Abstract
The majority of cystic fibrosis (CF) patients succumb to a chronic infection of the airway with Pseudomonas aeruginosa. Paradoxically, pathogenic strains are often susceptible to antibiotics, but the infection cannot be eradicated with antimicrobial therapy. We find that in a majority of patients with airway infections, late isolates of P. aeruginosa produce increased levels of drug-tolerant persister cells. The genomes of a clonal pair of early/late isolates from a single patient have been previously sequenced, and the late isolate (obtained at age 96 months) showed a 100-fold increase in persister levels. The 96-month isolate carries a large number of mutations, including a mutation in mutS that confers a hypermutator phenotype. There is also a mutation in the mexZ repressor controlling the expression of the MexXY-OprM multidrug pump, which results in a moderate increase in the ofloxacin, carbenicillin, and tobramycin MICs. Knocking out the mexXY locus restored the resistance to that of the parent strain but did not affect the high levels of persisters formed by the 96-month isolate. This suggests that the late isolate is a high-persister (hip) mutant. Increased persister formation was observed in exponential phase, stationary phase, and biofilm populations of the 96-month isolate. Analysis of late isolates from 14 additional patients indicated that 10 of them are hip mutants. Most of these hip mutants did not have higher drug resistance. Increased persister formation appears to be their sole mechanism for surviving chemotherapy. Taken together, these findings suggest a link between persisters and recalcitrance of CF infection and identify an overlooked culprit—high-persister mutants producing elevated levels of drug-tolerant cells. Persisters may play a similarly critical role in the recalcitrance of other chronic infections.
The majority of cystic fibrosis (CF) patients succumb to a chronic untreatable Pseudomonas aeruginosa infection of the airways (18). The strains responsible for these infections are largely clonal and show evidence of positive selection for mutations leading to adaptation to the airway environment (39). Mutants of P. aeruginosa resistant to antibiotics are selected in CF patients, which can contribute to therapy failure. These include overexpression of multidrug resistance pumps, target modification, and plasmid-mediated resistance (18). It is important to note, however, that isolates of P. aeruginosa from CF patients do not necessarily develop high-level antibiotic resistance (4, 19, 21). This creates a paradox: antibiotics that are effective in inhibiting the growth of the pathogen in vitro are unable to cure the infection. We suggested that persister cells are responsible for maintaining untreatable infections (36). Persisters are dormant phenotypic variants rather than mutants and have been produced by all microbial pathogens studied to date (28, 34, 44, 48). Persisters make up a small fraction of the population but are the only cells to survive treatment with high doses of bactericidal antibiotics. Persisters are distinct from both growing and stationary cells and are specialized survivors (36). Their tolerance to antibiotics is mechanistically distinct from resistance. Bactericidal antibiotics kill by corrupting active targets to produce toxic products, such as misfolded peptides formed in the presence of aminoglycosides (11) or DNA cleaved by topoisomerases bound to fluoroquinolone antibiotics (15, 30). Formation of reactive oxygen species after target corruption contributes to the lethal action of most bactericidal antibiotics (32). The persister state, in which targets are largely inactive, would then produce antibiotic tolerance.
Most of the work on persister formation has been carried out in Escherichia coli and has led to the identification of a number of candidate persister genes. These include toxin components of toxin/antitoxin (TA) modules (9, 13, 16, 29, 42, 44, 52), genes of glycerol metabolism (9, 49), nucleotide metabolism, and global regulators (23). In each case, overexpression of a persister gene shuts down important cellular functions. For example, overexpression of RelE produces tolerance (29) by inhibiting protein synthesis through cleaving mRNA (6), expression of HipA leads to persister formation (9) by inhibiting protein synthesis through phosphorylation of Ef-Tu (42), and expression of TisB creates persisters (13) by decreasing proton motive force (PMF) and ATP levels (51). P. aeruginosa has no experimentally confirmed homologs to these E. coli toxin genes. In P. aeruginosa, a screen of a transposon insertion library and subsequent genetic experiments in both PA01 and PA14 identified dinG (putative DNA helicase), pilH (type IV pilus response regulator), a predicted acetyl coenzyme A (acetyl-CoA) acetylase (PA3589), and a conserved hypothetical protein (PA5002) as candidate persister genes (12). The mechanisms of persister formation appear to be highly redundant, contributing to the difficulty of identifying genetic mechanisms of formation and eradicating these cells.
Persister levels rise in the stationary phase of growth (28) and are substantial in slow-growing/stationary biofilms (48). Biofilms do not present a strong barrier to antibiotics (37, 45, 55) but restrict penetration of the components of the immune system (1, 54). By harboring persisters tolerant to antibiotics, biofilms could then provide a reservoir of surviving pathogens responsible for a relapsing infection (2, 3, 36). P. aeruginosa has been reported to form biofilm-like microcolonies in the lungs of CF patients (2, 46, 56). However, late isolates of P. aeruginosa from CF patients often exhibit mutational loss of quorum-sensing genes involved in biofilm formation (10, 26, 33, 41, 47). P. aeruginosa reaches high densities in the CF lung, and a significant part of the population appears to be in stationary state, where persisters should be most abundant (58).
While persisters provide a plausible explanation for therapy failure, a simpler possibility is that the antibiotics fail to effectively reach at least some cells in the population, creating reservoirs of surviving cells that could cause a relapsing infection. The purpose of the present study was to differentiate between these hypotheses and test the possible causality between persisters and chronic infection.
Establishing potential causality between persisters and therapy failure is not trivial, since these cells form a small subpopulation with a temporary phenotype, which precludes introducing them into an animal model of infection. One approach to testing the persister hypothesis comes from in vitro studies of hip mutants. Periodic application of high doses of bactericidal antibiotics to a population of mutagenized cells leads to the selection of hip mutants that stably produce increased levels of persisters (40, 57). The hipA7 gain-of-function allele in E. coli was identified in these studies, showing that the level of persisters can be increased as a result of a mutation. While the persister state is a temporary one for a bacterial cell, the frequency with which cells enter this temporary state can be increased by mutations. A mutant that has an increased frequency of persister formation then is a hip mutant. Once these mutants are selected, the population stably forms more persisters than the wild-type strain from which it was derived. Because periodic application of high doses of antibiotics is the standard approach to treating the CF infection, we reasoned that hip mutants of P. aeruginosa would be selected during the course of treatment. In order to select for hip mutants, the drugs would have to effectively reach and kill the pathogen. Here we show that late isolates of P. aeruginosa from several CF patients are hip mutants.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The P. aeruginosa strains used in this study are described in Table 1. E. coli DH5α was used as a recipient for all cloning procedures, and E. coli HB101 harboring plasmid pRK2013 was used in triparental matings to mobilize constructs into P. aeruginosa (43). For routine maintenance and subculturing, cells were grown in LB medium. For killing experiments and MIC determination, cells were grown in Mueller-Hinton broth (MHB). The MIC was determined according to CLSI recommendations (7). The ofloxacin MIC of persister progeny was determined by recovering surviving persisters after 8 h of ofloxacin treatment (50× MIC), washing cells, and allowing surviving persisters to regrow in fresh medium to the stationary phase. These cells, the progeny of surviving persisters, were used in a new MIC test.
TABLE 1.
Properties of the P. aeruginosa strains used in this study
| Straina | Patient age (yr) when isolate was collected | Growth rate (doubling time in min) | MIC (μg/ml) of: |
||
|---|---|---|---|---|---|
| Ofloxacinb | Carbenicillin | Tobramycin | |||
| Parent (AMT0023-30) | 0.5 | 72.7 | 1 (1) | 64 | 1.0 |
| AMT0023-31 | 7.7 | 81.0 | 8 | 128 | 32.0 |
| AMT0023-31 | 7.7 | 74.3 | 8 | 128 | 32.0 |
| AMT0023-35 | 8.0 | 80.3 | 8 | 128 | 32.0 |
| 96-mo isolate (AMT0023-34) | 8.0 | 70.8 | 8 | 128 | 32.0 |
| ΔmexXY mutant (KLE2000) | 8.0 | 62.2 | 1 (2) | 128 | 2.0 |
| AMT0047-2 | 0.8 | 100.0 | 2 (1) | 1,600 | 0.8 |
| AMT0047-3 | 7.3 | 122.1 | 0.5 (1) | 400 | 0.4 |
| AMT0060-3 | 7.7 | 62.2 | 8 (14) | 800 | 4.0 |
| AMT0060-2 | 15.4 | 71.2 | 16 (16) | 100 | 1.0 |
| AMT0066-3 | 7.2 | 82.0 | 2 (0.25) | 200 | 3.125 |
| AMT0066-1 | 15.2 | 86.8 | 4 (4) | 800 | >50.0 |
| AMT0071-2 | 3.0 | 54.3 | 2 (2) | 400 | 0.8 |
| AMT0071-3 | 9.1 | 43.3 | 1 (2) | 400 | 0.8 |
| AMT0074-1 | 9.2 | 64.5 | 16 (16) | 100 | 3.125 |
| AMT0074-3 | 19.6 | 86.8 | 16 (16) | 200 | 1.56 |
| AMT0076-3 | 10.8 | 58.9 | 2 (1) | 1,600 | 6.25 |
| AMT0076-1 | 19.6 | 56.4 | 1 (1) | 800 | 0.4 |
| AMT0101-3 | 1.0 | 39.8 | 2 (2) | 400 | 1.56 |
| AMT0101-1 | 9.6 | 112.9 | 0.25 (0.25) | 200 | 1.56 |
| AMT0033-2 | 1.1 | 43.7 | 4 (2) | 800 | 1.56 |
| AMT0033-3 | 13.2 | 36.1 | 2 (2) | 800 | >50.0 |
| AMT0041-1 | 5.6 | 56.4 | 4 (4) | 800 | 3.125 |
| AMT0041-3 | 12.8 | 46.7 | 16 (16) | >1,600 | >50.0 |
| AMT0075-1 | 7.1 | 58.2 | 1 (1) | 400 | 0.8 |
| AMT0075-4 | 23.4 | 61.9 | 2 (2) | 400 | 0.8 |
Strain pairs from individual patients are grouped by spacing. Strain pairs in boldface indicate that a hip mutant emerged during the course of infection with no increase in antimicrobial resistance.
For ofloxacin, MICs for persister progeny are shown in parentheses.
Biofilms were grown by the hanging-peg model (5). The device used for biofilm formation in this study is a platform carrying 96 polystyrene pegs (Nunc no. 445497) with a peg hanging into each microtiter plate well (Nunc no. 269787). For biofilm formation, the device was placed in its original sterile tray filled with MHB and cells (104/ml) and incubated for 24 h at 37°C on a tilting shaker that provides a shearing force. After biofilms were formed on the pegs, they were washed in MHB and the device with intact biofilms was placed in a microtiter plate with fresh MHB for drug susceptibility testing. Following an 8-h incubation in the presence of an antimicrobial agent, the pegs were washed twice in MHB and the device was placed in a microtiter plate with MHB and incubated for 10 min in a water bath sonicator (Branson Ultrasonic Cleaner; Branson Cleaning Equipment Company). For each antimicrobial concentration tested, cells were collected from four parallel pegs and plated for colony counting.
Construction of strains and plasmids.
To construct the ΔmexXY strain, approximately 800-bp fragments upstream and downstream of mexXY were PCR amplified and used in an overlap extension reaction to create a single 1,600-bp product. The following primers were used: upstream primers oLM007 (5′ CCCAAGCTTTTCTCCCTGGGCAGTGCAAACAAGCGCAAC 3′) and oLM008 (5′ ATCGAGGGACACCCATGTGATGCCCCTAGCGAAACTCTCG 3′) and downstream primers oLM009 (5′ GTTTCGCTAGGGGCATCACATGGGTGTCCCTCGATTCG 3′) and oLM010 (5′ ATGCTCTAGATTGCCCCTGCTTCTCGAGCAGCTC 3′).
This product was cloned into plasmid pEX18Gm (25), using XbaI and HindIII sites, which was then transformed into DH5α. The plasmid was transferred into a recipient strain (the 96-month isolate from the index patient) by triparental mating that included the donor strain DH5α conjugation with strain HB101 and the helper HB101, followed by selection and screening for P. aeruignosa transconjugants with the deletion as previously described (43). Deletion of the mexXY genes was verified by PCR.
Persistence assay.
Persistence was measured by determining survival upon exposure to antibiotics. Time- or dose-dependent killing produces a distinct biphasic pattern with a plateau of surviving persisters (28). The concentration of antibiotics used was therefore within the persister plateau. To determine the number of persisters in the stationary phase, 3 ml of cell culture was incubated in a 10-ml culture tube with aeration for 16 h overnight in MHB medium. The cultures were exposed to ofloxacin for at least 8 h, and samples of cells were removed prior to and after ofloxacin exposure. Samples were washed, serially diluted, spot plated on MHB agar plates, and grown for at least 48 h before CFU determination.
RESULTS
Drug tolerance of longitudinal isolates from a single patient.
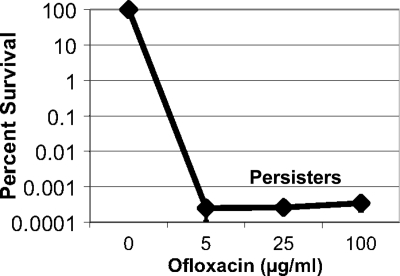
We previously reported that killing of P. aeruginosa PAO1 is biphasic, indicating the presence of drug-tolerant persister cells (48). In order to examine drug tolerance in a clinical isolate, a dose-dependent killing experiment was performed with an early isolate from a single CF patient (Fig. 1). Persisters are most abundant at the stationary state, and after overnight growth in rich medium, cells were exposed to increasing concentrations of ofloxacin. Testing in the stationary state provides an additional advantage of eliminating the differences in drug tolerance due to possible variation in growth rates among different isolates. The bulk of the population was effectively killed with 5 μg/ml ofloxacin, the maximal concentration achieved in the human plasma (22). A small fraction of 10−4 to 10−3% of cells were tolerant to 100 μg/ml ofloxacin (200× MIC), represented by the typical persister plateau in the biphasic killing plot. Applying an antibiotic at a concentration within the plateau followed by plating and counting colonies directly measures the amount of persisters formed. This approach was used to determine the level of persister cells in the clinical isolates examined in this study.
FIG. 1.
Dose-dependent killing of a clinical isolate of P. aeruginosa. Strain AMT0023-30 was isolated from an 8-month-old patient. Antibiotic was added at time zero at the indicated concentration to a stationary-phase culture, and after an 8-h incubation, surviving persister cells were plated for colony count (percent survival ± standard error of the mean [SEM]; n = 3).
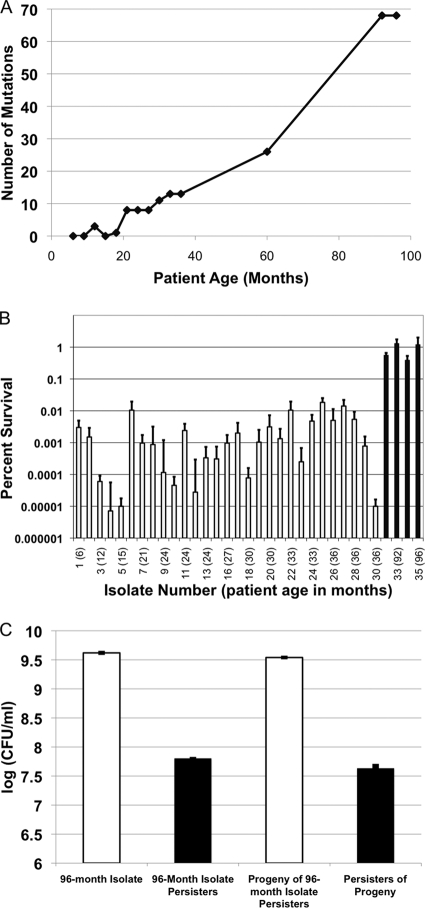
In order to test if hip mutants are selected in the course of a chronic CF infection, we examined a set of longitudinal isolates of P. aeruginosa from a single patient collected between the ages of 8 and 96 months. The set comprises 35 strains, and the genomes of the first and last isolates (AMT0023-30 and AMT0023-34, respectively) were sequenced (47). The 96-month isolate has 68 unique mutations, as compared to the first isolate. PCR amplification and sequencing of the putative mutant regions in the remaining 33 isolates showed that many of them were shared among the strains, and there was a progressive accumulation of genetic changes (Fig. 2A) (47). This showed that the infection was caused by a clonal expansion of the early isolate with potentially several lineages coevolving into a dominant infecting clone. Between months 60 and 92, a mutS mutation appeared, conferring a hypermutator phenotype on the clonal lineage (39), and there was a concomitant sharp increase in the number of mutations, from 26 to 68 in the last four isolates.
FIG. 2.
Persister levels in longitudinal clonal isolates of P. aeruginosa from a CF patient. (A) The number of mutations at the time a given strain was isolated (47); (B) persister levels (percent survival ± SEM; n = 3) of 35 longitudinal isolates of P. aeruginosa. Stationary-phase cultures were exposed to 100 μg/ml of ofloxacin for 8 h, and surviving cells were determined by colony count. hip mutants are indicated with black bars. (C) Persister progeny from the 96-month hip mutant form the same levels of persisters. The 96-month isolate was grown to the stationary phase and treated with 100× the MIC of ofloxacin for 8 h, and persister levels were determined (96-month isolate persisters). Persisters were regrown to the stationary phase (progeny of 96-month isolate persisters) and then treated with 100× the MIC of ofloxacin for 8 h, after which persister levels were determined were determined by colony count (persisters of progeny) (log [CFU/ml] ± SEM; n = 3).
The 35 isolates from the patient were cultured in MHB medium overnight, and the level of surviving persisters was determined after an 8-h incubation with ofloxacin added at 100 μg/ml. The persister level varied among the strains, but there was a dramatic, approximately 100-fold increase in surviving cells in the four late isolates, approaching 1% of the total population (Fig. 2B). This increase in persister formation suggested that the late isolates are hip mutants. One potential concern with this assay is that the late isolates, including the 96-month isolate, are hypermutators, and the hip mutants could be formed and selected for in the course of the experiment, rather than during evolution of these strains in the patient. To distinguish between these two possibilities, we tested the progeny of persisters from the 96-month isolate for their ability to form persisters. After treatement for 8 h with 100× MIC of ofloxacin, the surviving persister cells of the 96-month isolate were allowed to regrow to stationary phase in fresh medium, and subjected to a second round of killing with 100× the MIC of ofloxacin. Persister levels in the progeny did not increase, showing that hip mutants do not form in the course of the experiment. Rather, the late-term isolate is a persister mutant (Fig. 2C).
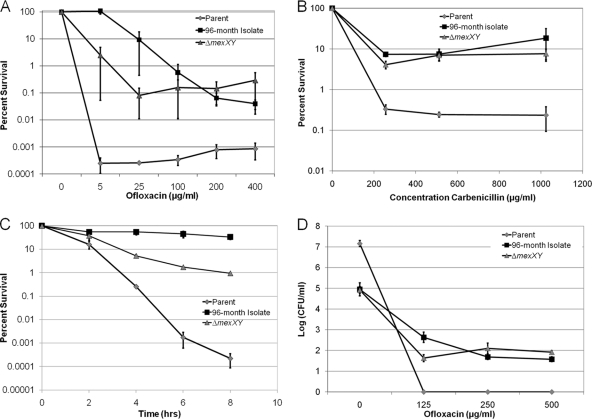
The late isolates also have an inactivating mutation in mexZ, encoding a repressor of the MexXY-OprM multidrug resistance (MDR) pump. This pump extrudes ofloxacin and other antimicrobials (38) and is frequently upregulated in isolates of P. aeruginosa from CF patients (53). The resistance of these strains to ofloxacin increased 8-fold compared to that in the early isolates (1 μg/ml) (Table 1). In order to establish whether the survival of late isolates was due to increased tolerance or increased resistance, the mexXY genes were deleted in the 96-month isolate. The resistance of the ΔmexXY knockout strain was restored to that of the early parent strain (ofloxacin MIC of 1 μg/ml) (Table 1). The survival of the parent, 96-month isolate, and isogenic ΔmexXY derivative of the 96-month isolate was then examined in a dose-dependent killing experiment with ofloxacin (Fig. 3A). The bulk of the cells of the parent isolate were highly susceptible to killing by ofloxacin, and surviving persisters formed a plateau at 10−4 to 10−3% of the total population. The 96-month isolate and its ΔmexXY derivative both showed an approximately 100-fold-higher plateau of surviving cells. The progeny of surviving persisters did not have increased resistance to ofloxacin (Table 1), indicating that surviving cells are not resistant mutants that form before or during antibiotic treatment. Since the ΔmexXY derivative and the early isolate have the same ofloxacin MIC, but ΔmexXY forms 100 times more persisters, we conclude that the 96-month isolate is a hip mutant.
FIG. 3.
Separating resistance from tolerance in a 96-month isolate of P. aeruginosa. In each experiment, the same set of strains was examined: the parent (AMT0023-30), 96-month isolate (AMT0023-34), and the ΔmexXY (KLE2000) derivative of the 96-month isolate. (A) Stationary-phase cultures were incubated for 8 h with ofloxacin, and surviving cells were enumerated by colony count (percent survival ± SEM; n = 3). (B) Exponential-phase cultures were treated for 8 h with carbenicillin. Surviving cells were enumerated by colony count (percent survival ± SEM; n = 3). (C) Stationary-phase cells were treated with tobramycin (20× MIC), and the surviving cells were enumerated by colony count (percent survival ± SEM; n = 4). (D) Twenty-four-hour biofilms were treated with ofloxacin for 8 h, and surviving cells were determined by colony count (log [CFU/ml] ± SEM; n = 4).
An important hallmark of persisters is multidrug tolerance (36). If the late isolate is indeed a hip mutant, then it should be tolerant to unrelated antibiotics. Carbenicillin, a β-lactam, only kills rapidly growing cells (20, 50) therefore a dose-dependent experiment was performed with exponentially growing cultures. The 96-month isolate had a 2-fold increase in carbenicillin MIC, not a significant change considering that the intrinsic variation in the MIC test is 100% (7). The parent strain was readily killed with carbenicillin and formed a distinct plateau of surviving persisters (Fig. 3B). The late isolate and its isogenic ΔmexXY mutant had a similar pattern of killing and formed a considerably higher number of surviving persisters than the parent. Increased carbenicillin tolerance could have resulted from a simple decrease in the growth rate of the ΔmexXY strain. However, the ΔmexXY strain actually grows slightly faster than the parent isolate (Table 1). The MexXY-OprM pump is especially effective in extruding aminoglycosides (27), and the 96-month isolate is resistant to tobramycin, with a MIC 32 times higher than that of the parent strain (Table 1). The parent strain was rapidly killed by tobramycin at the stationary phase, with little evidence for surviving persisters (Fig. 3C). The ΔmexXY strain, in contrast, showed little killing in this time-dependent experiment, despite its increased susceptibility to tobramycin. Taken together, these results demonstrate that the late isolates from this patient are hip mutants that produce increased levels of multidrug-tolerant persister cells.
Drug tolerance in biofilms.
Biofilm-like microcolonies of P. aeruginosa and quorum-sensing factors favoring biofilm development have been described in CF infection (46, 58). Biofilms also exhibit high levels of drug tolerance, largely due to the presence of persister cells (24, 48). At the same time, the involvement of biofilms in CF pathogenesis and drug tolerance remains controversial, since late isolates from CF patients often carry mutations affecting quorum sensing and biofilm formation. The late isolates (92 and 96 months) have a mutation in lasR and are reported to be deficient in biofilm formation (47). In order to investigate the effect of growth in a biofilm on drug tolerance of the hip mutants, biofilms were formed on prongs of the Calgary device (5), transferred to fresh medium, and exposed to ofloxacin. The parent strain formed a robust biofilm, as judged by the number of cells per peg (between 107 and 108 CFU/peg), but was highly susceptible to killing by ofloxacin (Fig. 3D). Under these conditions of high concentration of antibiotic and relatively long, 8-h incubation, no surviving persisters were observed. The 96-month isolate and its ΔmexXY derivative were unable to form a robust biofilm, and the number of cells per peg was approximately 100-fold lower than that of the parent strain (105 CFU/peg). Nevertheless, both the 96-month isolate and the ΔmexXY strain formed a distinct plateau of surviving persisters. This strongly suggests that it is the increased persister formation of the hip mutants rather than the ability to form biofilms that is responsible for elevated antibiotic tolerance in both the biofilm and planktonic phases of growth.
Screening P. aeruginosa isolates from different patients for hip mutants.
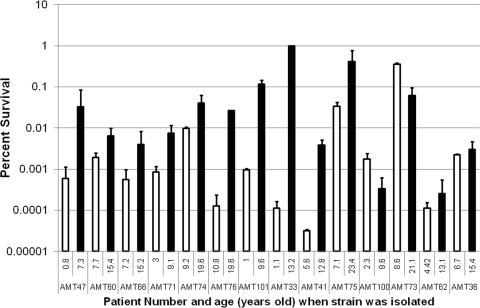
The results reported in the previous sections were obtained with a clonal lineage of P. aeruginosa strains isolated from a single CF patient. It was important to establish whether this was an isolated event or a common feature of the disease. Pairs of early/late isolates from 14 individual patients that were found to be clonal, based on multilocus sequence typing (47), were tested for their ability to form persisters. There was a wide variation in the absolute levels of persisters determined by exposure of stationary-phase cultures to ofloxacin at 50× MIC (Fig. 4). Of the 14 pairs, 10 showed an increased level of persisters in the later isolate and thus were deemed to be hip mutants. The increased frequency of persister formation was dramatic in some cases, ranging from a 100- to 10,000-fold increase. Such strong phenotypic changes are likely the result of stable mutations occurring in the late isolates, even though the complete spectrum of mutations within these strains is not known at present. These strains have a wide range of growth rates, with no correlation between growth rate and increased persister formation (Table 1). About half of the hip mutants grow faster than their clonal parental strain, while half grow slower. In addition, the high-persister phenotype is observed in the stationary phase where cells are not growing.
FIG. 4.
A screen of P. aeruginosa clinical isolates for hip mutants. Stationary-phase cultures of clonal early/late isolate pairs from 14 patients were exposed to ofloxacin (50× MIC) for 8 h, and the surviving cells were determined by colony count (percent survival ± SEM; n = 4). Early isolates are indicated with white bars, while late isolates are indicated with black bars. The patient number and age at which the tested isolates were obtained are displayed on the x axis. A hip mutant emerged in 10 of the 14 patients. A hip mutant did not emerge in the isolates from the last four patients displayed on the right side of the graph.
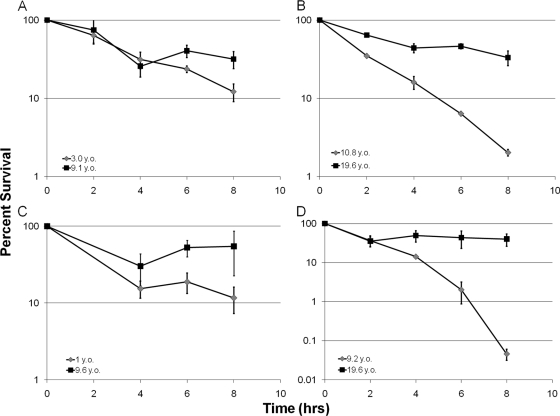
Interestingly, among the 10 hip mutants, 7 did not show increased resistance to the tested antibiotics—ofloxacin, tobramycin, and carbenicillin (Table 1). Similarly to the late isolate from patient 1, the progeny of recovered persisters from these additional patients did not have an increase in resistance to ofloxacin (Table 1). Increased persister formation due to the acquisition of hip mutations appears to be the sole mechanism of increased tolerance to killing by antimicrobials in these isolates. In order to further probe the nature of these additional hip mutants, four of the strains that did not show increased antibiotic resistance were tested for survival with a different antibiotic, tobramycin, in a time-dependent killing experiment with stationary-phase cells. In all cases, the hip mutant identified in the screen with ofloxacin showed increased survival when treated with 20× the MIC of tobramycin (Fig. 5A to D). This suggests that the hip mutants emerging in independent patients produce increased levels of multidrug-tolerant persister cells.
FIG. 5.
(A to D) Time-dependent killing of paired early/late isolates from different patients by tobramycin. Strains were grown to the stationary phase, treated with tobramycin (20× MIC), and the surviving cells were enumerated by colony count (percent survival ± SEM; n = 3). The age of the patient at the time the isolate was obtained (years old [y.o.]) is indicated.
DISCUSSION
Chronic infections present a formidable therapeutic challenge. The most difficult case is that of the incurable infection of CF patients. However, all chronic infections share a puzzling property—the causative pathogen is not necessarily resistant to antibiotics in vitro, while the infection is difficult and even impossible to eradicate.
We reasoned that periodic application of high doses of antibiotics, which is how CF patients are typically treated, would select for hip mutants of P. aeruginosa. Persister levels of longitudinal isolates of P. aeruginosa from a single patient were determined by measuring the number of cells surviving treatment with a high dose of ofloxacin. Indeed, there was a dramatic, 100-fold increase in persister cells in the last four isolates obtained at 92 and 96 months. The late isolates also acquired a mutation in the MexZ repressor, leading to overexpression of the MexXY-OprM MDR pump and a modest increase in the ofloxacin MIC (from 1 to 8 μg/ml). Knocking out the mexXY locus from the 96-month isolate restored the resistance levels to that of the parent isolate but had no effect on the level of persister cells surviving a challenge with high concentrations of antibiotic. Importantly, the progeny of surviving persisters in all strains examined in this study did not show increased antibiotic resistance as measured by the MIC assay. This excludes the possibility of selection for antibiotic resistance over the course of the experiments.
This means that the 96-month isolate is indeed a hip mutant, and its survival of antibiotic therapy is due primarily to the presence of persister cells and not classical resistance mediated by efflux pumps or other mechanisms. The persisters of the 96-month isolate and its isogenic ΔmexXY mutant showed high levels of tolerance not only to ofloxacin but also to carbenicillin and tobramycin. This demonstrates multidrug tolerance of hip persisters, a property previously documented for persisters of wild-type P. aeruginosa (48). Testing of paired strains from 14 additional patients showed that in most cases (10/14), there was a considerable increase in persister levels in the late isolate from a patient. Interestingly, most of the late hip mutants (7/10) had no change in susceptibility to ofloxacin, carbenicillin, and tobramycin when compared to their clonal parent strain, suggesting that classical acquired resistance plays little or no role in the increased tolerance of hip mutants.
We also examined the pattern of killing of the 96-month hip mutant of P. aeruginosa in a biofilm model. This isolate acquired a lasR mutation that has been reported to affect biofilm formation (47). Indeed, a biofilm of the 96-month hip mutant grown on a polystyrene peg had 100 times less cells than the parent strain. In spite of this, the hip mutant showed considerably better survival than the parent strain in the biofilm (Fig. 3D). However, this survival was considerably less than in a stationary culture of the same strain. A significant fraction of P. aeruginosa is likely to exist in a stationary state-like population in the CF airway (58). These findings suggest that persister formation, rather than the ability to form a biofilm, is the main cause of recalcitrant CF airway infection.
In the evolution of P. aeruginosa within an individual patient, the type of mutations selected provides a telling guide to factors that enable pathogen survival. In the clonal lineage from a single patient examined in this study, there was a gradual accumulation of mutations over time, as well as a jump between months 60 and 92, coinciding with the appearance of a mutS mutation conferring a hypermutator phenotype on the subsequent strains (47). Hypermutators are commonly found in clinical isolates, and they speed up the adaptive evolution that allows the pathogen to thrive in the CF airway (39). The genome sequence of the 96-month isolate we tested in this study showed that it had acquired 42 additional mutations compared to the last isolate with parental levels of persister cell formation. Those 42 mutations include mutS (DNA mismatch repair), mexZ (repressor of MexXY-OprM expression), lasR (quorum sensing and biofilm defect), and at least eight important virulence factors (47). The general themes of these changes are decrease in virulence, apparently to avoid provoking an immune response, decrease in biofilm formation, modest increase in resistance, and a dramatic increase in the ability to form multidrug-tolerant persisters. One or more of these 42 mutations will confer the hip phenotype.
Identifying the hip mutations and establishing the mechanism by which they increase persister formation are the next steps for this project. Among the 42 mutations of the late isolate, there are no obvious candidates for persister genes. Such candidates would include genes coding for proteins that can shut down cellular functions, causing dormancy and accompanying multidrug tolerance. Good examples from the study of E. coli persisters are the toxins HipA, (9, 42), RelE (29), and TisB (13). Of the 42 mutations in the 96-month isolate, 19 are in genes with unknown functions, and one is a 188-kb deletion that results in the complete loss of 139 genes. None of the mutated genes in the late isolate from patient 1 have been reported in previous studies of persisters. The best approach to identify candidate hip mutations then is to compare the genomes of several hip isolates and identify common mutations in genes or genetic pathways.
One of the potential benefits of finding the mechanism of drug tolerance is advancing development of therapy. Currently used therapies have limited activity against persisters. Failure of conventional treatments has led to application of very high doses of antibiotics in the form of aerosols: aerosolized tobramycin reaches peak concentrations of 1,237 μg/g of sputum, with levels that are ≥25 times higher than the MIC of P. aeruginosa isolates from patients (17). Inhalation levofloxacin (in post-phase II development) achieves up to 1,760 μg/ml of sputum (>50× MIC) at the site of infection (31). At these concentrations, the drugs will effectively kill resistant mutants but are still unable to eradicate the persister-forming pathogens. Another recent approach is the revival of colistin, an antimicrobial peptide that disrupts the outer membrane of Gram-negative species (8). This membrane-acting antimicrobial will kill all cells, including persisters, if given at a high enough concentration, but not surprisingly it is also toxic and has to be administered sparingly. Our work highlights the need to develop new strategies to treat persister cells that are tolerated by the patient.
Our findings suggest, for the first time, a link between persisters and the clinical manifestation of the disease. We recently discovered that Candida albicans isolates from patients with oropharyngeal candidiasis with extended carriage are high-persister strains (35). Emergence of hip mutants then may be a general feature of recalcitrant infectious diseases.
Acknowledgments
We thank Massimo Merighi for assistance with P. aeruginosa strain construction.
L.R.M. is supported by a fellowship from the Cystic Fibrosis Foundation. J.L.B. is supported by a core laboratory grant from the Cystic Fibrosis Foundation. S.L. is supported by National Institutes of Health (NIH) grant AI021451. K.L. is supported by NIH grants R01 GM061162-05A1 and T1R01AI085585-01 and by ARO grant W9911NF-09-1-0265.
Footnotes
Published ahead of print on 8 October 2010.
REFERENCES
- 1.Alhede, M., T. Bjarnsholt, P. O. Jensen, R. K. Phipps, C. Moser, L. Christophersen, L. D. Christensen, M. van Gennip, M. Parsek, N. Hoiby, T. B. Rasmussen, and M. Givskov. 2009. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155:3500-3508. [DOI] [PubMed] [Google Scholar]
- 2.Bjarnsholt, T., P. Ø. Jensen, M. J. Fiandaca, J. Pedersen, C. R. Hansen, C. B. Andersen, T. Pressler, M. Givskov, and N. Høiby. 2009. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44:547-558. [DOI] [PubMed] [Google Scholar]
- 3.Bjarnsholt, T., K. Kirketerp-Moller, P. O. Jensen, K. G. Madsen, R. Phipps, K. Krogfelt, N. Hoiby, and M. Givskov. 2008. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 16:2-10. [DOI] [PubMed] [Google Scholar]
- 4.Burns, J. L., J. M. Van Dalfsen, R. M. Shawar, K. L. Otto, R. L. Garber, J. M. Quan, A. B. Montgomery, G. M. Albers, B. W. Ramsey, and A. L. Smith. 1999. Effect of chronic intermittent administration of inhaled tobramycin on respiratory microbial flora in patients with cystic fibrosis. J. Infect. Dis. 179:1190-1196. [DOI] [PubMed] [Google Scholar]
- 5.Ceri, H., M. E. Olson, C. Stremick, R. R. Read, D. Morck, and A. Buret. 1999. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, S. K., and K. Gerdes. 2003. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 48:1389-1400. [DOI] [PubMed] [Google Scholar]
- 7.CLSI. 2008. Performance standards for antimicrobial susceptibility testing: 18th informational supplement, vol. CLSI M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Conway, S. P., K. G. Brownlee, M. Denton, and D. G. Peckham. 2003. Antibiotic treatment of multidrug-resistant organisms in cystic fibrosis. Am. J. Respir. Med. 2:321-332. [DOI] [PubMed] [Google Scholar]
- 9.Correia, F. F., A. D'Onofrio, T. Rejtar, L. Li, B. L. Karger, K. Makarova, E. V. Koonin, and K. Lewis. 2006. Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J. Bacteriol. 188:8360-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Argenio, D. A., M. Wu, L. R. Hoffman, H. D. Kulasekara, E. Déziel, E. E. Smith, H. Nguyen, R. K. Ernst, T. J. Larson Freeman, D. H. Spencer, M. Brittnacher, H. S. Hayden, S. Selgrade, M. Klausen, D. R. Goodlett, J. L. Burns, B. W. Ramsey, and S. I. Miller. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 64:512-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, B. D., L. L. Chen, and P. C. Tai. 1986. Misread protein creates membrane channels: an essential step in the bactericidal action of aminoglycosides. Proc. Natl. Acad. Sci. U. S. A. 83:6164-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Groote, V., N. Verstraeten, M. Fauvart, C. Kint, A. Verbeeck, S. Beullens, P. Cornelis, and J. Michiels. 2009. Novel persistence genes in Pseudomonas aeruginosa identified by high-throughput screening. FEMS Microbiol. Lett. 297:73-79. [DOI] [PubMed] [Google Scholar]
- 13.Dorr, T., M. Vulic, and K. Lewis. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8:e1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reference deleted.
- 15.Fournier, B., X. Zhao, T. Lu, K. Drlica, and D. C. Hooper. 2000. Selective targeting of topoisomerase IV and DNA gyrase in Staphylococcus aureus: different patterns of quinolone-induced inhibition of DNA synthesis. Antimicrob. Agents Chemother. 44:2160-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fricke, W. F., T. J. Welch, P. F. McDermott, M. K. Mammel, J. E. LeClerc, D. G. White, T. A. Cebula, and J. Ravel. 2009. Comparative genomics of the IncA/C multidrug resistance plasmid family. J. Bacteriol. 191:4750-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geller, D. E., W. H. Pitlick, P. A. Nardella, W. G. Tracewell, and B. W. Ramsey. 2002. Pharmacokinetics and bioavailability of aerosolized tobramycin in cystic fibrosis. Chest 122:219-226. [DOI] [PubMed] [Google Scholar]
- 18.Gibson, R. L., J. L. Burns, and B. W. Ramsey. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918-951. [DOI] [PubMed] [Google Scholar]
- 19.Gibson, R. L., J. Emerson, S. McNamara, J. L. Burns, M. Rosenfeld, A. Yunker, N. Hamblett, F. Accurso, M. Dovey, P. Hiatt, M. W. Konstan, R. Moss, G. Retsch-Bogart, J. Wagener, D. Waltz, R. Wilmott, P. L. Zeitlin, B. Ramsey, and Cystic Fibrosis Therapeutics Development Network Study Group. 2003. Significant microbiological effect of inhaled tobramycin in young children with cystic fibrosis. Am. J. Respir. Crit. Care Med. 167:841-849. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert, P., P. J. Collier, and M. R. Brown. 1990. Influence of growth rate on susceptibility to antimicrobial agents: biofilms, cell cycle, dormancy, and stringent response. Antimicrob. Agents Chemother. 34:1865-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilligan, P. H. 2006. Is there value in susceptibility testing of Pseudomonas aeruginosa causing chronic infection in patients with cystic fibrosis? Expert Rev. Anti Infect. Ther. 4:711-715. [DOI] [PubMed] [Google Scholar]
- 22.Guay, D. R., J. A. Opsahl, F. G. McMahon, R. Vargas, G. R. Matzke, and S. Flor. 1992. Safety and pharmacokinetics of multiple doses of intravenous ofloxacin in healthy volunteers. Antimicrob. Agents Chemother. 36:308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen, S., K. Lewis, and M. Vulic. 2008. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob. Agents Chemother. 52:2718-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison, J. J., R. J. Turner, and H. Ceri. 2005. Persister cells, the biofilm matrix and tolerance to metal cations in biofilm and planktonic Pseudomonas aeruginosa. Environ. Microbiol. 7:981-994. [DOI] [PubMed] [Google Scholar]
- 25.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman, L. R., H. D. Kulasekara, J. Emerson, L. S. Houston, J. L. Burns, B. W. Ramsey, and S. I. Miller. 2009. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J. Cyst. Fibros. 8:66-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Islam, S., H. Oh, S. Jalal, F. Karpati, O. Ciofu, N. Hoiby, and B. Wretlind. 2009. Chromosomal mechanisms of aminoglycoside resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Clin. Microbiol. Infect. 15:60-66. [DOI] [PubMed] [Google Scholar]
- 28.Keren, I., N. Kaldalu, A. Spoering, Y. Wang, and K. Lewis. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13-18. [DOI] [PubMed] [Google Scholar]
- 29.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khodursky, A. B., and N. R. Cozzarelli. 1998. The mechanism of inhibition of topoisomerase IV by quinolone antibacterials. J. Biol. Chem. 273:27668-27677. [DOI] [PubMed] [Google Scholar]
- 31.King, P., O. Lomovskaya, D. C. Griffith, J. L. Burns, and M. N. Dudley. 2010. In vitro pharmacodynamics of levofloxacin and other aerosolized antibiotics under multiple conditions relevant to chronic pulmonary infection in cystic fibrosis. Antimicrob. Agents Chemother. 54:143-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohanski, M. A., D. J. Dwyer, and J. J. Collins. 2010. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8:423-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Köhler, T., A. Buckling, and C. van Delden. 2009. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc. Natl. Acad. Sci. U. S. A. 106:6339-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaFleur, M. D., C. A. Kumamoto, and K. Lewis. 2006. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob. Agents Chemother. 50:3839-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaFleur, M. D., Q. Qi, and K. Lewis. 2010. Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob. Agents Chemother. 54:39-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis, K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48-56. [DOI] [PubMed] [Google Scholar]
- 37.LiPuma, J. J., S. Rathinavelu, B. K. Foster, J. C. Keoleian, P. E. Makidon, L. M. Kalikin, and J. R. Baker. 2009. In vitro activities of a novel nanoemulsion against Burkholderia and other multidrug-resistant cystic fibrosis-associated bacterial species. Antimicrob. Agents Chemother. 53:249-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mena, A., E. E. Smith, J. L. Burns, D. P. Speert, S. M. Moskowitz, J. L. Perez, and A. Oliver. 2008. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J. Bacteriol. 190:7910-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moyed, H. S., and K. P. Bertrand. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaber, J. A., N. L. Carty, N. A. McDonald, E. D. Graham, R. Cheluvappa, J. A. Griswold, and A. N. Hamood. 2004. Analysis of quorum sensing-deficient clinical isolates of Pseudomonas aeruginosa. J. Med. Microbiol. 53:841-853. [DOI] [PubMed] [Google Scholar]
- 42.Schumacher, M. A., K. M. Piro, W. Xu, S. Hansen, K. Lewis, and R. G. Brennan. 2009. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323:396-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schweizer, H. P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 44.Shah, D., Z. Zhang, A. Khodursky, N. Kaldalu, K. Kurg, and K. Lewis. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shigeta, M., G. Tanaka, H. Komatsuzawa, M. Sugai, H. Suginaka, and T. Usui. 1997. Permeation of antimicrobial agents through Pseudomonas aeruginosa biofilms: a simple method. Chemotherapy 43:340-345. [DOI] [PubMed] [Google Scholar]
- 46.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 47.Smith, E. E., D. G. Buckley, Z. Wu, C. Saenphimmachak, L. R. Hoffman, D. A. D'Argenio, S. I. Miller, B. W. Ramsey, D. P. Speert, S. M. Moskowitz, J. L. Burns, R. Kaul, and M. V. Olson. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spoering, A. L., M. Vulic, and K. Lewis. 2006. GlpD and PlsB participate in persister cell formation in Escherichia coli. J. Bacteriol. 188:5136-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuomanen, E., R. Cozens, W. Tosch, O. Zak, and A. Tomasz. 1986. The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J. Gen. Microbiol. 132:1297-1304. [DOI] [PubMed] [Google Scholar]
- 51.Unoson, C., and E. G. Wagner. 2008. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 70:258-270. [DOI] [PubMed] [Google Scholar]
- 52.Vázquez-Laslop, N., H. Lee, and A. A. Neyfakh. 2006. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J. Bacteriol. 188:3494-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vettoretti, L., P. Plésiat, C. Muller, F. El'garch, G. Phan, I. Attrée, A. Ducruy, and C. Llanes. 2009. Efflux unbalance in cystic fibrosis isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:1987-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vuong, C., J. M. Voyich, E. R. Fischer, K. R. Braughton, A. R. Whitney, F. R. DeLeo, and M. Otto. 2004. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 6:269-275. [DOI] [PubMed] [Google Scholar]
- 55.Walters, M. C., III, F. Roe, A. Bugnicourt, M. J. Franklin, and P. S. Stewart. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 57.Wolfson, J. S., D. C. Hooper, G. L. McHugh, M. A. Bozza, and M. N. Swartz. 1990. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and β-lactam antimicrobial agents. Antimicrob. Agents Chemother. 34:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, L., J. A. Haagensen, L. Jelsbak, H. K. Johansen, C. Sternberg, N. Hoiby, and S. Molin. 2008. In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections. J. Bacteriol. 190:2767-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]