Abstract
In this work we characterize the function of the flagellar protein FliL in Rhodobacter sphaeroides. Our results show that FliL is essential for motility in this bacterium and that in its absence flagellar rotation is highly impaired. A green fluorescent protein (GFP)-FliL fusion forms polar and lateral fluorescent foci that show different spatial dynamics. The presence of these foci is dependent on the expression of the flagellar genes controlled by the master regulator FleQ, suggesting that additional components of the flagellar regulon are required for the proper localization of GFP-FliL. Eight independent pseudorevertants were isolated from the fliL mutant strain. In each of these strains a single nucleotide change in motB was identified. The eight mutations affected only three residues located on the periplasmic side of MotB. Swimming of the suppressor mutants was not affected by the presence of the wild-type fliL allele. Pulldown and yeast two-hybrid assays showed that that the periplasmic domain of FliL is able to interact with itself but not with the periplasmic domain of MotB. From these results we propose that FliL could participate in the coupling of MotB with the flagellar rotor in an indirect fashion.
Bacterial flagellar motility is dependent on a complex motor that is embedded in the inner membrane (IM) (for reviews, see references 9, 37, 43, and 66). This motor can be divided into two functionally distinctive units: the rotor and the stator. The stator is a proton channel formed by MotA4-MotB2 that uses the proton motive force to provide the energy for flagellar rotation (12, 57, 64). The flagellar MotB protein has a short cytoplasmic N terminus, a single transmembrane (TM) helix, and a large periplasmic domain (16). It has been proposed that the single TM domain of MotB together with TM3 and TM4 of MotA forms a functional proton channel (13, 14). The invariant residue Asp32 in MotB is essential for rotation, and it has been shown that this residue is involved in proton conductance (15, 30, 73). In addition, mutations in this residue alter the conformation of MotA (36). From these results, it has been proposed that rounds of protonation and deprotonation of Asp32 produce conformational changes in the stator, which drives flagellar rotation (36, 68, 73). A maximum of 11 subunits of the MotA4-MotB2 complex are arranged around the flagellar rotor (11, 12, 54).
The rotor can be divided into three different regions: the basal body, the hook, and the filament (for a review, see reference 42). In Escherichia coli and Salmonella, the filament is formed by subunits of a single protein, FliC, which is assembled as a helical structure that drives cell movement when it rotates (3, 35, 45, 55, 60). The hook connects the filament with the rod that expands from the outer membrane through the peptidoglycan layer to the periplasmic space (18, 19, 39). The rod is assembled on the periplasmic-side surface of the MS ring (44) that is formed by 26 subunits of the FliF protein (32). FliF has two TM regions (22, 32, 63, 69), and it has been shown that the C terminus is located in the cytoplasm. This region of FliF interacts with FliG, which together with FliM and FliN forms the C ring that is involved in the control of flagellar rotation (20, 22, 34, 65) and in the stabilization of the export apparatus required for the formation of the extracytoplasmic structures of the flagellum (27, 38).
FliG is also involved in generating rotation of the flagellum through its interaction with the MotA subunits of the stator (25, 26, 65). Specifically, it has been shown that well-conserved charged residues located in the C-terminal domain of FliG (41) interact with charged residues of MotA that are located in the cytoplasmic loop between TM helices 2 and 3 (71). In E. coli, Sinorhizobium meliloti, and Vibrio alginolyticus, it has been demonstrated that electrostatic interactions between these residues promote and control flagellar rotation (4, 41, 70, 72).
Rhodobacter sphaeroides has a single subpolar flagellum that promotes swimming at velocities of up to 100 μm/s. In E. coli flagella rotate in a counterclockwise (CCW) direction when the cell is swimming and clockwise (CW) when the cell is reorienting (9). In contrast, the R. sphaeroides flagellum rotates only in the CW direction and reorientation occurs when the flagellum stops or reduces its speed and the filament helix relaxes into a short-wavelength, high-amplitude coil (1). It has been proposed that reorientation is achieved by Brownian motion and slow rotation of the coiled filament (1, 2). Under regular laboratory conditions, this is the only flagellum present. However, under particular conditions R. sphaeroides assembles a second functional flagellum (47). The above-mentioned features, and others reviewed elsewhere (1), make of this bacterium an interesting model to study the flagellar system.
Of the approximately 40 flagellar genes present in most flagellated bacteria, the functions of only a few still remain unknown (reviewed in reference 46); this is the case for the fliL gene. FliL is an integral membrane protein that was copurified with the flagellar basal body of Salmonella, and from this result it was proposed that FliL might be localized around the basal body close to the MotA-MotB complexes (58). In contrast to other flagellar genes, different phenotypes have been reported for fliL mutants in different bacteria. Although in E. coli and Salmonella enterica serovar Typhimurium a fliL mutant is not affected in its swimming ability (52), it was recently shown that in these species FliL is essential for swarming, and a role in strengthening the rod was proposed (5). In Proteus mirabilis FliL was involved in swarming differentiation in response to the density of the growth medium (8). The absence of fliL in Silicibacter TM1040 (7) and Caulobacter crescentus (31) elicits a Mot− phenotype (the mutant has a flagellum, but it does not rotate). In C. crescentus FliL was also implicated in the regulation of the stability of FliF (31), whereas in Pseudomonas putida the absence of fliL was associated with a Fla− phenotype (absence of flagella) (52). Even though these reports suggest a role for FliL in flagellar rotation and/or biogenesis, the molecular mechanisms that allow FliL to accomplish any of its functions have not been characterized.
In order to get a better insight into the molecular role of FliL in flagellar formation and/or functioning, in this work we studied the role of this protein in R. sphaeroides. We observed that FliL is essential for flagellar rotation in this bacterium but that its absence can be compensated for by secondary mutations in motB. We found that this protein is located not only at the base of the flagellum but also as a minor population of laterally located foci that show dynamic movement. The implications of these results are discussed.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
Plasmids and bacterial strains used in this work are listed in Table 1. R. sphaeroides WS8 (62) was grown at 30°C in Sistrom's minimal medium (61) under heterotrophic conditions. Swimming assays were carried out with bacteria grown in liquid medium or on swimming plates prepared with Sistrom's medium and 0.25% agar. When required, antibiotics were added at the following concentrations: 25 μg/ml kanamycin, 50 μg/ml spectinomycin, and 1 μg/ml tetracycline. E. coli was grown in LB at 37°C. Antibiotics for E. coli cultures were used at the following concentrations: 100 μg/ml ampicillin, 50 μg/ml kanamycin, 10 μg/ml tetracycline, 30 μg/ml gentamicin, and 50 μg/ml spectinomycin. Saccharomyces cerevisiae was grown at 30°C in YPDA culture medium (1% yeast extract, 2% peptone, 2% dextrose, and 0.003% adenine) or in synthetic defined (SD) minimal medium (Clontech) complemented with the appropriate supplements (dropout supplements from Clontech).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17-1 | recA endA thi hsdR RP4-2-Tc::Mu::Tn7; Tpr Smr | 50 |
| LMG194 | Protein expression strain | Invitrogen |
| TOP10 | Cloning strain | Invitrogen |
| Rosetta | Protein expression strain, Cmr | Novagen |
| R. sphaeroides | ||
| WS8 | Wild-type strain, spontaneous Nalr | 62 |
| SP7 | WS8 derivative ΔrpoN2::Kan | 49 |
| SP13 | WS8 derivative ΔfleQ1::Kan | 48 |
| SP18 | WS8 derivative flgC1::Kan | 47 |
| FS3 | WS8 derivative ΔfliL3::aadA | This study |
| FS4 | WS8 derivative ΔmotB1::Kan | This study |
| FS5 | WS8 derivative ΔfliL3::aadA ΔmotB1::Kan | This study |
| SUP1 | WS8 derivative ΔfliL3::aadA motB (A67E) | This study |
| SUP2 | WS8 derivative ΔfliL3::aadA motB (A67E) | This study |
| SUP3 | WS8 derivative ΔfliL3::aadA motB (A56E) | This study |
| SUP4 | WS8 derivative ΔfliL3::aadA motB (F63L) | This study |
| SUP5 | WS8 derivative ΔfliL3::aadA motB (A67D) | This study |
| SUP6 | WS8 derivative ΔfliL3::aadA motB (A67T) | This study |
| SUP7 | WS8 derivative ΔfliL3::aadA motB (F63L) | This study |
| SUP8 | WS8 derivative ΔfliL3::aadA motB (A67G) | This study |
| S. cerevisiae AH109 | Yeast reporter strain, for HIS3, ADE2, and lacZ | Clontech |
| Plasmids | ||
| pTZ19R | Cloning vector, Apr, pUC derivative | Pharmacia |
| pRK415 | pRK404 derivative | 33 |
| pJQ200mp18 | Mobilizable suicide vector for R. sphaeroides | 51 |
| pBAD/HisB | Expression vector of six-His-tagged proteins | Invitrogen |
| pBAD/HisC | Expression vector of six-His-tagged proteins | Invitrogen |
| pBAD-fliL | pBAD/HisB expressing His6-FliLp | This study |
| pGEX-4T-2 | Expression vector for GST gene fusion | Amersham |
| pEGFP | Expression vector for GFP gene fusion | Clontech |
| pUC4K | Source of the Kanr cassette | Pharmacia |
| pPIRL | Vector that encodes tRNAs for rare codons | 6 |
| pRK-fliL | pRK415 derivative, fliL gene | Laboratory collection |
| pRK-motB | pRK415 derivative, expressing motB+ | This study |
| pRK-motBsup1 | pRK415 derivative, expressing motBsup1 | This study |
| pRK-motBsup2 | pRK415 derivative, expressing motBsup2 | This study |
| pRK-motBsup3 | pRK415 derivative, expressing motBsup3 | This study |
| pRK-motBsup4 | pRK415 derivative, expressing motBsup4 | This study |
| pRK-motBsup5 | pRK415 derivative, expressing motBsup5 | This study |
| pRK-motBsup6 | pRK415 derivative, expressing motBsup6 | This study |
| pRK-motBsup7 | pRK415 derivative, expressing motBsup7 | This study |
| pRK-motBsup8 | pRK415 derivative, expressing motBsup8 | This study |
| pRK-fliLΔ15 | pRK415 derivative, expressing fliLΔ15 gene | This study |
| pRK_FliL-GFP | pRK415 derivative, expressing fliL-gfp | This study |
| pRK_GFP-FliL | pRK415 derivative, expressing gfp-fliL | This study |
| pGBKT7 | GAL4 DNA binding domain, TRP1 | Clontech |
| pGADT7 | GAL4 activation domain, LEU2 | Clontech |
| pGBD-fliLp | pGBKT7 derivative expressing GAL4 BD-FliLp | This study |
| pGAD-fliLp | pGADT7 derivative expressing GAL4 AD-FliLp | This study |
| pGBD-motBp | pGBKT7 derivative expressing GAL4 BD-MotBp | This study |
Oligonucleotides.
The oligonucleotides used in this work are shown in Table S1 in the supplemental material.
Isolation of mutant strains.
The FS3 mutant strain was obtained by a double recombination event between the WS8 wild-type strain and the allele ΔfliL3::aadA cloned in the suicide vector pJQ200mp18 (51). The ΔfliL3::aadA allele was generated by cloning together two independent PCR products, the first one containing 770 bp upstream from the stop codon of fliK and 72 bp downstream from the start codon of fliL and the second one containing 193 bp upstream from the stop codon of fliL and 839 bp downstream from the start codon of fliM (oligonucleotides upfliL1 and upfliL3 for the first PCR and fliL950 and downfliL3 for the second one). These products were triple ligated in pTZ19R by taking advantage of the restriction sites that were included in the oligonucleotides. The resultant plasmid was named pTZΔfliL3. The aadA gene encoding aminoglycoside 3′-adenyltransferase was obtained by PCR as an internal portion of the omega-Spcr cassette, which removed the known transcriptional termination signals. The PCR product containing the aadA gene was cloned into the EcoRV restriction site of pTZΔfliL3. The fragment carrying the ΔfliL3::aadA allele was subcloned into pJQ200mp18 and introduced into WS8 by conjugation with the S17-1 strain (50). The double recombination events were selected as described previously (49).
The FS4 mutant strain carrying the allele ΔmotB1::Kan was isolated by following the general procedure described above. In this case, the first PCR product contained 745 bp upstream from the stop codon of motA and 5 bp downstream from the start codon of motB and the second PCR product contained 15 bp upstream from the stop codon of motB and 929 bp downstream (oligonucleotides motBFup1 and motBRup2 and oligonucleotides motBFdown1 and motBRdown2, respectively). The resultant plasmid was named pTZΔmotB1. A Kanr cassette obtained from pUC4K was cloned into the EcoRV site of pTZΔmotB1. The resulting ΔmotB1::Kan allele was subcloned into pJQ200mp18 and introduced into WS8. The mutant strain was selected as described previously (49).
Motility assays.
A 5-μl sample of a culture in stationary phase was placed on the surface of plates containing Sistrom's minimal medium with 0.25% agar. Swimming was evaluated after 30 h of incubation at 30°C.
Analysis of the primary sequence of FliL and MotB.
The sequences of FliL and MotB were analyzed using the TOPCONS web server (http://topcons.cbr.su.se/) (10). The predicted topology for MotB was the inverse of what the experimental results suggest.
Plasmid constructs used in this work.
Plasmids that were used for complementation tests and topology analysis are described in this paragraph; other plasmids are described in the proper section. In all cases, the pRK415 plasmid was used as an expression vector for R. sphaeroides; this plasmid allows the expression of the cloned genes from the plac and/or ptet promoter (33). pRK-fliL was obtained by subcloning a 1.3-kb BamHI-EcoRI fragment from the pRS401 plasmid (24) into pRK415. pRK-fliLΔ15 was generated by cloning the PCR product obtained using the oligonucleotides fliLΔfw and GSTfliLdel1rv into pRK415.
pRK-motB was generated by cloning into pRK415 a 1.7-kb fragment obtained as a PCR product by using the oligonucleotides motBfwXba and motB.
pRK_FliL-GFP (green fluorescent protein) was generated by cloning a 924-bp DNA fragment obtained by PCR with the oligonucleotides fliLfw and fliLgfp2 as forward and reverse primers, respectively. The PCR product was cloned into pEGFP (Clontech). The resultant plasmid was purified and digested with HindIII and EcoRI, and the DNA fragment carrying fliL-gfp was subcloned into pRK415.
pRK_GFP-FliL was obtained by cloning the coding region of fliL without the initiation codon, in frame with the coding region of gfp. For this, fliL was amplified by PCR using the oligonucleotides FliLNHgfp and FliLGST2. The resultant product was cloned in pTZ19R as an XbaI-EcoRI fragment, and the coding region of gfp was cloned upstream of fliL as a HindIII-XbaI fragment. gfp was obtained by PCR using the oligonucleotides GFP-67(SD-GFP) and GFP-67rvs(stop)Xba. The first oligonucleotide includes the Shine-Dalgarno consensus sequence and the recognition site for HindIII, whereas the second oligonucleotide carries the recognition site for XbaI. The DNA fragment carrying this fusion was subcloned into pRK415.
Microscopy.
An aliquot of an exponentially growing culture was placed on a slide with an agarose pad containing Sistrom medium. Epifluorescence images were taken using a Nikon Eclipse 600 equipped with a Hamamatsu Orca-ER cooled charge-coupled device (CCD) camera, and images were acquired for 5 s. For time-lapse observations, an image was acquired each minute.
Protease assay.
Five milliliters of exponentially growing R. sphaeroides cultures was harvested at 12,000 × g for 5 min. Cells were washed twice with Tris-HCl (10 mM, pH 7) and resuspended in 500 μl of 10 mM Tris-20% sucrose. At this point, a sample was taken as control. Spheroplasts were obtained by the addition of EDTA and lysozyme (final concentrations of 50 μM and 0.5 mg/ml, respectively). After 15 min of incubation at 37°C, the suspension was divided into two, and buffer or 1 mg/ml of proteinase K was added. These reaction mixtures were incubated at 37°C for 15 min. To stop the reaction, phenylmethylsulfonyl fluoride (PMSF; 2 μM) was added and the samples were further incubated for 5 min before being boiled in 1× Laemmli sample buffer. The samples were analyzed by immunoblotting with polyclonal antibodies against the His6-FliLp protein obtained as described below. Monoclonal anti-GFP antibody was purchased from Clontech.
Protein overexpression and purification.
The DNA region encoding the periplasmic domain of FliL (FliLp) (residues 49 to 190) was obtained by PCR using the oligonucleotides His-fliLfw and His-fliLrv. The product of this reaction was cloned into pBAD/HisB. E. coli LMG174/pPIRL (6) was transformed with pBAD-His/fliLp. An overnight culture of this strain was diluted 1:100 in fresh medium and incubated at 37°C until it reached an optical density at 600 nm (OD600) of 0.5. At this point, 2% arabinose was added, and incubation was allowed to proceed for 2 h. Cells were harvested and resuspended in phosphate-buffered saline (PBS) with 20% glycerol, pH 7.4 (1/100 volume). Lysozyme was added (1-mg/ml final concentration), and the mixture was incubated for 15 min on ice. The cell suspension was sonicated on ice by using three cycles of 10-s bursts/min. Cell debris was removed by at least three steps of centrifugation (14,000 rpm for 5 min). The supernatant was mixed with nickel nitrilotriacetic acid (Ni2+) agarose beads and incubated for 1 h on ice, with the tube being inverted sporadically. The mixture was used to load a polypropylene column and washed with 10 volumes of buffer (PBS-20 mM imidazole). The protein was eluted using PBS-200 mM imidazole-20% glycerol, pH 7.4.
To obtain GST (glutathione S-transferase)-FliLp, a PCR product (oligonucleotides fliLGST3 and fliLGST2) containing the coding region of FliLp corresponding to residues 49 to 190 (without the TM domain) was cloned into pGEX-4T-2. To obtain GST-FliLpΔ15, the PCR product obtained using the oligonucleotides fliLGST3 and GSTfliLdel1RV was cloned in pGEX-4T-2. The resultant plasmids were transformed into the Rosetta strain. To purify these proteins, the following procedure was used. The cells were grown in LB at 37°C until the culture reached an OD600 of 0.5; at this point expression of the protein was induced using 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After 3 h of incubation in the presence of the inducer, the cells were harvested and resuspended in a 1/100 volume by using PBS (pH 7.4)-20% glycerol. The cells were lysed as described above. The soluble fraction was mixed with glutathione-agarose beads (Sigma) and incubated on ice for 1 h, with the tube being inverted sporadically. The mixture was used to load a polypropylene column and washed with 50 volumes of washing buffer (PBS, pH 7.4). The protein was eluted with 10 mM reduced glutathione (Sigma) dissolved in 50 mM Tris-HCl, pH 8.
To obtain GST-MotBp, a PCR product (oligonucleotides motBsDTM2fw and motBsDTM2rv) containing the periplasmic region of MotB (MotBp) corresponding to residues 55 to 366 (without the TM domain) was cloned into pGEX-4T-2. The protein was purified by following the procedure described above.
FliL antibodies.
Polyclonal antibodies were raised in female BALB/c mice against His6-FliLp protein, accordingly to previously reported protocols (28).
GST pulldown assays.
According to previously reported protocols (40), 40 μg of protein-bound glutathione agarose beads, i.e., GST-FliLp, GST-MotBp, or GST, was mixed with His6-FliLp to yield a 2:1 molar ratio of GST-FliLp, GST-MotBp, or GST to His6-FliLp. The total volume was adjusted to 100 μl with PBS, pH 7.4. The mixture was incubated for 1 h at 4°C with constant agitation. The beads were collected by centrifugation (1 min at 3,500 rpm), and the supernatant was removed carefully. The beads were washed five times with 1 volume of PBS (pH 7.4) and boiled in SDS loading buffer, and an aliquot of the sample was loaded on a 12% SDS-PAGE gel. After electrophoresis, the proteins were transferred to a nitrocellulose membrane as described elsewhere (28). The membrane was incubated with an anti-His (Qiagen) or anti-GST (Pierce/GE) polyclonal antibody. The blot was developed using the Western Star immunodetection system (Applied Biosystems).
Yeast two-hybrid assays.
The Matchmaker GAL4 two-hybrid system 3 (Clontech) was used to test FliLp-FliLp and FliLp-MotBp interactions. The region encoding FliLp was cloned into pGBKT7 (encoding the DNA binding domain [BD] of GAL4) and pGADT7 (encoding the activation domain [AD] of GAL4) by using the oligonucleotides FliL-BDfw and FliL-BDrev. MotBp was cloned into pGBKT7 by using the oligonucleotides BDMotB1 and BDMotB3. The yeast strain AH109 was cotransformed with either pGBKT7-FliLp (BD-FliLp) and pGADT7-FliLp (AD-FliLp) or pGADT7-FliLp (AD-FliLp) and pGBKT7-MotBp (BD-MotBp). The double transformants were selected as tryptophan and leucine prototrophs. After the initial selection, the two different transformant strains were grown overnight in 3 ml of synthetic defined (SD) minimal medium lacking tryptophan and leucine but supplemented with histidine and adenine. Aliquots of the cultures were washed once with SD minimal medium lacking Trp, Leu, His, and Ade and then normalized to an OD600 of 0.5, and 10-fold serial dilutions were made in the same medium. From these dilutions, 10-μl aliquots were seeded on selection plates lacking Trp, Leu, and His or lacking Trp, Leu, His, and Ade. A control plate lacking Trp and Leu was also included. AH109 was transformed with the plasmids included in the kit that represent positive and negative protein-protein interactions. In order to rule out spurious activation mediated by FliLp, AH109 was transformed only with pGBKT7-FliLp (BD-FliLp), which expresses a fusion of the DNA binding domain of GAL4 with FliLp.
RESULTS
Isolation and characterization of a fliL mutant strain.
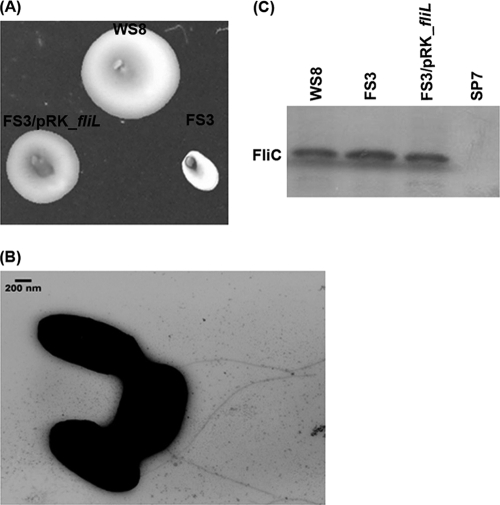
To characterize the function of FliL in R. sphaeroides motility, the FS3 strain carrying the ΔfliL3::aadA allele was isolated as described in Materials and Methods. As shown in Fig. 1A, this strain was unable to swim in soft-agar plates; however, when pRK-fliL plasmid was introduced into FS3, motility was recovered (Fig. 1A), indicating that FliL is essential for swimming in R. sphaeroides. When cells of the fliL mutant strain were examined by electron microscopy, flagellar filaments were observed (Fig. 1B). In agreement with this result, the amount of extracellular flagellin detected by Western blotting for this strain was similar to that detected for the WS8 wild-type strain (Fig. 1C). Observation of FS3 cells by light microscopy revealed that although the majority of the population was nonmotile, a few cells under the field showed swimming periods of variable duration (see movies S1 and S2 in the supplemental material). From these results we conclude that FliL is required for proper motor rotation.
FIG. 1.
Characterization of the fliL mutant strain. (A) Swimming of FS3 (ΔfliL3::aadA) and FS3 carrying the wild-type fliL allele cloned in pRK451. WS8 was included as a positive control. (B) Electron micrograph of FS3 cells showing the presence of flagella. (C) Western blot analysis of the supernatant fraction obtained after strong vortexing of WS8, FS3, and FS3/pRK-fliL cultures. A polyclonal anti-FliC antibody was used for detection. Strain SP7 (ΔrpoN2::Kan) was included as a negative control.
Determination of FliL topology in R. sphaeroides.
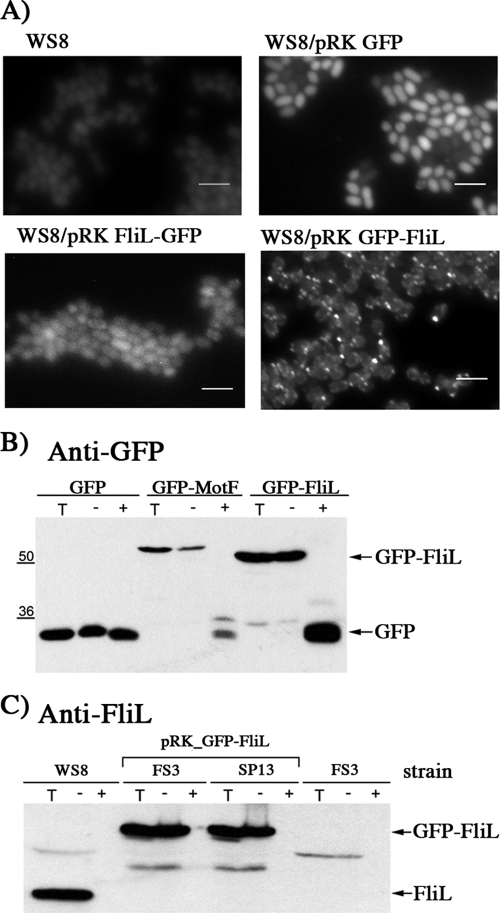
The fliL gene of R. sphaeroides encodes a protein of 190 amino acids. Analysis of the primary sequence of this protein suggested the presence of a TM segment between residues 21 and 44 (see Materials and Methods for details). Recently, it was shown that the C terminus of the E. coli FliL protein is located in the periplasm (5). However, given the different phenotypes of the E. coli and R. sphaeroides ΔfliL strains, we decided to corroborate this result in our system. For this purpose, the GFP was fused to the N- or C-terminal region of FliL. Observation of WS8 cells expressing these proteins from the lac promoter of pRK415 revealed the presence of fluorescent foci generated by the N-terminal GFP-FliL fusion but not by the C-terminal FliL-GFP fusion (Fig. 2A). It is known that GFP is fluorescent only in the cytoplasm (17, 21) or when it is translocated to the periplasm in its mature state by the twin-arginine pathway (56, 67). Since this last situation does not apply to FliL, our results are in agreement with the suggestion that the C terminus of FliL is localized on the periplasmic side of the membrane.
FIG. 2.
Determination of FliL topology. (A) Representative images of WS8 expressing FliL-GFP or GFP-FliL. Strains WS8 and WS8 expressing GFP from pRK415 were used as controls. Bars, 3 μm. (B) Western blot analysis of WS8 cells expressing GFP, GFP-MotF, and GFP-FliL. MotF is an inner membrane protein related to flagellar rotation in R. sphaeroides that has a C-terminal domain in the periplasmic side of the IM (V. F. Ramirez-Cabrera and L. Camarena, unpublished results). Lanes: +, spheroplasts treated with proteinase K; −, spheroplasts without proteinase K; T, total cell extract. (C) Western blot analysis of WS8 wild-type, FS3 (ΔfliL3::aadA), and SP13 (ΔfleQ1::Kan) cells expressing GFP-FliL. Lanes: +, spheroplasts treated with proteinase K; −, spheroplasts without proteinase K; T, total cell extract. As a control, the FS3 strain was included.
To rule out a possible effect of GFP on the normal topology of FliL, a protease accessibility assay was carried out to compare the protease sensitivity of GFP-FliL with that of wild-type FliL (Fig. 2B and C). The results obtained from these experiments demonstrate that GFP-FliL in WS8 and FS3 cells is properly inserted in the inner membrane.
Localization of GFP-FliL in different flagellar mutant strains.
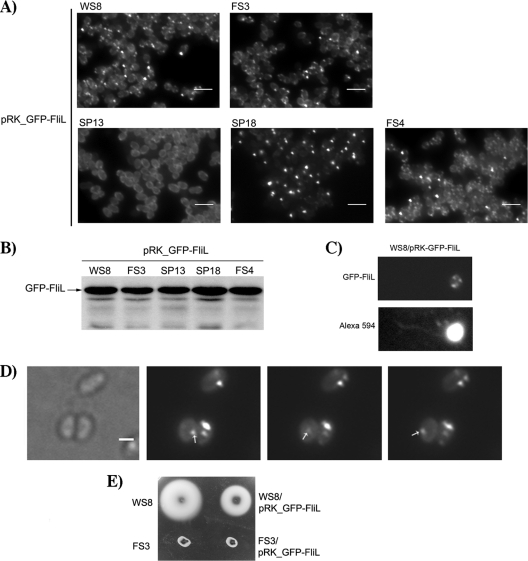
To determine if the fluorescent foci formed by the GFP-FliL protein were dependent on the presence of other flagellar proteins, we introduced the plasmid encoding GFP-FliL into the SP13 strain, which lacks the master regulator FleQ, preventing the expression of the flagellar regulon (48). No fluorescent foci were observed in this strain; instead, a diffused fluorescent signal was observed throughout the cell (Fig. 3A). The amount of GFP-FliL detected by Western blotting for the SP13 strain was similar to that detected for WS8 (Fig. 3B), and a protease accessibility assay showed that in SP13 cells, GFP-FliL is still properly inserted in the membrane (Fig. 2C). These results suggest that clustering of GFP-FliL requires another flagellar protein(s). Given that the absence of FliL has been mainly associated with Mot− and rod fragility phenotypes, we tested the localization of GFP-FliL in FS4 (ΔmotB1::Kan) and SP18 (flgC1::Kan) strains. GFP-FliL generated fluorescent foci similar to those detected in WS8 in these two strains, indicating that neither the flagellar rod nor the stator protein MotB is required for clustering (Fig. 3A).
FIG. 3.
Characterization of GFP-FliL localization. (A) Representative images of different strains expressing GFP-FliL: WS8 (wild type), FS3 (ΔfliL3::aadA), SP13 (ΔfleQ1::Kan), SP18 (flgC1::Kan), FS4 (ΔmotB1::Kan). Bars, 3 μm. (B) Western blot of GFP-FliL expressed in different strains. Total cell extracts of the indicated strains were analyzed by Western blotting using an anti-GFP antibody. (C) Example of a characteristic WS8 wild-type cell expressing GFP-FliL. Cells were stained with Alexa 594 to covisualize flagella and the signal from GFP-FliL. (D) Representative images of WS8 cells exemplifying the motion of the lateral foci of GFP-FliL. A moving lateral focus is indicated by an arrow. Bar, 1 μm. (E) Swimming plate of WS8 and FS3 strains expressing GFP-FliL.
A closer examination of WS8 and FS3 cells expressing GFP-FliL showed that this protein forms a highly fluorescent polar focus and less intense lateral foci (Fig. 3C). Frequently, both types of patterns were found simultaneously on the same cell. The intense foci commonly correlate with what seems to be the base of the flagellum (Fig. 3C), whereas the less intense aggregates are randomly distributed. A time-lapse observation revealed that the less intense foci move along the cellular body whereas the bright foci do not change their position (Fig. 3D). Since no GFP-FliL was detected after the protease treatment (Fig. 2B), we conclude that all the foci detected by fluorescence microscopy are generated by a properly inserted protein in the inner membrane (IM).
Interestingly, similar mobile and immobile foci were observed in E. coli cells expressing a GFP-FliG protein fusion (23).
It should be noted that GFP-FliL is unable to complement the swimming defect of the FS3 strain, indicating that the GFP moiety interferes with the proper functioning of FliL. In line with this idea, we noticed that the expression of GFP-FliL results in a 30% reduction of the swarm ring of wild-type WS8 cells (Fig. 3E).
Suppressor mutants of the ΔfliL3::aadA allele.
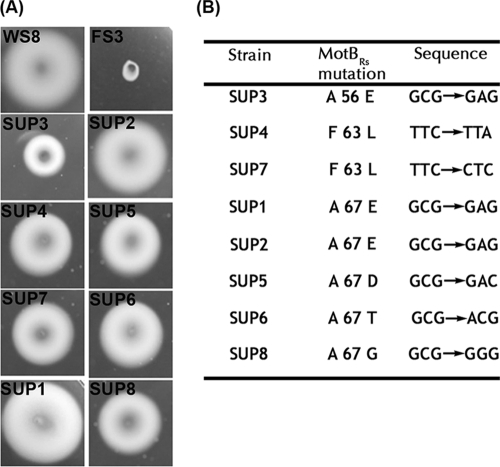
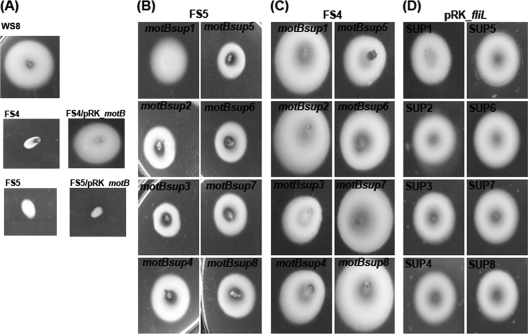
To obtain a better insight into the role of FliL in the function of the flagellar motor, we isolated eight pseudorevertants from the FS3 mutant. For this, swarm plates inoculated with independent cultures of this strain were incubated for 5 to 7 days, after which a small halo around the inoculation point was occasionally detected. Purification and subcultivation of the cells taken from these halos yielded pure strains that were able to swim to different degrees (Fig. 4A). For one of these pseudorevertants (SUP1), we determined the nucleotide sequence of the flagellar genes previously reported to produce a Mot− phenotype in E. coli after being mutated (fliG, fliM, fliN, motA, and motB). Comparison of the obtained sequences with those of the wild-type strain showed a single change in motB. This point mutation replaces an alanine residue at position 67 for a glutamic acid (E). The motB sequences from the rest of the pseudorevertants revealed that a single change in motB occurred in each of them. As shown in Fig. 4B, all these changes are confined to a small region of MotB located at the beginning of the periplasmic domain of the protein that comprises residues 56 to 67.
FIG. 4.
Characterization of the second-site suppressors isolated from the FS3 strain. (A) Swimming of the different suppressor strains. WS8 and FS3 (ΔfliL3::aadA) were included as positive and negative controls, respectively. (B) Nucleotide and residue changes present in motB for each suppressor mutant.
To corroborate the idea that each of the single amino acid substitutions found in the suppressor strains is sufficient to allow flagellar rotation in the absence of FliL, we carried out a complementation analysis. For this, wild-type and suppressor motB alleles (motBsup) were cloned in pRK415. These plasmids were then introduced into the nonmotile FS4 (ΔmotB1::Kan) and FS5 (ΔmotB1::Kan ΔfliL3::aadA) mutant strains, and motility was tested on soft-agar plates. As expected, the plasmid carrying the wild-type motB allele was able to complement FS4 but not the motB fliL double mutant (FS5) (Fig. 5A). Notably, FS5 (ΔmotB1::Kan ΔfliL3::aadA) recovered its swimming ability only when it was transformed with any of the plasmids that express the motBsup alleles (Fig. 5B).
FIG. 5.
Swimming characterization of different strains expressing the motBsup alleles. (A) Swimming assay of the control strains WS8, FS4 (ΔmotB1::Kan), and FS5 (ΔmotB1::Kan ΔfliL3::aadA). Also included are FS4 and FS5 expressing the motB wild-type allele from pRK415. (B) Swimming of the double mutant FS5 (ΔmotB1::Kan ΔfliL3::aadA) expressing the motBsup alleles from the lac promoter of pRK415. (C) Swimming of FS4 (ΔmotB1::Kan) expressing the motBsup alleles from pRK415. (D) Swimming of the suppressor mutants expressing the fliL wild-type allele from pRK415.
To evaluate the possibility that the motBsup alleles would not function properly in the presence of wild-type FliL, we introduced the plasmids that express the motBsup alleles into the mutant strain FS4 (ΔmotB1::Kan) (Fig. 5C) as well as introducing the plasmid that expresses fliL+ into the original suppressor strains (Fig. 5D). All the resultant strains were able to swim independently of the allele combination. These results indicate that the motBsup alleles support swimming in the presence or absence of FliL.
FliL-FliL and FliL-MotB interactions tested by pulldown and yeast two-hybrid assays.
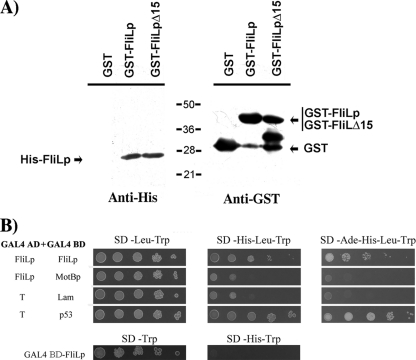
To test if FliL forms a complex with the periplasmic domain of the stator protein MotB, we carried out a pulldown assay using His6-FliLp as prey and GST-MotBp as bait. It should be noted that only the periplasmic domain of FliL (amino acids 40 to 190) was present in the His6-FliLp fusion, given that overexpression of the complete coding region of fliL could not be achieved. Among the FliL protein sequences annotated in the public database, the C terminus of FliL is the most conserved part of the protein. For this reason, we also tested if His6-FliLp could interact with GST-FliLp and GST-FliLpΔ15, which lacks the last 15 residues of FliL.
These assays revealed that GST-MotBp does not interact with FliLp (data not shown). However, His6-FliLp was retained by GST-FliLp and GST-FliLpΔ15 (Fig. 6A). As negative control, it is shown that GST was unable to interact with His6-FliLp (Fig. 6A). These results indicate that the periplasmic region of FliL interacts with itself but not with the periplasmic region of MotB. Unfortunately, our efforts to test in vivo the relevance of the C-terminal region of FliL were unsuccessful due to the instability of FliLΔ15 protein (data not shown).
FIG. 6.
(A) Interaction of His6-FliLp with FliLp. Coprecipitation of His6-FliLp, using GST-FliLp, GST-FliLpΔ15, or GST alone as bait. After the coprecipitation assay, the sample was divided in two and probed with anti-His and anti-GST antibodies. The proteins observed with anti-GST confirm a proper concentration of the bait, whereas the presence of the His-tagged protein indicates a positive interaction. (B) Double-hybrid assay to test protein-protein interactions. Yeast cells were transformed with the plasmids indicated at the left margin; GAL4AD-T and GAL4BD-Lam and GAL4AD-T and GAL4BD-p53 plasmid pairs served as negative and positive controls, respectively. Transformant cells were seeded on the medium indicated at the top. To rule out spurious activation of the reporter gene (HIS3) mediated by FliLp, AH109 transformed with GAL4BD-FliLp was also tested (lower part of the figure). Pictures were taken after 4 days of incubation for culture plates lacking Trp and Leu and plates lacking Trp, Leu, and His or 10 days for plates lacking Trp, Leu, His, and Ade.
These results were corroborated in a yeast double-hybrid assay. For this, the periplasmic domains of FliL and MotB were fused to the GAL4 DNA binding domain and FliLp was also fused to the GAL4 activation domain (BD-FliLp, BD-MotBp, and AD-FliLp, respectively). The plasmids encoding these fusions were introduced into AH109, and the expression of the reporter genes HIS3 and ADE2 was tested. The strain coexpressing AD-FliLp and BD-MotBp failed to grow in the absence of histidine or adenine. In contrast, AH109 expressing AD-FliLp and BD-FliLp was able to grow under these conditions, indicating a positive interaction between the periplasmic domains of FliL (Fig. 6B).
DISCUSSION
The role of FliL in bacterial motility has been difficult to establish given that different phenotypes have been associated with the absence of this protein, from the absence of phenotype in E. coli and Salmonella (52), to the Mot− phenotype in C. crescentus (31) and Silicibacter TM1040 (7), to the Fla− phenotype in P. putida (59). Moreover, the absence of FliL has also been associated with swarming defects in P. mirabilis and Salmonella (5, 8). Although in all these reports FliL is related to flagellar rotation or biogenesis, the molecular mechanisms that allow FliL to accomplish its particular function remain unknown.
In this work we show that in R. sphaeroides the absence of FliL affects swimming negatively. From our results, we propose that in this bacterium FliL modulates the function of the flagellar motor through MotB.
A direct interaction between FliL and MotB was reported for Campylobacter jejuni (53). However, we were unable to detect a positive interaction between the periplasmic domains of these two proteins by using two different methods (pulldown and yeast two-hybrid assays). The possibility remains that the interaction between FliL and MotB could be dependent on the presence of the TM domains or on the formation of the MotA-MotB complex or that it could be indirect.
In spite of these negative results, the isolation of eight independent second-site suppressors of the ΔfliL3::aadA allele in the motB gene supports the idea that FliL exerts its action on the flagellar motor through MotB.
But how FliL could affect MotB functioning? We consider that the answer could be related to the function of a MotB region that acts as the plug of the proton channel.
A recent study carried out in E. coli showed that a region of MotB acts as a plug to prevent proton flow before the MotA-MotB complex associates with the flagellar structure. It was shown that this region extends from K53 to T64 and that the hydrophobic residues I58, Y61, and F62 are essential for plug function. A defective plug combined with the overexpression of MotA inhibits growth. However, if these proteins were expressed at normal levels, the effect on motility or growth was nondetectable (30).
MotB from R. sphaeroides has 366 residues, and remarkable differences were detected when it was compared to the E. coli MotB sequence (29). In fact, from the alignment of the MotB sequences of these bacteria, it can be observed that the similarity is restricted mainly to the C-terminal region while the region proposed as the MotB channel plug is not conserved in R. sphaeroides. Nevertheless, we think that the region of MotB that lies between residues 56 and 67 could be equivalent to the plug region of MotB in E. coli, given that the two regions are located in the periplasmic space immediately after the transmembrane segment and that both are predicted to form an amphipathic alpha-helix (see Fig. S1 in the supplemental material).
It should be emphasized that each one of the suppressor strains isolated in this work carries a single mutation in motB affecting the short segment of the protein that lies between residues 56 and 67. Moreover, in E. coli the function of the plug was compromised when two hydrophobic residues, I58 and F63, were affected; likewise, the changes detected in seven out of eight fliL pseudorevertants affected the hydrophobic residues F63 and A67 (Fig. 4; see also Fig. S1 in the supplemental material). Among the suppressor strains, the only change that did not lie in this region is A56E (SUP3); this residue is at the end of the TM helix of MotB. However, it is possible that this change could also affect the plug indirectly by shifting the transmembrane helix or inducing a conformational change of MotB.
It is tempting to speculate that in R. sphaeroides in the absence of FliL, the mechanism that triggers the release of the plug could be hindered. Interestingly, the motB suppressor alleles are insensitive to the presence of FliL, suggesting that FliL is required only for the activation of the channel. This could occur in a single event, after which the channel remains constantly open, or FliL could be required to repeatedly activate the MotA-MotB channel, perhaps in coordination with the rotation of the flagellum. This last possibility seems particularly interesting since it could allow the bacterium to regulate the function of the MotA-MotB channel in response to particular stimuli.
With the idea that the localization of FliL is dependent on its interaction with the flagellar structure, we followed the formation of GFP-FliL fluorescent foci in different flagellar mutant strains. Our hypothesis was supported by the absence of GFP-FliL foci in the mutant of the flagellar master transcriptional activator FleQ. In contrast, we observed localization of GFP-FliL in a mutant that does not assemble the rod and also in a motB mutant, indicating that neither the motor nor the rod is required for FliL recruitment to the flagellar structure. Further research will be required to establish the identity of the flagellar components required for the stable clustering of FliL.
Supplementary Material
Acknowledgments
We thank Teresa Ballado, Javier de la Mora, and Georgina Diaz Herrera for technical assistance. We also thank the IFC Molecular Biology Unit for sequencing facilities as well as the Microscopy Unit for the electron micrographs.
This work was supported in part by grants from Consejo Nacional de Ciencia y Tecnología (SEP-CONACYT 106081 and DGAPA/UNAM IN213408).
Footnotes
Published ahead of print on 1 October 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Armitage, J. P., and R. M. Macnab. 1987. Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J. Bacteriol. 169:514-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armitage, J. P., T. P. Pitta, M. A. Vigeant, H. L. Packer, and R. M. Ford. 1999. Transformations in flagellar structure of Rhodobacter sphaeroides and possible relationship to changes in swimming speed. J. Bacteriol. 181:4825-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asakura, S., G. Eguchi, and T. Iino. 1964. Reconstitution of bacterial flagella in vitro. J. Mol. Biol. 10:42-56. [DOI] [PubMed] [Google Scholar]
- 4.Attmannspacher, U., B. Scharf, and R. Schmitt. 2005. Control of speed modulation (chemokinesis) in the unidirectional rotary motor of Sinorhizobium meliloti. Mol. Microbiol. 56:708-718. [DOI] [PubMed] [Google Scholar]
- 5.Attmannspacher, U., B. E. Scharf, and R. M. Harshey. 2008. FliL is essential for swarming: motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Mol. Microbiol. 68:328-341. [DOI] [PubMed] [Google Scholar]
- 6.Bao, K., and S. N. Cohen. 2001. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev. 15:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belas, R., E. Horikawa, S. Aizawa, and R. Suvanasuthi. 2009. Genetic determinants of Silicibacter sp. TM1040 motility. J. Bacteriol. 191:4502-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belas, R., and R. Suvanasuthi. 2005. The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J. Bacteriol. 187:6789-6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg, H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19-54. [DOI] [PubMed] [Google Scholar]
- 10.Bernsel, A., H. Viklund, A. Hennerdal, and A. Elofsson. 2009. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37:W465-W468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blair, D. F., and H. C. Berg. 1988. Restoration of torque in defective flagellar motors. Science 242:1678-1681. [DOI] [PubMed] [Google Scholar]
- 12.Blair, D. F., and H. C. Berg. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439-449. [DOI] [PubMed] [Google Scholar]
- 13.Braun, T. F., L. Q. Al-Mawsawi, S. Kojima, and D. F. Blair. 2004. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 43:35-45. [DOI] [PubMed] [Google Scholar]
- 14.Braun, T. F., and D. F. Blair. 2001. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H(+) channels in the stator complex. Biochemistry 40:13051-13059. [DOI] [PubMed] [Google Scholar]
- 15.Che, Y. S., S. Nakamura, S. Kojima, N. Kami-ike, K. Namba, and T. Minamino. 2008. Suppressor analysis of the MotB(D33E) mutation to probe bacterial flagellar motor dynamics coupled with proton translocation. J. Bacteriol. 190:6660-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chun, S. Y., and J. S. Parkinson. 1988. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science 239:276-278. [DOI] [PubMed] [Google Scholar]
- 17.Daley, D. O., M. Rapp, E. Granseth, K. Melen, D. Drew, and G. von Heijne. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321-1323. [DOI] [PubMed] [Google Scholar]
- 18.DePamphilis, M. L., and J. Adler. 1971. Attachment of flagellar basal bodies to the cell envelope: specific attachment to the outer, lipopolysaccharide membrane and the cyoplasmic membrane. J. Bacteriol. 105:396-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePamphilis, M. L., and J. Adler. 1971. Fine structure and isolation of the hook-basal body complex of flagella from Escherichia coli and Bacillus subtilis. J. Bacteriol. 105:384-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Driks, A., and D. J. DeRosier. 1990. Additional structures associated with bacterial flagellar basal body. J. Mol. Biol. 211:669-672. [DOI] [PubMed] [Google Scholar]
- 21.Feilmeier, B. J., G. Iseminger, D. Schroeder, H. Webber, and G. J. Phillips. 2000. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 182:4068-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francis, N. R., V. M. Irikura, S. Yamaguchi, D. J. DeRosier, and R. M. Macnab. 1992. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc. Natl. Acad. Sci. U. S. A. 89:6304-6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukuoka, H., Y. Sowa, S. Kojima, A. Ishijima, and M. Homma. 2007. Visualization of functional rotor proteins of the bacterial flagellar motor in the cell membrane. J. Mol. Biol. 367:692-701. [DOI] [PubMed] [Google Scholar]
- 24.Garcia, N., A. Campos, A. Osorio, S. Poggio, B. Gonzalez-Pedrajo, L. Camarena, and G. Dreyfus. 1998. The flagellar switch genes fliM and fliN of Rhodobacter sphaeroides are contained in a large flagellar gene cluster. J. Bacteriol. 180:3978-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garza, A. G., R. Biran, J. A. Wohlschlegel, and M. D. Manson. 1996. Mutations in motB suppressible by changes in stator or rotor components of the bacterial flagellar motor. J. Mol. Biol. 258:270-285. [DOI] [PubMed] [Google Scholar]
- 26.Garza, A. G., L. W. Harris-Haller, R. A. Stoebner, and M. D. Manson. 1995. Motility protein interactions in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 92:1970-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez-Pedrajo, B., T. Minamino, M. Kihara, and K. Namba. 2006. Interactions between C ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol. Microbiol. 60:984-998. [DOI] [PubMed] [Google Scholar]
- 28.Harlow, E., and D. Lane. 1988. Antibodies. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Hosking, E. R., and M. D. Manson. 2008. Clusters of charged residues at the C terminus of MotA and N terminus of MotB are important for function of the Escherichia coli flagellar motor. J. Bacteriol. 190:5517-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosking, E. R., C. Vogt, E. P. Bakker, and M. D. Manson. 2006. The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 364:921-937. [DOI] [PubMed] [Google Scholar]
- 31.Jenal, U., J. White, and L. Shapiro. 1994. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J. Mol. Biol. 243:227-244. [DOI] [PubMed] [Google Scholar]
- 32.Jones, C. J., R. M. Macnab, H. Okino, and S. Aizawa. 1990. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J. Mol. Biol. 212:377-387. [DOI] [PubMed] [Google Scholar]
- 33.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 34.Khan, I. H., T. S. Reese, and S. Khan. 1992. The cytoplasmic component of the bacterial flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 89:5956-5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi, T., J. N. Rinker, and H. Koffler. 1959. Purification and and chemical properties of flagellin. Arch. Biochem. Biophys. 84:342-362. [DOI] [PubMed] [Google Scholar]
- 36.Kojima, S., and D. F. Blair. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041-13050. [DOI] [PubMed] [Google Scholar]
- 37.Kojima, S., and D. F. Blair. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int. Rev. Cytol. 233:93-134. [DOI] [PubMed] [Google Scholar]
- 38.Konishi, M., M. Kanbe, J. L. McMurry, and S. Aizawa. 2009. Flagellar formation in C-ring-defective mutants by overproduction of FliI, the ATPase specific for flagellar type III secretion. J. Bacteriol. 191:6186-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kubori, T., N. Shimamoto, S. Yamaguchi, K. Namba, and S. Aizawa. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 226:433-446. [DOI] [PubMed] [Google Scholar]
- 40.Lane, M. C., P. W. O'Toole, and S. A. Moore. 2006. Molecular basis of the interaction between the flagellar export proteins FliI and FliH from Helicobacter pylori. J. Biol. Chem. 281:508-517. [DOI] [PubMed] [Google Scholar]
- 41.Lloyd, S. A., and D. F. Blair. 1997. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J. Mol. Biol. 266:733-744. [DOI] [PubMed] [Google Scholar]
- 42.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 43.Minamino, T., K. Imada, and K. Namba. 2008. Molecular motors of the bacterial flagella. Curr. Opin. Struct. Biol. 18:693-701. [DOI] [PubMed] [Google Scholar]
- 44.Minamino, T., S. Yamaguchi, and R. M. Macnab. 2000. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J. Bacteriol. 182:3029-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morgan, D. G., C. Owen, L. A. Melanson, and D. J. DeRosier. 1995. Structure of bacterial flagellar filaments at 11 Å resolution: packing of the alpha-helices. J. Mol. Biol. 249:88-110. [DOI] [PubMed] [Google Scholar]
- 46.Pallen, M. J., and N. J. Matzke. 2006. From the origin of species to the origin of bacterial flagella. Nat. Rev. Microbiol. 4:784-790. [DOI] [PubMed] [Google Scholar]
- 47.Poggio, S., C. Abreu-Goodger, S. Fabela, A. Osorio, G. Dreyfus, P. Vinuesa, and L. Camarena. 2007. A complete set of flagellar genes acquired by horizontal transfer coexists with the endogenous flagellar system in Rhodobacter sphaeroides. J. Bacteriol. 189:3208-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poggio, S., A. Osorio, G. Dreyfus, and L. Camarena. 2005. The flagellar hierarchy of Rhodobacter sphaeroides is controlled by the concerted action of two enhancer-binding proteins. Mol. Microbiol. 58:969-983. [DOI] [PubMed] [Google Scholar]
- 49.Poggio, S., A. Osorio, G. Dreyfus, and L. Camarena. 2002. The four different sigma(54) factors of Rhodobacter sphaeroides are not functionally interchangeable. Mol. Microbiol. 46:75-85. [DOI] [PubMed] [Google Scholar]
- 50.Priefer, U. B., R. Simon, and A. Pühler. 1985. Extension of the host range of Escherichia coli vectors by incorporation of RSF1010 replication and mobilization functions. J. Bacteriol. 163:324-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 52.Raha, M., H. Sockett, and R. M. Macnab. 1994. Characterization of the fliL gene in the flagellar regulon of Escherichia coli and Salmonella typhimurium. J. Bacteriol. 176:2308-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rajagopala, S. V., B. Titz, J. Goll, J. R. Parrish, K. Wohlbold, M. T. McKevitt, T. Palzkill, H. Mori, R. L. Finley, Jr., and P. Uetz. 2007. The protein network of bacterial motility. Mol. Syst. Biol. 3:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reid, S. W., M. C. Leake, J. H. Chandler, C. J. Lo, J. P. Armitage, and R. M. Berry. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. U. S. A. 103:8066-8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samatey, F. A., K. Imada, S. Nagashima, F. Vonderviszt, T. Kumasaka, M. Yamamoto, and K. Namba. 2001. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410:331-337. [DOI] [PubMed] [Google Scholar]
- 56.Santini, C. L., A. Bernadac, M. Zhang, A. Chanal, B. Ize, C. Blanco, and L. F. Wu. 2001. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J. Biol. Chem. 276:8159-8164. [DOI] [PubMed] [Google Scholar]
- 57.Sato, K., and M. Homma. 2000. Functional reconstitution of the Na(+)-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:5718-5722. [DOI] [PubMed] [Google Scholar]
- 58.Schoenhals, G. J., and R. M. Macnab. 1999. FliL is a membrane-associated component of the flagellar basal body of Salmonella. Microbiology 145:1769-1775. [DOI] [PubMed] [Google Scholar]
- 59.Segura, A., E. Duque, A. Hurtado, and J. L. Ramos. 2001. Mutations in genes involved in the flagellar export apparatus of the solvent-tolerant Pseudomonas putida DOT-T1E strain impair motility and lead to hypersensitivity to toluene shocks. J. Bacteriol. 183:4127-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silverman, M., and M. Simon. 1974. Flagellar rotation and the mechanism of bacterial motility. Nature 249:73-74. [DOI] [PubMed] [Google Scholar]
- 61.Sistrom, W. R. 1962. The kinetics of the synthesis of photopigments in Rhodopseudomonas spheroides. J. Gen. Microbiol. 28:607-616. [DOI] [PubMed] [Google Scholar]
- 62.Sockett, R. E., J. C. A. Foster, and J. P. Armitage. 1990. Molecular biology of the Rhodobacter sphaeroides flagellum. FEMS Symp. 53:473-479. [Google Scholar]
- 63.Sosinsky, G. E., N. R. Francis, D. J. DeRosier, J. S. Wall, M. N. Simon, and J. Hainfeld. 1992. Mass determination and estimation of subunit stoichiometry of the bacterial hook-basal body flagellar complex of Salmonella typhimurium by scanning transmission electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 89:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stolz, B., and H. C. Berg. 1991. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J. Bacteriol. 173:7033-7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang, H., T. F. Braun, and D. F. Blair. 1996. Motility protein complexes in the bacterial flagellar motor. J. Mol. Biol. 261:209-221. [DOI] [PubMed] [Google Scholar]
- 66.Terashima, H., S. Kojima, and M. Homma. 2008. Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270:39-85. [DOI] [PubMed] [Google Scholar]
- 67.Thomas, J. D., R. A. Daniel, J. Errington, and C. Robinson. 2001. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 39:47-53. [DOI] [PubMed] [Google Scholar]
- 68.Togashi, F., S. Yamaguchi, M. Kihara, S. I. Aizawa, and R. M. Macnab. 1997. An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB. J. Bacteriol. 179:2994-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ueno, T., K. Oosawa, and S. Aizawa. 1992. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J. Mol. Biol. 227:672-677. [DOI] [PubMed] [Google Scholar]
- 70.Yakushi, T., J. Yang, H. Fukuoka, M. Homma, and D. F. Blair. 2006. Roles of charged residues of rotor and stator in flagellar rotation: comparative study using H+-driven and Na+-driven motors in Escherichia coli. J. Bacteriol. 188:1466-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou, J., and D. F. Blair. 1997. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J. Mol. Biol. 273:428-439. [DOI] [PubMed] [Google Scholar]
- 72.Zhou, J., S. A. Lloyd, and D. F. Blair. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 95:6436-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou, J., L. L. Sharp, H. L. Tang, S. A. Lloyd, S. Billings, T. F. Braun, and D. F. Blair. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 180:2729-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.