukaryotic cells. Establishment of cell type specific gene expression is a fundamental aspect of cellular differentiation that begins in embryogenesis and continues throughout fetal development. By working together DNA methylation and histone modifications regulate cell type specific gene expression, thus giving rise to the formation of tissues and organs that Waddington noted when he first described epigenetics.
Fig. 1.
Conrad Waddington’s depiction of an epigenetic landscape during development. “A symbolic representation of the developmental potentialities of a genotype in terms of a surface, sloping towards the observer, down which there run balls each of which has a bias corresponding to the particular initial conditions in some part of the newly fertilised egg. The sloping surface is grooved, and the balls will run into one or other of these channels, finishing at a point corresponding to some typical organ”. Figure and legend from Waddington, 1956 [20].
DNA methylation
Genetic information is commonly composed of four bases: adenine, thymine, guanine and cytosine. However, epigenetic control of gene expression by DNA methylation gives rise to a fifth base in DNA, 5-methyl-cytosine (5-MeC). Methylation of cytosine is found almost exclusively at CpG di-nucleotides in animals [23]. These CpG di-nucleotides occur at an unusual frequency in the human genome, approximately one-fifth of their expected value. Moreover, CpG di-nucleotides cluster together to form what epigenesists refer to as “CpG islands” [24]. These CpG islands play a critical role in controlling gene expression. The completion of the human genome revealed that approximately half of all human genes contain CpG islands within their regulatory elements [25]. Thus, it seems likely that DNA methylation at CpG islands might constitute one means by which cell type specific gene expression is achieved during development. As a general rule epigenetic silencing of gene expression is associated with the hypermethylation of CpG islands. The exact influence of CpG hypermethylation on gene expression will be discussed below.
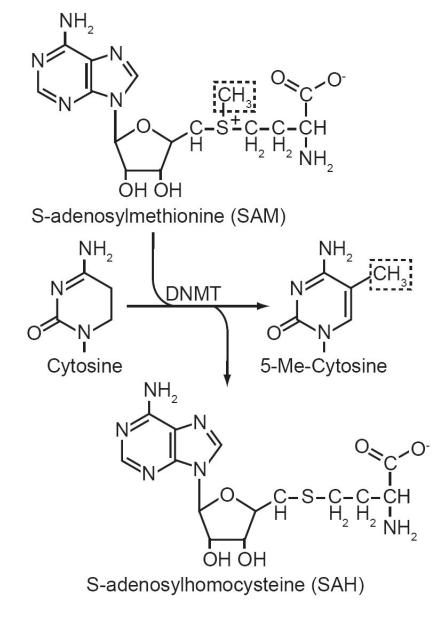
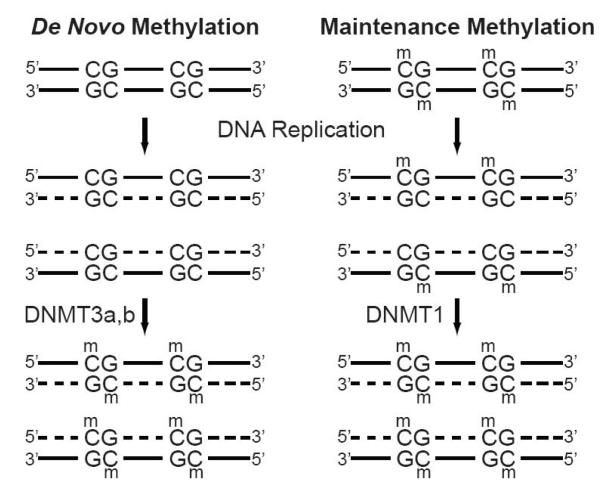
Epigenetic control of gene expression by cytosine methylation is facilitated by the activity of DNA methyltransferases (DNMTs). These enzymes recognize CpGs within double stranded DNA as substrates. DNMTs catalyze the transmethylation of cytosine by transferring methyl groups from S-adenosylmethionine (SAM) to position 5 of the pyrimidine ring (Fig. 2). This reaction results in the production of 5-MeC in DNA and the spent cofactor S-adenosylhomocysteine (SAH). The methylation of cytosine in mammalian genomes is predominantly carried out by three DNMTs: DNMT1, DNMT3a and DMNT3b. These enzymes are subdivided into two classes based on their fundamental differences in CpG substrate specificity in vivo [26]. The first, maintenance DNA methylation, is catalyzed by DNMT1 and occurs rapidly following DNA replication. This gives DNMT1 the primary role of passing on epigenetic control of gene expression to daughter cells [27]. The maintenance DNA methylation activity of DNMT1 can is prescribed to its high affinity for hemimethylated DNA [28]. DNA replication results in the formation of hemimethylated DNA, where only the parent strand contains methylated CpGs. During DNA synthesis DNMT1 rides the DNA replication fork through protein/protein interactions with PCNA [29]. This interaction puts DNMT1 in close proximity to newly synthesized hemimethylated CpG di-nucleotides. The second class is referred to de novo DNA methylation, and is primarily responsible for initiating new epigenetic events that regulate gene expression (Fig. 3). De novo methylation is catalyzed by DNMT3a and DNMT3b. The timing of de novo methylation is more puzzling, it can occur anytime following DNA replication to initiate new epigenetic events that can be passed on during future cell divisions. These elusive de novo methyltransferases were identified in 1998 with the cloning of DNMT3a and DNMT3b by Masaki Okano in 1998 [30-32]. DNMT3a and DNMT3b are encoded by different genes [33, 34]. Their initial characterization revealed a crucial difference between their activities and that of DNMT1, they do not exhibit a substrate preference between hemimethylated and unmethylated DNA [30]. This high affinity for unmethylated CpG di-nucleotides is critical for assigning the role of de novo methylation to DNMT3a and DNMT3b [30]. These two classes of enzymes compose an enzymatic toolkit to generate and perpetuate epigenetic control gene expression during development, gametogenesis, and imprinting.
Fig. 2.
The transmethylation reaction catalyzed DNA methyltransferases (DNMTs). DNMTs catalyze the transfer of the methyl donor group from S-adenosylmethionine (dashed box) to the 5 position of the pyrimidine ring of cytosines within CpG di-nucleotides in genomic DNA. The reaction results in the production of 5-Me-Cytosine and S-adenosylhomocysteine.
Fig. 3.
De novo and maintenance methylation of genomic DNA. De novo methylation of CG doublets by DNA methyltransferases 3a and 3b (DNMT3a,b). DNMT3a and b initiate new epigenetic marks by methylating previously unmethylated CG doublets in genomic DNA after the replication of new DNA daughter strands (dashed lines). DNA methyltransferase 1 (DNMT1) recognizes hemimethylated CpG doublets after DNA replication and methylates the newly synthesized daughter strands to perpetuate epigenetic events to daughter cells.
DNA methylation in development
The idea that DNA methylation controls gene expression was first proposed by Robin Holliday and John Pugh in 1975 [35]. They proposed that the methylation of DNA could account for developmental changes by controlling the activity of genes. There are three major ways in which DNA methylation can influence development: imprinting, X-inactivation, and cell type specific gene expression. We will discuss the role of DNA methylation in each briefly below. The influence of the maternal and paternal genetic information passed on to progeny is not equivalent. Imprinting events can regulate whether the maternal or paternal allele of a specific gene is expressed in offspring. These imprinting events can be directly controlled by DNA methylation [36]. During gametogenesis, and after fertilization, the level of DNA methylation in genomes becomes decreased [37, 38]. This act of global DNA demethylation is not a passive event, meaning it is not based on loss of DNMT activity [39]. The rate at which the DNA in gametes becomes demethylated exceeds that which can be accounted for based on cell division alone in the absence of maintenance methylation. Therefore the global demethylation during early embryogenesis is an active process. After gametes are formed, their level of DNA methylation increases [38, 40] . At this time potential imprinting events are set in oocytes and spermatozoa. Imprinting by epigenetics also gives insight into the altered phenotype of animals produced from somatic cell cloning. These animals are often larger than the original animal from which they were cloned. This phenotype most likely manifests itself due to the epigenetic instability caused by aberrant DNA methylation in their genomes [41].
Imprinting events are initiated and perpetuated by DNMTs. Inhibitors of DNMTs such as 5-aza-2′-deocytidine induce DNA hypomethylation and elicit cell differentiation [42, 43]. However, these treatments do not distinguish between maintenance and de novo DNMTs. During gametogenesis and embryogenesis de novo DNA methylation predominates over maintenance methylation [30]. This is supported by the pattern of expression of the enzymes responsible for each. The expression of DNMT1 is lowest during gametogenesis, while DNMT3a and DNMT3b expression is at their highest. Knockout mice models of DNMT3a and DNMT3b have been generated and single and double knockouts of these enzymes and are embryonic lethal [30]. The observed lethality can attributed to the loss de novo DNA methylation which is essential for genetic imprinting. Even though these enzymes are closely related on a sequence level, the fact that each knockout is lethal suggests they have exclusive activities in vivo. The greatest difference between these two enzymes is their apparent substrate preference. DNMT3a prefers to methylate CpGs that are packed closely together, while DNMT3b is more efficient at methylating isolated CpGs [44, 45]. Loss of DNMT3b activity is also responsible for immunodeficiency, centromere instability and facial anomalies (ICF) syndrome (For review see [46]). ICF patients are immunosuppressed and often have facial abnormalities associated with their disease. Losing both alleles of DNMT3b activity leads to hypomethylation DNA at the juxtacentromeric heterochromatin regions of chromosomes 1 and 16 in humans [47]. This leads to the acquisition of pericentric chromosomal aberrations as part of the manifestation of the disease during development.
DNA methylation also influences the expression of specific genes once the fetus has started to develop. This is accomplished by DNA methylation during its role in X-inactivation, and cell type specific gene expression. The two X chromosomes in females presents a gene expression dosage problem. To compensate for this females use the Xist mechanism to silence one of their X chromosomes. Xist is a large untranslated RNA that coats the X-chromosome from which it is expressed [48]. Both X chromosomes initially express the Xist gene at a low level. Eventually, during a random process, one of the Xist genes becomes silenced by DNA methylation, while expression of the other is increased [49]. The X chromosome with a silenced Xist houses the genes that will become expressed in future progeny cells, while the other X chromosome becomes coated with the high levels of Xist RNA it is expressing [50, 51]. Cell type specific gene expression by DNA methylation has been linked to methylation of CpG islands within the promoter of specific genes. For example, work in our laboratory has revealed that maspin and 14-3-3σ can both be silenced in association with DNA methylation in a cell type specific manner [52]. While these examples are interesting they may be the exception and not the rule, and the precise mechanisms responsible for establishing epigenetic inactivation of specific genes at other loci remain unknown.
Histones
Genetic information is packaged into higher order structures by nucleosomes. Nucleosomes package approximately 146 base pairs of DNA wrapped around an octamer of histone proteins. Each nucleosome is an octamer of two of histone H2A, H2B, H3, and H4 proteins. Besides effectively organizing genetic material in the nucleus, nucleosomes also play an important role in regulating gene activity by controlling their accessibility. When the structure of the nucleosome was first solved by Karolin Luger and colleges in 1997 it was noticed that the N and C terminuses of histone proteins protrude form the central axis of the nucleosome [53]. These protrusions are referred to as “histone tails”. Because histone tails project from the main structure of the nucleosome they can modulate the interactions between DNA and nucleosomes, or between nucleosomes. Changing the interactions between nucleosomes is carried out by a complex array of post-translational modifications at histone tails.
Histone modifications are a multifaceted epigenetic process that plays a critical role to dynamically regulate gene expression. The study of histone modifications began with the discovery of lysine acetylation in histones by Phillips in 1963 [54]. This was quickly followed by the observation that lysines in histones are also methylated [55]. Early work also noted that histone modifications occur in a post-translational manner and directly influence the synthesis of RNA [56, 57]. Today we know that histone tails are the beneficiary, or victim, of various types of post-translation modifications. These modifications are found at various amino acids within histones. This list has grown to include: acetylation, methylation, phosphorylation, ubiquitination, poly-ADP-ribosylation, biotinylation, and sumoylation (For review of these topics see [58-60]). These different modifications interact in a concerted manner to form a “histone code” to regulate gene expression [61]. For the purposes of this review pertaining to development, we will confine our discussion to histone methylation and acetylation.
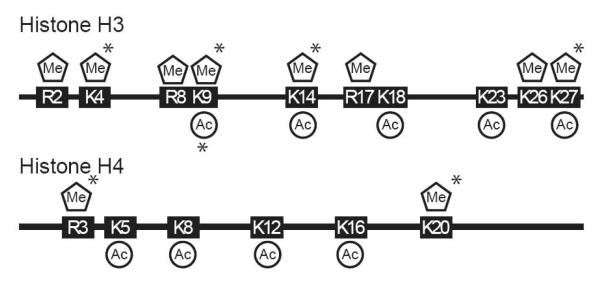
The addition and removal of histone modifications is an enzymatically driven process. Acetylation of histones H3 and H4 occur primarily at lysines located within histone tails. This process is catalyzed by histone acetyltransferases (HATs) that utilize acetyl-CoA as a cofactor [62, 63]. Several histone acetyltransferases exist in mammalian cells. Recruitment of these HATs to promoters is generally associated with activated transcription. Histone acetylation occurs at many lysine residues in both histones H3 and H4 (Fig. 4). An example of this is acetylation of histone H3 lysine 9 (H3-K9). Genes that are transcriptionally active usually have an abundance of H3-K9 acetylated histones at their regulatory regions. Unlike histone acetylation, histone methylation can activate or inhibit transcription depending on where the modification occurs. The methylation of histones is carried out by a large family of histone methyltransferases (HMTs). Members of HMT family can methylate lysine and arginines in histone tails (Fig. 4). Just like DNMTs, HMTs utilize SAM as a cofactor to methylate their target amino acids and produce SAH as a byproduct. The activity and substrate specificity of HMTs is centralized in their SET domains and other surrounding motifs [64]. Histone methylation by specific HMTs generates an increased complexity of epigenetic control of gene expression through the modification of different amino acids in histone tails [65]. An example of this is the dichotomy existing in the methylation histone H3. Methylation of lysine four (Me-H3K4) is usually associated with active transcription, while lysine 9 (Me-H3K9) methylation exhibits and inhibitory affect. Modification of each lysine within histone H3 can block the modification of the other [66]. The di-methylation of H3K9 by SUV39H1 can lead to the formation of heterochromatin by generating a substrate for heterochromatin protein 1 (HP-1) binding [67]. Once HP-1 binding has occurred, it can recruit additional HMTs through its chromoshadow domain to facilitate the spread of heterochromatin across a gene [68].
Fig. 4.
Location of acetylated (Ac) and methylated (Me) lysines (K) and methylated arginines (R) within the N-terminal histone tails of histones H3 and H4. (*) denotes locations of mono, di, or tri acetylation and methylation.
Acetylation and methylation of histone tails are not permanent modifications. Histone deacetylases (HDACs) and histone demethylases remove acetylation and methylation from histones providing a plasticity to the epigenetic control of gene expression [69, 70]. The presence of HDACs within gene regulatory regions is consistent with epigenetic gene silencing. By removing acetyl groups form histones an epigenetically silenced state can be maintained. HDACs and histone demethylases can also work in conjunction with HATs and HMTs to silence gene expression. For example, HDACs can remove acetyl groups from H3-K9 to allow for its methylation by HMTs to initialize the formation of heterochromatin [71].
Histone modifications in development
Histone modifications are also involved in regulating gene expression during development. The pattern of histone methylation changes during development and is indicative of its role in regulating gene expression. Tri-methylation of H3K9, and monomethylation of H4K20 change dynamically during neural tube development in mice [5]. Histone methylation and chromatin remodeling machinery also play a vital role during imprinting. As we have discussed above DNA methylation is one mechanism by which imprinting takes place; however, histone methylation can also imprint genes independent of DNA methylation. Imprinting center 2 and Kcnq1 are both imprinted in mouse placenta by increased di-methylated H3K9 and tri-methylated H3K27 [72, 73].
Cooperation between epigenetic processes
DNA methylation and histone modifications work in a cooperative manner to regulate gene expression. The role of each in initializing epigenetic control of gene activity has received much attention; however both are commonly found working in concert to control gene expression in mammals. Early studies demonstrated the common occurrence of hypermethylated DNA with condensed transcriptionally inactive heterochromatin [74, 75]. At this time it is uncertain which process begets the other, but it is widely accepted that DNA methylation and histone modifications can initialize each other. The factor that connects these two processes is Methyl-CpG binding domain proteins (MeCP). These proteins bind methylated CpG di-nucleotides and assist in gene silencing by recruiting HDACs and HMTs to initialize the formation of heterochromatin at genes [75, 76]. DNA methylation and histone modifications can also be linked independent of MeCPs. DNA methyltransferases can recruit chromatin remodeling machinery to initialize epigenetic repression of gene expression [77, 78]. Through these protein/protein interactions DNA methylation and histone modifications are able to fashion the building blocks required for the formation heterochromatin and gene silencing.
Metabolic control of epigenetic mechanisms
In the past five years a push towards understanding the influence of diet on epigenetic processes has revealed that metabolism can influence gene expression. These studies have centered around two metabolites involved in one carbon metabolism, folate and SAM. Both cofactors are essential to maintaining epigenetic control of gene expression by influencing the methylation of DNA and histones. Folate and SAM are also biochemically linked by the interconnection between the folate and methionine cycles (Fig. 5). In mammalian cells dietary folate is the primary dietary donor to one carbon metabolism. Folate is not directly used in cells for methylation reactions. Prior to its use it is converted to tetrahydrofolate. Inhibiting dihydrofolate reductase with methotrexate blocks the conversion of deoxyuridylic acid to thymidylic acid [79, 80]. However, methotrexate also influences the production of SAM [81]. From this a direct link can be made between one carbon metabolism and epigenetic control by DNA and histone methylation. The overlying concept is that if folate status is interrupted cells will no longer be able to maintain epigenetic control of gene expression. This is supported by an increasing body of work outlining the impact of folate on development and the relationship it has with DNA methylation [82-84]. Recently work from Lorraine Young’s laboratory has shown that influencing the folate cycle of embryonic stem cells affects the function of epigenetic mechanisms in pre-implanted embryos [85]. Other studies from Randy Jirtle’s group have shown that supplementing diet of pregnant mice with metabolites that influence one-carbon metabolism impact phenotype of their unborn offspring. In these pioneering studies genetically identical offspring have different coat colors by changing the level of DNA methylation at the agouti locus [86-88]. Mutations in genes that metabolize folate to prior to the synthesis of SAM also globally disrupt genomic DNA methylation [82]. These studies illustrate the importance of maintaining the levels of these metabolites to generate differential epigenotypes during development. By what means could free radicals change the level of these metabolites to affect epigenetics processes during development?
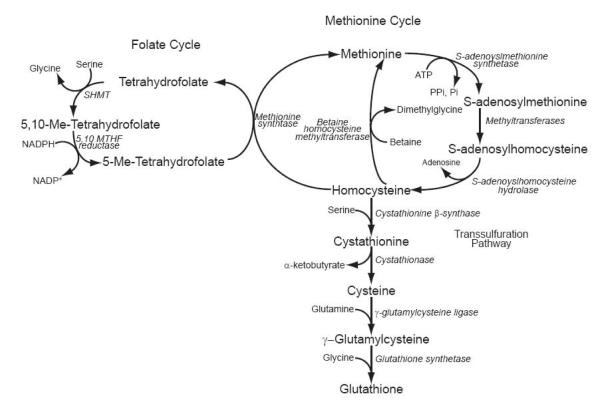
Fig. 5.
Schematic representation of the intermediary biochemistry of one-carbon metabolism. Depicted are the connections between the folate cycle, methionine cycle, transsulfuration pathway, and glutathione synthesis. and the enzymes (shown in italics) that connect them.
The production of glutathione (GSH) is biochemically linked to cofactors influencing epigenetic processes. We have discussed above the relationship between GSH synthesis and development. How can GSH synthesis be linked to epigenetic changes during development? Altering the production of glutathione directly impacts cellular sulfur pools. One such inlet for sulfur into glutathione biosynthesis is via the flux of homocysteine through the transsulfuration pathway (Fig. 5) [89]. Homocysteine is derived from the hydrolysis of SAH, the byproduct of DNA and histone methyltransferases utilizing SAM, and stands at the critical crossroads between the methionine cycle and transsulfuration pathway. The most common route for homocysteine is to remain in the methionine cycle where it is converted back into methionine by methionine synthase or betaine homocysteine methyltransferase. Methionine can then be used to resynthesize SAM for future transmethylation reactions. Under these conditions the amount of homocysteine entering the transsulfuration pathway is limited. However, when increased glutathione production is required during times of oxidative stress, or in the case of development, homocysteine’s entry into the transsulfuration pathway would be favored. This can mostly be attributed to increased activity of cystathione β-synthase, the enzyme located at entry point of homocysteine into the transsulfuration pathway [90]. In some tissues upwards of 50% of their GSH is produced via the transsulfuration pathway [91]. During development the activity and expression of GSH synthesizing enzymes dynamically changes [12]. When these changes in gene expression are coupled to increased demands on sulfur pools the methionine cycle would be affected. Such a case would lead to a migration of metabolites away from the methionine cycle into the transsulfuration pathway for the eventual production of GSH. By favoring homocysteine’s entry into the transsulfuration pathway the levels of methionine and SAM would become decreased. A more direct relationship between increased glutathione production and epigenetic processes is seen when increased GSH production is required after GSH depletion. Depleting GSH decreases the level of SAM in cells and leads to genome wide DNA hypomethylation [89, 92, 93]. Taken together these studies demonstrate that altering the level or synthesis of GSH in cells can directly impact DNA methylation by altering SAM pools. While these studies have focused on DNA methylation, we reason that a similar effect would be seen for histone methylation. Therefore, we can speculate that changing GSH production can influence the forming epigenotype of developing tissues by impacting both DNA and histone methylation. A linkage between GSH production and DNA methylation is best illustrated in gametogenesis and embryogenesis. Glutathione synthesis changes during these stages of development to prevent reactive oxygen species (ROS) induced damage [94]. Is it possible that increasing GSH synthesis might also be influencing epigenetic processes? The global decrease in genomic methylation in gametes and embryos is inversely correlated with their increased production of GSH [37, 94]. Once a blastocyst has formed, GSH synthesis tapers off and occurs in conjunction with increased genomic DNA methylation [14-16]. Therefore, it is tantalizing to speculate that GSH synthesis during gametogenesis and embryogenesis is mechanistically linked to epigenetic control of gender and cell type specific gene expression.
As we have discussed above, supplementing the diet of pregnant mice with metabolites that influence one-carbon metabolism impacts the phenotype of their unborn offspring. Thus far almost all of the metabolites tested in this system enhance the flux of metabolites through the methionine cycle to increase the production of SAM. Conversely, as we propose here, oxidative stress would decrease this flux by shunting metabolites towards GSH synthesis. Thus, the migration of metabolites away from the methionine cycle and into the transsulfuration pathway would influence the epigenotype of gametes and developing embryos by creating a deficit of SAM. This deficit of SAM might be accompanied by decreased de novo DNA methylation by DNMT3a and DNMT3b. This would in turn influence the epigenetic imprinting of genes and cell type specific gene expression. It would be of great interest to analyze how SAM content is affected by the over expression of GCLc and GCLm. Such a system already exists for these studies. The aforementioned Drosophila lines that over express GCLc and GCLm would be an excellent model to study an association among GSH synthesis, SAM levels, and epigenetic processes during development. Drosophila doesn’t utilize DNA methylation as a process to control gene expression, but their use of histone methylation and acetylation during morphogenesis is well characterized. Establishing such a link would open new avenues to explore the role of redox status in controlling the development of higher organisms.
Redox regulation of SAM synthetase
The redox state of a cell can also afflict the level of SAM by changing the activity of SAM synthetase, the enzyme producing SAM. SAM synthetase, also known as methionine adenosyltransferase (MAT), catalyzes the enzymatic addition of methionine to adenosine to produce SAM. In mammals three forms of MAT are known to exist: MATI, MATII and MATIII. Both MATI and MATIII are encoded by the gene mat1α and are predominantly expressed in the liver [95]. The only apparent difference between these two isoforms of MAT is the multimeric complexes they form. MATI functions as a homotetramer, while MATIII a homodimer [96]. MATII is the predominant form of SAM synthetase found in all other tissues and is encoded by the gene mat2α. MATII exists in a multimeric complex containing one enzymatic α subunit and two regulatory ß subunits encoded by the mat2ß gene [97]. Redox influences MAT activity by influencing its tertiary structure and activity [98]. Altering the redox status by inhibiting the synthesis of GSH with BSO reduces MAT activity and the level of SAM in hepatocytes. However, if animals are pretreated with exogenous SAM, the inhibition of MAT activity is lifted [99]. This influence of SAM on MAT activity is most likely not a direct effect but attributable to downstream metabolites formed after its use in transmethylation reactions [100]. These observations reveal an interesting link between SAM levels, MAT activity, and redox biology. The redox buffering capacity in cells also influences MAT activity. Increasing the level of GSH enhances the activity of purified MATI and MATIII [101]. The ratio of GSH/GSSG also influences MAT activity. High GSH/GSSG ratios maintain the enzyme in a reduced state and allow it to achieve its maximum activity, however at ratios below 10:1 MAT activity becomes markedly decreased [101]. This fundamental change in MAT activity can be attributed to the oxidation state of particular cysteine within MATI and MATIII. Pajares and others have identified cysteine 150 (C150) in MAT I and III as a critical cysteine redox switch controlling their MAT activity. Oxidation of C150 in MATI and III decreases enzyme activity and is believed to be a contributing factor in the development of liver cirrhosis [102, 103]. Nitric oxide (NO?) also affects the activity of MAT through S-nitrosylation of C150 [104]. These studies have focused primary on MATI and III, however, C150 is conserved in MATII and therefore we speculate that redox modifications should influence its activity as well. Taken together these findings suggest that MAT activity would be altered by changes in glutathione production in tissues as they develop. The GSH/GSSG ratio changes seen in developing organisms would directly influence MAT activity, and in turn influence the epigenotype of these tissues by altering the level of SAM as we have described above. Nitric oxide can also elicit developmental changes in cells [105, 106]. Changes in SAM content by the S-nitrosylation of MAT could be one mechanism by which NO• could exert its influence on development. From the points above it would be of great interest to study how the activities of the SAM synthetases influence gametogenesis and development. For example, diminishing MAT activity by either pharmacological or molecular means would be expected to alter the epigenetic program of developing cells by decreasing the global levels of DNA and histone methylation. These changes would alter gene imprinting and cell type specific gene expression, both of which are controlled by DNA methylation, and would most likely result in developmental failure.
The influence of oxygen on development
Oxygen gradients in developing tissues act as a morphogen to determine the differentiation pattern of cells. Several studies have dealt with how oxygen concentration influences development. Developing Drosophila larvae are sensitive to high oxygen concentrations, and have an altered eye shape and color [107]. Others have noted that oxygen concentration can influence the differentiation of cells forming human tissues [108-110]. We can generally attribute differentiation to alterations in gene activity. The concentration of oxygen in cells can influence the production of ROS such as O2•− and H2O2. The level of SOD, CAT and GPx activity in cells changes dynamically during development [7]. According to the free radical theory of development it is the balance in production and removal of ROS that influences development. Through this theoretical construct we can connect oxygen concentration to development by an altered redox state. This classical influence of oxygen level on development would be directly related to our discussion above regarding how an oxidative state influences the link between epigenetic cofactors and GSH production. However, since the initial rendering of the free radical theory of development we know that cells have mechanisms capable of directly perceiving oxygen that influence gene expression. By what means do cells directly perceive the level of oxygen they are receiving and transmit this into altered gene activity during development? Two mechanisms exist that could potentially facilitate the transmission of oxygen levels in gene expression: the hypoxia inducible factor (HIF) family of transcription factors, and the histone modifying enzymes. We will discuss each below and how they potentially influence gene expression during development.
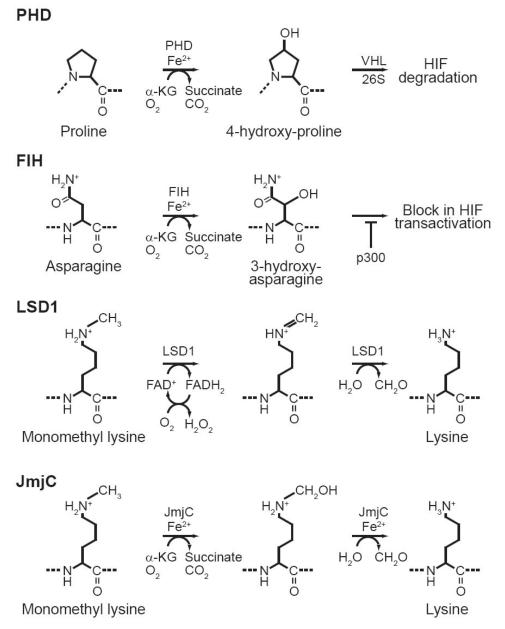
Many hypoxic genes are targets for regulation by the HIF family of transcription factors. This family of transcription factors contains three members, HIF-1a, HIF-2a, and HIF-3a, all of which form hetorodimers with ARNT to bind hypoxia response elements within gene promoters (For review see [111]). HIF stability and transactivation is negatively regulated by the hydroxylation of specific proline and asparagine residues within its sequence. The first regulatory mechanism is based on the hydroxylation of prolines within HIF by prolyl hydroxylates (PHDs). The transcription of HIF is generally constant within cells. Hydroxylation of prolines within HIFs controls their stability to regulate them in a post-translational manner. Once HIF is hydroxylated it becomes a target for the E3 ubiquitin ligase Von Hippel-Lindau (VHL), which marks HIF for degradation by the 26S proteasome. HIF mediated transcription is further negatively regulated by the asparaginyl hydroxylase FIH (factor inhibiting HIF). This hydroxylation event prevents the interaction of HIF with transcriptional coactivator p300, leaving HIF unable to activate gene expression. The enzyme activities of PHDs and FIH both require oxygen, a-ketoglutarate, Fe2+ and ascorbate as cofactors (Fig. 6). Under low oxygen concentrations HIF is released from the negative regulation of PHDs and FIH. Since HIFs are not hydroxylated in low oxygen by PHDs and FIH they are no longer degraded by the 26S proteasome and can interact with their transcriptional co-activators. The oxygen concentration gradient that exists across developing tissues would directly influence the activity of both PHDs and FIH (Fig. 7). This means the transcriptional activity of HIFs would be inversely related to the concentration of oxygen received in developing tissue. Moreover low oxygen concentration would allow for the increased transcription of HIF responsive genes. Several target genes contain hypoxic responsive elements (HREs) within their promoters. Some of the classical examples we will discuss in this review are vasculature endothelial growth factor (VEGF), and the VEGF receptors Flt1 and Flk1 [112]. Some hypoxia responsive genes are preferentially targeted by one HIF family member over another. Therefore, the three HIF family members do not demonstrate redundancy in function [112, 113].
Fig 6.
The reactions catalyzed by proline hydroxylases (PHD), factor inhibiting HIF (FIH), lysine specific demethylase 1 (LSD1), and Jumonji C domain containing proteins (JmjC). All these reactions require oxygen as a cofactor, therefore their activities during development are expected to be regulated by oxygen gradients
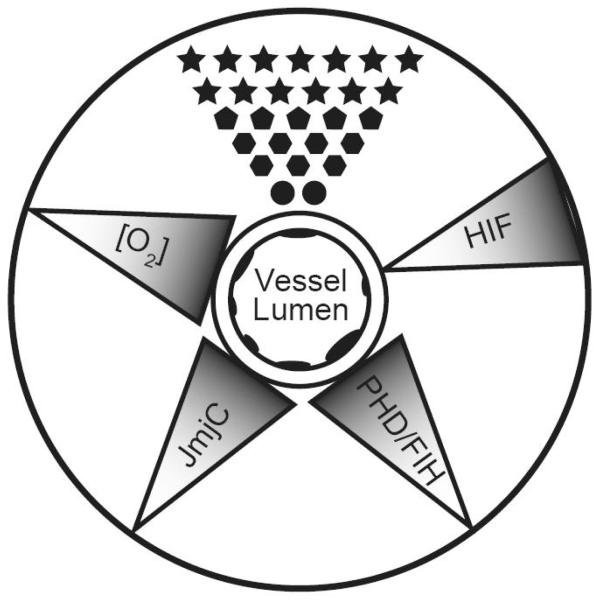
Fig 7.
Oxygen concentration gradient ([O2]) influences the phenotype of differentiating cells (represented by different shaped polygons) by changing the activity of oxygen sensing mechanisms that control gene expression. In developing tissues oxygen concentration in differentiation cells decreases as the distance from a vessel increases. This leads to decreases in the activities of proline hydroxylases (PHD), factor inhibiting HIF (FIH), and jumonji demethylases (JmjC), leading to activation of transcription of hypoxic response genes through hypoxia inducible factors (HIF) and chromatin remodeling. Shading of triangles represents the gradient of O2 concentration or enzyme activity.
Oxygen might also play a critical role in setting up and maintaining the epigenotype of cells. Recently two new classes of histone demethylating enzymes that utilize oxygen have been discovered. Two members of this emerging class of enzymes include lysine specific demethylase 1 (LSD1) and the jumonji C (JmjC) family of histone demethylases [114, 115]. LSD1 was initially identified as a member of several transcriptional repression complexes including CtBP, CoREST, and NRD [116-118]; Further characterization of LSD1 by Yang Shi’s group has demonstrated that it represses transcription by demethylating lysine 4 of histone H3 (H3-K4), a post-translational modification associated with active transcription [114]. The ability of LSD1 to perceive oxygen during development can be related to its flavin dependent amine oxidase activity. Histone demethylation by LSD1 is believed to utilize oxygen as an electron acceptor to reduce methylated lysine to form lysine, formaldehyde and hydrogen peroxide (Fig. 6) [114]. The second class of histone demethylases is jumonji C (JmjC), a large family of proteins that continues to grow [115]. Each has a preferred lysine it targets for demethylation. The JmjC histone demethylases function similar to PHDs and FIH; they utilize oxygen, a-ketoglutarate, Fe2+ and ascorbate to directly hydroxylate mono, di or tri methyl lysines in histones [115, 119]. After hydroxylation formaldehyde is liberated, resulting in unmethylated lysine (Fig. 6). Since LSD1 and JmjC histone demethlyases both require oxygen, the concentration of oxygen would directly influence their activities and the epigenotype of developing tissues. Connecting oxygen levels to the epigenotype of developing tissues is potentially astounding. The oxygen gradient induced change in epigenotype could drive development by directing cell type specific gene expression. Furthermore, selectively demethylating specific histones allows for the dynamic control of gene expression that is essential during development. Altering histone methylation is also a means by which the level of oxygen in cells can influence DNA methylation. As we have discussed above histone methylation and DNA methylation are closely linked. Because of this DNA methylation in developing organisms would be indirectly influenced by oxygen.
The role of oxygen gradients in modulating HIF and chromatin remodeling machinery is perhaps best explained in the context of the development of highly vascularized organs. Proteins like VEGF and its receptors Flt1 and Flk1 are essential to signaling the formation of new blood vessels [112]. The decreased level of oxygen drives vasculogenesis in embryos by increasing HIF mediated transcription of VEGF and VEGF receptors [120]. Because of this, vascular organs such as the kidney, lung, and brain develop in low oxygen environments [112]. At the same time the activity of jumonji proteins would also be affected. Jumonji family members seem to have an emerging role in development (for review see [121]). Truncation of the jumonji family member PHF8 leads to diminished mental capacity and the formation of cleft lip and palate in humans [122]. Another member of the jumonji family is RBP2 and was recently characterized to specifically demethylate di and tri-methyl K4 of histone H3. This same study also outlined the role of RBP2 in controlling the expression of the HOX gene cluster during mouse development by changing the level of tri-methylated histones located at the HOX cluster [123]. These studies outline the importance of maintaining the activity of these enzymes during development. At this time it is believed that different members of the JmjC family have specific genes on which they remove histone methylation. Therefore changing the concentration of oxygen in systems would have dramatic effects on their activity, and alter the expression of their target genes. Furthermore, these mechanisms could work in concerted process to activate or repress gene expression as the concentration of oxygen changes. For example, under times of high oxygen genes with HREs would be silenced by jumonji family members actively demethyalting di-methylated K4H3 nucleosomes at their promoters. When the level of oxygen becomes decreased, the activity of jumonji proteins, PHDs and FIH would become decreased, resulting in the stabilization of HIF and the effective transcriptional activation of such hypoxia responsive genes. This dynamic control of gene expression and its influence on cell phenotype is outlined in Figure 7. This model suggests that oxygen concentration would dynamically regulate gene expression by HIF and histone demethylases. If oxygen concentration is playing a role to dynamically regulate gene expression during development, then blocking the activity of these enzymes would alter the differentiation program of cells. Genes that are negatively regulated by jumonji demethylases would be actively transcribed and have increased methylation of histones at their promoters. This would leave these cells either unable to differentiate, or favor becoming one cell type specific lineage differentiation.
An oxygen gradient also influences the manner in which cells produce energy. In low oxygen environments cells would rely more on glycolysis rather than oxidative respiration to generate ATP. How can the manner in which cells produce energy be related to epigenetic control of gene expression? Such a link to metabolism and development can be made through the sirtuins family of protein deacetylases. Sirtuins are a class of NAD+ dependent protein deacetylases that are commonly known for their role in increasing lifespan of yeast, and C. elegans during caloric restriction (CR) [124]. Thus far seven members of this family have been identified in humans: SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6 and SIRT7. Each family member has the same NAD+ dependent activity, but differ in their placements and functions in cells during CR (for review see [125]). It is believed that one manner by which CR activates sirtuin activity is by increasing the NAD+/NADH ratio. With increased NAD+ sirtuins can more readily deacetylate transcription factors to increase the activity of some genes, and repress the expression of other genes by deacetylating histones [126, 127]. In the context of this review we will focus on the HDAC activity of sirtuins as it relates to development. SIRT1 is the human homologue of the yeast histone deacetylase SIR2 [128]. The role of SIRT1 in mouse development has been characterized. SIRT1 knockout mice survive until birth, but have decreased body mass and most die soon after. The SIRT1 null animals that do survive to adulthood are sterile [129]. Enhancing SIRT1 activity by resveratrol causes mesenchymal stem cells to preferentially differentiate into osteoblasts [130]. By contrast, abrogating SIRT1 function in conditional knockout mice disrupts the formation of mammary glands and renders them unable to lactate [131]. The authors attributed this phenotypic change to estrogen and insulin-like growth factor 1 signaling in mammary tissue. However, they did not determine what role histone deacetylation by SIRT1 might be playing during mammary development. The linkage between metabolism and sirtuin activity is best illustrated in muscle development. Work from Vittorio Sartorelli’s group has shown that the NAD+/NADH ratio decreases as muscle cell differentiate. This decreased NAD+/NADH ratio is accompanied by increased expression of muscle genes. However, when the NAD+/NADH ratio is maintained at a high level muscle differentiation does not occur and can be attributed to differential histone acetylation [132]. The increased expression of muscle specific genes is attributable to histone hyperacetylation after sirtuin deacetylation activity is lost because NAD+ levels are too low. These studies illustrate the role of sirtuins during development. We speculate that as oxygen content changes in developing tissues, so will the activity of sirtuins. As organisms develop, the manner in which they produce energy is dependent upon the level of oxygen their developing tissues receive. For example, poorly oxygenated tissues would rely more heavily on glycolysis. High glycolytic activity is associated with a low NAD+/NADH ratio, and therefore the histone deacetylase activity of some sirtuins would be minimal during these conditions. However, once these same tissues become well oxygenated, a switch to oxidative phosphorylation would occur and increase the NAD+/NADH ratio. This condition would favor the deacetylase activity of sirtuins in these developing tissues and thus silence the expression of sirtuin target genes.
Summary
Epigenetic processes are a major force controlling gene expression during development. We have outlined here a potential role for glutathione production and O2 concentration in controlling gene expression by shaping the epigenotype of developing cells. This role relies upon the altered availability of cofactors such as SAM and O2, which power the enzymes responsible for initiating and perpetuating epigenetic control of gene expression. The dynamic changes in oxygen concentration and redox state can be directly related to Waddington’s epigenetic canalization of developing cells by creating what he called “a bias corresponding to the particular initial conditions in some part of the newly fertilised egg” [20] (Fig. 1). When it was first postulated that oxygen, ROS, and GSH production influence gene expression during development it was aptly hypothesized that they alter chromatin structure [2, 4]. Epigenetic control of gene expression through DNA methylation and histone modifications directly influences chromatin structure [74, 76]. Thus, free radicals and oxygen influencing cofactor availability to DNA and histone methyltransferases during development provides a more direct link to gene expression changes during development. Such a linkage between these epigenetic cofactors and the redox state of cells further substantiates the free radical theory of development by prescribing novel morphogenic properties to free radicals and oxygen [1-4]. This epigenetic perspective on the free radical theory of development opens new avenues of research to understand the role of epigenetics in development.
Acknowledgements
This work was supported by NIH R01 CA73612 to FED. MJH received salary support from T32 CA078585.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Sohal RS, Allen RG. Relationship between metabolic rate, free radicals, differentiation and aging: a unified theory. In: Woodhead AD, Blackett AD, Hollander A, editors. Molecular biology of aging, Basic Life Sciences. volume 35. Plenum Press; New York: 1985. pp. 75–104. [DOI] [PubMed] [Google Scholar]
- [2].Sohal RS, Allen RG, Nations C. Oxygen free radicals play a role in cellular differentiation: an hypothesis. J Free Radic Biol Med. 1986;2:175–181. doi: 10.1016/s0748-5514(86)80067-8. [DOI] [PubMed] [Google Scholar]
- [3].Sohal RS, Allen RG. Relationship between Oxygen-Metabolism, Aging and Development. Advances in Free Radical Biology and Medicine. 1986;2:117–160. [Google Scholar]
- [4].Allen RG, Balin AK. Oxidative influence on development and differentiation: an overview of a free radical theory of development. Free Radic Biol Med. 1989;6:631–661. doi: 10.1016/0891-5849(89)90071-3. [DOI] [PubMed] [Google Scholar]
- [5].Biron VL, McManus KJ, Hu N, Hendzel MJ, Underhill DA. Distinct dynamics and distribution of histone methyl-lysine derivatives in mouse development. Dev Biol. 2004;276:337–351. doi: 10.1016/j.ydbio.2004.08.038. [DOI] [PubMed] [Google Scholar]
- [6].Mavelli I, Autuori F, Dini L, Spinedi A, Ciriolo MR, Rotilio G. Correlation between superoxide dismutase, glutathione peroxidase and catalase in isolated rat hepatocytes during fetal development. Biochem Biophys Res Commun. 1981;102:911–916. doi: 10.1016/0006-291x(81)91624-7. [DOI] [PubMed] [Google Scholar]
- [7].Mavelli I, Rigo A, Federico R, Ciriolo MR, Rotilio G. Superoxide dismutase, glutathione peroxidase and catalase in developing rat brain. Biochem J. 1982;204:535–540. doi: 10.1042/bj2040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, Wallace DC, Epstein CJ. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- [9].Tsan MF, White JE, Caska B, Epstein CJ, Lee CY. Susceptibility of heterozygous MnSOD gene-knockout mice to oxygen toxicity. Am J Respir Cell Mol Biol. 1998;19:114–120. doi: 10.1165/ajrcmb.19.1.3066. [DOI] [PubMed] [Google Scholar]
- [10].Takahashi S, Zeydel M. gamma-Glutamyl transpeptidase and glutathione in aging IMR-90 fibroblasts and in differentiating 3T3 L1 preadipocytes. Arch Biochem Biophys. 1982;214:260–267. doi: 10.1016/0003-9861(82)90029-7. [DOI] [PubMed] [Google Scholar]
- [11].Warshaw JB, Wilson CW, 3rd, Saito K, Prough RA. The responses of glutathione and antioxidant enzymes to hyperoxia in developing lung. Pediatr Res. 1985;19:819–823. doi: 10.1203/00006450-198508000-00008. [DOI] [PubMed] [Google Scholar]
- [12].Stover SK, Gushansky GA, Salmen JJ, Gardiner CS. Regulation of gamma-glutamate-cysteine ligase expression by oxidative stress in the mouse preimplantation embryo. Toxicol Appl Pharmacol. 2000;168:153–159. doi: 10.1006/taap.2000.9030. [DOI] [PubMed] [Google Scholar]
- [13].Orr WC, Radyuk SN, Prabhudesai L, Toroser D, Benes JJ, Luchak JM, Mockett RJ, Rebrin I, Hubbard JG, Sohal RS. Overexpression of glutamate-cysteine ligase extends life span in Drosophila melanogaster. J Biol Chem. 2005;280:37331–37338. doi: 10.1074/jbc.M508272200. [DOI] [PubMed] [Google Scholar]
- [14].Brad AM, Bormann CL, Swain JE, Durkin RE, Johnson AE, Clifford AL, Krisher RL. Glutathione and adenosine triphosphate content of in vivo and in vitro matured porcine oocytes. Mol Reprod Dev. 2003;64:492–498. doi: 10.1002/mrd.10254. [DOI] [PubMed] [Google Scholar]
- [15].Luciano AM, Goudet G, Perazzoli F, Lahuec C, Gerard N. Glutathione content and glutathione peroxidase expression in in vivo and in vitro matured equine oocytes. Mol Reprod Dev. 2006;73:658–666. doi: 10.1002/mrd.20469. [DOI] [PubMed] [Google Scholar]
- [16].Perreault SD, Barbee RR, Slott VL. Importance of glutathione in the acquisition and maintenance of sperm nuclear decondensing activity in maturing hamster oocytes. Dev Biol. 1988;125:181–186. doi: 10.1016/0012-1606(88)90070-x. [DOI] [PubMed] [Google Scholar]
- [17].Oberley LW, Buettner GR. Role of superoxide dismutase in cancer: a review. Cancer Res. 1979;39:1141–1149. [PubMed] [Google Scholar]
- [18].Oberley LW, Oberley TD, Buettner GR. Cell division in normal and transformed cells: the possible role of superoxide and hydrogen peroxide. Med Hypotheses. 1981;7:21–42. doi: 10.1016/0306-9877(81)90018-9. [DOI] [PubMed] [Google Scholar]
- [19].Waddington CH. The epigenetics of birds. University Press; Cambridge [Eng.]: 1952. [Google Scholar]
- [20].Waddington CH. Principles of embryology. Macmillan; New York: 1956. [Google Scholar]
- [21].Holliday R. Epigenetics: A Historical Overview. Epigenetics. 2006;1:76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- [22].Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- [23].Browne MJ, Turnbull JF, McKay EL, Adams RL, Burdon RH. The sequence specificity of a mammalian DNA methylase. Nucleic Acids Res. 1977;4:1039–1045. doi: 10.1093/nar/4.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Russell GJ, Walker PM, Elton RA, Subak-Sharpe JH. Doublet frequency analysis of fractionated vertebrate nuclear DNA. J Mol Biol. 1976;108:1–23. doi: 10.1016/s0022-2836(76)80090-3. [DOI] [PubMed] [Google Scholar]
- [25].Venter JC, Adams MD, Myers EW, Li PW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- [26].Bacolla A, Pradhan S, Larson JE, Roberts RJ, Wells RD. Recombinant human DNA (cytosine-5) methyltransferase. III. Allosteric control, reaction order, and influence of plasmid topology and triplet repeat length on methylation of the fragile X CGG.CCG sequence. J Biol Chem. 2001;276:18605–18613. doi: 10.1074/jbc.M100404200. [DOI] [PubMed] [Google Scholar]
- [27].Kautiainen TL, Jones PA. DNA methylation in mammalian nuclei. Biochemistry. 1985;24:5575–5581. doi: 10.1021/bi00341a043. [DOI] [PubMed] [Google Scholar]
- [28].Bolden AH, Nalin CM, Ward CA, Poonian MS, Weissbach A. Primary DNA sequence determines sites of maintenance and de novo methylation by mammalian DNA methyltransferases. Mol Cell Biol. 1986;6:1135–1140. doi: 10.1128/mcb.6.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Beaulieu N, Morin S, Chute IC, Robert MF, Nguyen H, MacLeod AR. An essential role for DNA methyltransferase DNMT3B in cancer cell survival. J Biol Chem. 2002;277:28176–28181. doi: 10.1074/jbc.M204734200. [DOI] [PubMed] [Google Scholar]
- [30].Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- [31].Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- [32].Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, Okumura K, Li E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- [33].Weisenberger DJ, Velicescu M, Preciado-Lopez MA, Gonzales FA, Tsai YC, Liang G, Jones PA. Identification and characterization of alternatively spliced variants of DNA methyltransferase 3a in mammalian cells. Gene. 2002;298:91–99. doi: 10.1016/s0378-1119(02)00976-9. [DOI] [PubMed] [Google Scholar]
- [34].Liu K, Wang YF, Cantemir C, Muller MT. Endogenous assays of DNA methyltransferases: Evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol Cell Biol. 2003;23:2709–2719. doi: 10.1128/MCB.23.8.2709-2719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- [36].Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- [37].Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- [38].Brandeis M, Kafri T, Ariel M, Chaillet JR, McCarrey J, Razin A, Cedar H. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. Embo J. 1993;12:3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kapoor A, Agius F, Zhu JK. Preventing transcriptional gene silencing by active DNA demethylation. FEBS Lett. 2005;579:5889–5898. doi: 10.1016/j.febslet.2005.08.039. [DOI] [PubMed] [Google Scholar]
- [40].Tada T, Tada M, Hilton K, Barton SC, Sado T, Takagi N, Surani MA. Epigenotype switching of imprintable loci in embryonic germ cells. Dev Genes Evol. 1998;207:551–561. doi: 10.1007/s004270050146. [DOI] [PubMed] [Google Scholar]
- [41].Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout WM, 3rd, Biniszkiewicz D, Yanagimachi R, Jaenisch R. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- [42].Christman JK, Price P, Pedrinan L, Acs G. Correlation between hypomethylation of DNA and expression of globin genes in Friend erythroleukemia cells. Eur J Biochem. 1977;81:53–61. doi: 10.1111/j.1432-1033.1977.tb11926.x. [DOI] [PubMed] [Google Scholar]
- [43].Creusot F, Acs G, Christman JK. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2′-deoxycytidine. J Biol Chem. 1982;257:2041–2048. [PubMed] [Google Scholar]
- [44].Gowher H, Jeltsch A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J Mol Biol. 2001;309:1201–1208. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- [45].Gowher H, Jeltsch A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J Biol Chem. 2002;277:20409–20414. doi: 10.1074/jbc.M202148200. [DOI] [PubMed] [Google Scholar]
- [46].Ehrlich M. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin Immunol. 2003;109:17–28. doi: 10.1016/s1521-6616(03)00201-8. [DOI] [PubMed] [Google Scholar]
- [47].Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- [49].Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 1995;9:2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- [50].Sheardown SA, Duthie SM, Johnston CM, Newall AE, Formstone EJ, Arkell RM, Nesterova TB, Alghisi GC, Rastan S, Brockdorff N. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell. 1997;91:99–107. doi: 10.1016/s0092-8674(01)80012-x. [DOI] [PubMed] [Google Scholar]
- [51].Panning B, Dausman J, Jaenisch R. X chromosome inactivation is mediated by Xist RNA stabilization. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- [52].Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, Domann FE. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet. 2002;31:175–179. doi: 10.1038/ng886. [DOI] [PubMed] [Google Scholar]
- [53].Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- [54].Phillips DM. The presence of acetyl groups of histones. Biochem J. 1963;87:258–263. doi: 10.1042/bj0870258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Murray K. The Occurrence Of Epsilon-N-Methyl Lysine In Histones. Biochemistry. 1964;3:10–15. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- [56].Allfrey VG, Faulkner R, Mirsky AE. Acetylation And Methylation Of Histones And Their Possible Role In The Regulation Of Rna Synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Pogo BG, Allfrey VG, Mirsky AE. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc Natl Acad Sci U S A. 1966;55:805–812. doi: 10.1073/pnas.55.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- [59].Prigent C, Dimitrov S. Phosphorylation of serine 10 in histone H3, what for? J Cell Sci. 2003;116:3677–3685. doi: 10.1242/jcs.00735. [DOI] [PubMed] [Google Scholar]
- [60].Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- [61].Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- [62].Racey LA, Byvoet P. Histone acetyltransferase in chromatin. Evidence for in vitro enzymatic transfer of acetate from acetyl-coenzyme A to histones. Exp Cell Res. 1971;64:366–370. doi: 10.1016/0014-4827(71)90089-9. [DOI] [PubMed] [Google Scholar]
- [63].Noland BJ, Hardin JM, Shepherd GR. Histone acetyltransferase activity in synchronized mammalian cells. Biochim Biophys Acta. 1971;246:263–268. doi: 10.1016/0005-2787(71)90136-5. [DOI] [PubMed] [Google Scholar]
- [64].Wilson JR, Jing C, Walker PA, Martin SR, Howell SA, Blackburn GM, Gamblin SJ, Xiao B. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell. 2002;111:105–115. doi: 10.1016/s0092-8674(02)00964-9. [DOI] [PubMed] [Google Scholar]
- [65].Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, Cheng X. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- [67].Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- [68].Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- [69].Libby PR. Activity of histone deacetylase in rat liver and Novikoff hepatoma. Biochim Biophys Acta. 1970;213:234–236. doi: 10.1016/0005-2787(70)90027-4. [DOI] [PubMed] [Google Scholar]
- [70].Hay CW, Candido EP. Histone deacetylase. Association with a nuclease resistant, high molecular weight fraction of HeLa cell chromatin. J Biol Chem. 1983;258:3726–3734. [PubMed] [Google Scholar]
- [71].Bartova E, Pachernik J, Harnicarova A, Kovarik A, Kovarikova M, Hofmanova J, Skalnikova M, Kozubek M, Kozubek S. Nuclear levels and patterns of histone H3 modification and HP1 proteins after inhibition of histone deacetylases. J Cell Sci. 2005;118:5035–5046. doi: 10.1242/jcs.02621. [DOI] [PubMed] [Google Scholar]
- [72].Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- [73].Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- [74].Antequera F, Macleod D, Bird AP. Specific protection of methylated CpGs in mammalian nuclei. Cell. 1989;58:509–517. doi: 10.1016/0092-8674(89)90431-5. [DOI] [PubMed] [Google Scholar]
- [75].Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- [76].Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- [77].Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- [78].Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Roberts D, Hall TC. The reduction of folate and of dihydrofolate by homogenates of leukocytes from patients with leukemia or with myeloid metaplasia. Cancer Res. 1967;27:994–999. [PubMed] [Google Scholar]
- [80].Jenh CH, Rao LG, Johnson LF. Regulation of thymidylate synthase enzyme synthesis in 5-fluorodeoxyuridine-resistant mouse fibroblasts during the transition from the resting to growing state. J Cell Physiol. 1985;122:149–154. doi: 10.1002/jcp.1041220122. [DOI] [PubMed] [Google Scholar]
- [81].Davis CD, Uthus EO. Dietary folate and selenium affect dimethylhydrazine-induced aberrant crypt formation, global DNA methylation and one-carbon metabolism in rats. J Nutr. 2003;133:2907–2914. doi: 10.1093/jn/133.9.2907. [DOI] [PubMed] [Google Scholar]
- [82].Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, Corrocher R, Selhub J. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Hoffman DR, Cornatzer WE, Duerre JA. Relationship between tissue levels of S-adenosylmethionine, S-adenylhomocysteine, and transmethylation reactions. Can J Biochem. 1979;57:56–65. doi: 10.1139/o79-007. [DOI] [PubMed] [Google Scholar]
- [84].Balaghi M, Wagner C. DNA methylation in folate deficiency: use of CpG methylase. Biochem Biophys Res Commun. 1993;193:1184–1190. doi: 10.1006/bbrc.1993.1750. [DOI] [PubMed] [Google Scholar]
- [85].Steele W, Allegrucci C, Singh R, Lucas E, Priddle H, Denning C, Sinclair K, Young L. Human embryonic stem cell methyl cycle enzyme expression: modelling epigenetic programming in assisted reproduction? Reprod Biomed Online. 2005;10:755–766. doi: 10.1016/s1472-6483(10)61120-0. [DOI] [PubMed] [Google Scholar]
- [86].Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- [87].Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Lertratanangkoon K, Savaraj N, Scimeca JM, Thomas ML. Glutathione depletion-induced thymidylate insufficiency for DNA repair synthesis. Biochem Biophys Res Commun. 1997;234:470–475. doi: 10.1006/bbrc.1997.6623. [DOI] [PubMed] [Google Scholar]
- [90].Prudova A, Bauman Z, Braun A, Vitvitsky V, Lu SC, Banerjee R. S-adenosylmethionine stabilizes cystathionine beta-synthase and modulates redox capacity. Proc Natl Acad Sci U S A. 2006;103:6489–6494. doi: 10.1073/pnas.0509531103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Mosharov E, Cranford MR, Banerjee R. The quantitatively important relationship between homocysteine metabolism and glutathione synthesis by the transsulfuration pathway and its regulation by redox changes. Biochemistry. 2000;39:13005–13011. doi: 10.1021/bi001088w. [DOI] [PubMed] [Google Scholar]
- [92].Lertratanangkoon K, Orkiszewski RS, Scimeca JM. Methyl-donor deficiency due to chemically induced glutathione depletion. Cancer Res. 1996;56:995–1005. [PubMed] [Google Scholar]
- [93].Lertratanangkoon K, Wu CJ, Savaraj N, Thomas ML. Alterations of DNA methylation by glutathione depletion. Cancer Lett. 1997;120:149–156. doi: 10.1016/s0304-3835(97)00300-5. [DOI] [PubMed] [Google Scholar]
- [94].Luberda Z. The role of glutathione in mammalian gametes. Reprod Biol. 2005;5:5–17. [PubMed] [Google Scholar]
- [95].Stipanuk MH. Sulfur amino acid metabolism: pathways for production and removal of homocysteine and cysteine. Annu Rev Nutr. 2004;24:539–577. doi: 10.1146/annurev.nutr.24.012003.132418. [DOI] [PubMed] [Google Scholar]
- [96].Cai J, Sun WM, Hwang JJ, Stain SC, Lu SC. Changes in S-adenosylmethionine synthetase in human liver cancer: molecular characterization and significance. Hepatology. 1996;24:1090–1097. doi: 10.1002/hep.510240519. [DOI] [PubMed] [Google Scholar]
- [97].Halim AB, LeGros L, Geller A, Kotb M. Expression and functional interaction of the catalytic and regulatory subunits of human methionine adenosyltransferase in mammalian cells. J Biol Chem. 1999;274:29720–29725. doi: 10.1074/jbc.274.42.29720. [DOI] [PubMed] [Google Scholar]
- [98].Pajares MA, Corrales F, Duran C, Mato JM, Alvarez L. How is rat liver S-adenosylmethionine synthetase regulated? FEBS Lett. 1992;309:1–4. doi: 10.1016/0014-5793(92)80726-w. [DOI] [PubMed] [Google Scholar]
- [99].Corrales F, Ochoa P, Rivas C, Martin-Lomas M, Mato JM, Pajares MA. Inhibition of glutathione synthesis in the liver leads to S-adenosyl-L-methionine synthetase reduction. Hepatology. 1991;14:528–533. [PubMed] [Google Scholar]
- [100].Corrales F, Gimenez A, Alvarez L, Caballeria J, Pajares MA, Andreu H, Pares A, Mato JM, Rodes J. S-adenosylmethionine treatment prevents carbon tetrachloride-induced S-adenosylmethionine synthetase inactivation and attenuates liver injury. Hepatology. 1992;16:1022–1027. doi: 10.1002/hep.1840160427. [DOI] [PubMed] [Google Scholar]
- [101].Pajares MA, Duran C, Corrales F, Pliego MM, Mato JM. Modulation of rat liver S-adenosylmethionine synthetase activity by glutathione. J Biol Chem. 1992;267:17598–17605. [PubMed] [Google Scholar]
- [102].Pajares MA, Corrales FJ, Ochoa P, Mato JM. The role of cysteine-150 in the structure and activity of rat liver S-adenosyl-L-methionine synthetase. Biochem J. 1991;274(Pt 1):225–229. doi: 10.1042/bj2740225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Mato JM, Alvarez L, Ortiz P, Mingorance J, Duran C, Pajares MA. S-adenosyl-L-methionine synthetase and methionine metabolism deficiencies in cirrhosis. Adv Exp Med Biol. 1994;368:113–117. doi: 10.1007/978-1-4615-1989-8_11. [DOI] [PubMed] [Google Scholar]
- [104].Ruiz F, Corrales FJ, Miqueo C, Mato JM. Nitric oxide inactivates rat hepatic methionine adenosyltransferase In vivo by S-nitrosylation. Hepatology. 1998;28:1051–1057. doi: 10.1002/hep.510280420. [DOI] [PubMed] [Google Scholar]
- [105].Magrinat G, Mason SN, Shami PJ, Weinberg JB. Nitric oxide modulation of human leukemia cell differentiation and gene expression. Blood. 1992;80:1880–1884. [PubMed] [Google Scholar]
- [106].Shami PJ, Moore JO, Gockerman JP, Hathorn JW, Misukonis MA, Weinberg JB. Nitric oxide modulation of the growth and differentiation of freshly isolated acute non-lymphocytic leukemia cells. Leuk Res. 1995;19:527–533. doi: 10.1016/0145-2126(95)00013-e. [DOI] [PubMed] [Google Scholar]
- [107].Nickla H, Anderson J, Palzkill T. Enzymes involved in oxygen detoxification during development of Drosophila melanogaster. Experientia. 1983;39:610–612. doi: 10.1007/BF01971122. [DOI] [PubMed] [Google Scholar]
- [108].Bassett CA, Herrmann I. Influence of oxygen concentration and mechanical factors on differentiation of connective tissues in vitro. Nature. 1961;190:460–461. doi: 10.1038/190460a0. [DOI] [PubMed] [Google Scholar]
- [109].Shaw JL, Bassett CA. The effects of varying oxygen concentrations on osteogenesis and embryonic cartilage in vitro. J Bone Joint Surg Am. 1967;49:73–80. [PubMed] [Google Scholar]
- [110].Erkell LJ. Differentiation of mouse neuroblastoma cells under increased oxygen tension. Exp Cell Biol. 1980;48:374–380. doi: 10.1159/000163002. [DOI] [PubMed] [Google Scholar]
- [111].Kaelin WG. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74:115–128. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- [112].Freeburg PB, Abrahamson DR. Hypoxia-inducible factors and kidney vascular development. J Am Soc Nephrol. 2003;14:2723–2730. doi: 10.1097/01.asn.0000092794.37534.01. [DOI] [PubMed] [Google Scholar]
- [113].Jain S, Maltepe E, Lu MM, Simon C, Bradfield CA. Expression of ARNT, ARNT2, HIF1 alpha, HIF2 alpha and Ah receptor mRNAs in the developing mouse. Mech Dev. 1998;73:117–123. doi: 10.1016/s0925-4773(98)00038-0. [DOI] [PubMed] [Google Scholar]
- [114].Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- [115].Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- [116].Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- [117].You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci U S A. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- [119].Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- [120].Lee YM, Jeong CH, Koo SY, Son MJ, Song HS, Bae SK, Raleigh JA, Chung HY, Yoo MA, Kim KW. Determination of hypoxic region by hypoxia marker in developing mouse embryos in vivo: a possible signal for vessel development. Dev Dyn. 2001;220:175–186. doi: 10.1002/1097-0177(20010201)220:2<175::AID-DVDY1101>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- [121].Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev Dyn. 2006;235:2449–2459. doi: 10.1002/dvdy.20851. [DOI] [PubMed] [Google Scholar]
- [122].Laumonnier F, Holbert S, Ronce N, Faravelli F, et al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J Med Genet. 2005;42:780–786. doi: 10.1136/jmg.2004.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- [124].Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- [125].Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- [126].Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- [127].Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- [128].Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- [129].McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Backesjo CM, Li Y, Lindgren U, Haldosen LA. Activation of Sirt1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. J Bone Miner Res. 2006;21:993–1002. doi: 10.1359/jbmr.060415. [DOI] [PubMed] [Google Scholar]
- [131].Li H, Rajendran GK, Liu N, Ware C, Rubin BP, Gu Y. SirT1 modulates the estrogen-insulin-like growth factor-1 signaling for postnatal development of mammary gland in mice. Breast Cancer Res. 2007;9:R1. doi: 10.1186/bcr1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]