Abstract
GOBASE (http://megasun.bch.umontreal.ca/gobase/) is a network-accessible biological database, which is unique in bringing together diverse biological data on organelles with taxonomically broad coverage, and in furnishing data that have been exhaustively verified and completed by experts. So far, we have focused on mitochondrial data: GOBASE contains all published nucleotide and protein sequences encoded by mitochondrial genomes, selected RNA secondary structures of mitochondria-encoded molecules, genetic maps of completely sequenced genomes, taxonomic information for all species whose sequences are present in the database and organismal descriptions of key protistan eukaryotes. All of these data have been integrated and organized in a formal database structure to allow sophisticated biological queries using terms that are inherent in biological concepts. Most importantly, data have been validated, completed, corrected and standardized, a prerequisite of meaningful analysis. In addition, where critical data are lacking, such as genetic maps and RNA secondary structures, they are generated by the GOBASE team and collaborators, and added to the database. The database is implemented in a relational database management system, but features an object-oriented view of the biological data through a Web/Genera-generated World Wide Web interface. Finally, we have developed software for database curation (i.e. data updates, validation and correction), which will be described in some detail in this paper.
INTRODUCTION
The biological context: organelles and their genomes
Mitochondria and chloroplasts are well defined, subcellular compartments (organelles) of the eukaryotic cell that contain their own, distinct genetic material. Mitochondrial (mt) and chloroplast (cp) DNAs code for only a small portion of the organellar components that are involved in the organellar translation machinery and in respiration/oxidative phosphorylation or photosynthesis. The large majority of organellar proteins are encoded by nuclear genes, translated in the cytoplasm and imported into the organelles (1). A few structural RNAs are also nuclear-encoded in certain species, and targeted to the organelles.
Research on mitochondria and chloroplasts covers a variety of topics. The bacterial origins of organelles raise evolutionary issues bearing on the transition from an endosymbiotic bacterium to a subcellular compartment, the functional and phylogenetic relationships between organellar and nuclear genomes, and the diversification of organelle genomes, genes and modes of gene expression. Particularly the rapidly evolving mitochondria have invented a number of intriguing molecular mechanisms such as post-transcriptional RNA editing by nucleotide deletion, insertion and modification, and trans-splicing of pre-messenger RNA. From a more biochemical point of view, research topics include the role of organelles in energy production (i.e. oxidative phosphorylation and photosynthesis), the mechanisms of protein import, the assembly process of multimeric membrane-bound enzyme complexes, and secondary and tertiary protein and RNA structure. Another array of questions is centered around the involvement of mitochondria in human diseases, the genetic variation of the mitochondrial genome within populations and the rules of extrachromosomal inheritance. This by far incomplete list of topics demonstrates the broad diversity of organelle research.
For many years, concerted efforts have been made to sequence complete organelle genomes on a large scale, e.g. by the Organelle Genome Megasequencing Program [OGMP (2); http://megasun.bch.umontreal.ca/ogmpproj.html], the Fungal Mitochondrial Genome Project [FMGP (3); http://megasun.bch.umontreal.ca/People/lang/FMGP/FMGP.html] and the Mitochondrial Genomics Workgroup [(4); http://biology.lsa.umich.edu/~jboore/index.html]. Currently, 116 complete mtDNA and 17 cpDNA sequences are available in public domain repositories. With the exception of viruses, organelle DNAs constitute the largest set of completely sequenced genomes, which makes them ideal for comparative genome studies.
The body of currently available organelle data includes detailed information about the structures of organellar enzyme complexes and their catalytic functions, protein import and processing pathways, DNA replication and transcription mechanisms, ultrastructural architecture, and genetic make-up and inheritance. However, this information is widely dispersed over numerous data sources such as books, journals, theses and electronic data repositories, so that, even for experts in the field, searching for relevant information has become increasingly difficult and time-consuming. For these reasons, a well structured and fully integrated database has become a crucial asset for tapping into this hard-to-exploit wealth of information.
A formal framework for organelle data
In order to take full advantage of the large body of dispersed organelle data and to integrate it with information about the organisms (taxonomy, morphology, etc.) that harbor these organelles, the organelle genome database project, GOBASE, was initiated in 1995. This database has been operational and publicly accessible via the Internet (World Wide Web, WWW) since 1996, and due to its fully validated contents, intuitive layout and sophisticated searching capabilities, it has been used frequently by the mitochondrial research community.
The first version of GOBASE was portrayed in an earlier issue of this journal (5), including a detailed description of the database front-end, the structure of the database and its implementation. In the present report, we will focus on how GOBASE compares with other databases, and then discuss software tools that assist in a central concern of database curation: data validation and updates.
SEQUENCE REPOSITORIES VERSUS QUERIABLE DATABASES
Public-domain biological sequence repositories such as GenBank (6) (now maintained by the National Center for Biotechnology Information, NCBI), the DNA DataBank of Japan (DDBJ) (7) and the European Molecular Biology Laboratory (EMBL) (8) are important assets in molecular biology research. However, these repositories are archival in nature, i.e. deposited sequence data are only validated to a limited degree (e.g. protein translation), and the nomenclature of genes and gene products is not standardized. Furthermore, the data retrieval systems (e.g. Entrez; 6) only support queries of moderate complexity.
A few observations will suffice to demonstrate the limitations of queries in public sequence repositories. First, submitters of sequence records may apply various names to gene homologs in different species, or even to the same gene of a given species (e.g. 5S, rrn5 or rrf is used to designate genes coding for 5S rRNA, a component of the ribosome). This inconsistency, and the fact that searches on the basis of gene product names are not supported, make it virtually impossible to directly identify gene homologs in the public data repositories. Sequence similarity searches may alleviate this shortcoming to some degree, but this approach is not only time-consuming, but also yields ambiguous results for poorly conserved genes such as 5S rRNA. Second, records are often released despite incomplete or inaccurate sequence feature annotations (e.g. lacking gene names or unspecified cellular location of the genome from which the sequences were obtained). Third, a number of important biological features such as cellular location and general functions of gene products are not searchable fields in public sequence repositories. The only way to zero in on these crucial features is to perform text searches in all fields, but this procedure often returns numerous false positives. For instance, the query for mitochondrial components involved in translation, using the advanced Entrez search clause
[mitochondr*[cellular_location]] AND [ translation [All fields]]
not only yields a fraction of the expected genes but also hundreds of false hits.
DISTINCT FEATURES OF GOBASE
GOBASE eliminates many of the aforementioned limitations. In order to reflect basic biological concepts, its database scheme classifies the biological information into 10 fundamental categories or entities (SEQUENCE, GENE, PROTEIN, SIGNAL, TAXON, etc.) with numerous well defined attributes (Table 1). It is important to point out that we did not adopt the classification scheme of the International Nucleotide Sequence Database Consortium (NCBI, EMBL, DDBJ), because it does not sufficiently distinguish between high-level and low-level biological categories (i.e. general categories such as CDS, EXON, INTRON, as well as specific ones such as iDNA, D-loop, CAAT_signal, are equally represented as ‘Feature Keys’). A second distinctive feature of GOBASE is that it cross-links the information contained in all categories, thus enabling complex biological queries that are currently not feasible in the public-domain databases. For illustration, we list below several example queries that can be formulated in GOBASE, but not in other publically accessible databases: (i) retrieve all mitochondrial-encoded 5S rRNAs; (ii) retrieve all proteins that are involved in animal mitochondrial translation; (iii) retrieve all complete coding sequences (DNA) for cytochrome c oxidase from protists; (iv) retrieve all complete protein sequences encoded by fungal mitochondrial plasmids; (v) retrieve all identified genes (not ORFs) located in mitochondrial introns; (vi) retrieve all ORFs of liverwort mtDNA, except intron ORFs; (vii) retrieve all organisms using a mitochondrial translation code with TGA=tryptophan.
Table 1. The GOBASE data scheme.
| Category |
Attributes |
| Sequence | Type, Species name, Taxon name, Taxon division, Completeness, Plasmid, Topology, Map availability, Seq. Length, Sub. date, Last update date, GBK, PIR, SWISS-PROT, Entrez and EMBL acc.#, GOBASE ID. |
| Gene | Gene, Product, Species and Taxon names, ORF, Product type, Taxon division, Gene included in intron, Genetic code, Pseudo, Partialness, Trans-spliced, Chloroplast origin, Contains intron(s), Plasmid encoded, Gene location on sequence, Upstream genes, Downstream genes, Entrez and GBK acc.#, GOBASE gene and seq. ID. |
| Protein | Product, Gene, Species and Taxon names, General function, Enzyme complex, EC no., Taxon division, Partialness, Plasmid, Seq. length, SWISS-PROT and Entrez acc.#, GOBASE ID. |
| RNA | Gene, Product, Species and Taxon name, RNA type, Taxon division, Partialness, Secondary structure availability, Entrez and GBK acc.#, GOBASE ID. |
| Exon | Gene, Species and Taxon names, Taxon division, Exon number, Partialness, Location on seq., Entrez and GBK acc.#, GOBASE ID. |
| Signal | Promoter, Processing site, Stem loop, Translations initiation, Replication origin, D-loop, Location on seq., Entrez and GBK acc.#, GOBASE ID. |
| Intron | Gene, Species and Taxon names, Taxon division, Intron number, Partialness, Contains genes or ORFs, Secondary structure availability, Location on seq., Entrez and GBK acc.#, GOBASE ID. |
| Gene and Product Class | Gene and product names, Product type, Product general function, Enzyme complex, EC#, GOBASE ID. |
| Map | Species name, PID record availability, Sequence availability. |
| Taxonomy | Division, Rank, Scientific name, Synonym, Mitochondrial genetic code, Map, PID record availability, GOBASE ID. |
Biological information is classified into 10 fundamental categories with numerous attributes designed specifically for comparative genomics. The category names correspond to the names of query pages in the database. GBK, GenBank; acc.#, accession number; Seq. length, sequence length; Sub. date, submission date; ID, identifier; PID, Protist Image Database, see text.
It is obvious that a complex query capability is only meaningful when data are complete, correct and up-to-date, which is another important mission of GOBASE. Data extracted from GenBank are fully validated with regard to gene and gene product nomenclature, genetic code, cellular location and more, before being presented to the public. Since this requires significant inputs from our biological experts, numerous helper utilities have been developed and will be described below.
In addition to the information available from GenBank (sequence and taxonomy data), a variety of other data types have been integrated into GOBASE. For instance, for all sequences present in the database, relevant information on the gene function can be retrieved (query page ‘Gene and Products’, Table 1), biochemical pathways can be inspected via web links to specialized enzyme databases, organismal information about key protistan eukaryotes can be requested via links to the Protist Image Database (http://megasun.bch.umontreal.ca/protists/protists.html), and selected RNA secondary structure diagrams and genetic mtDNA maps are available. A sizable portion of these latter data have been generated by the GOBASE team in collaboration with M. W. Gray and M. Schnare (Dalhousie University, Halifax, NS, Canada). Finally, GOBASE has adopted the four-kingdom scheme of eukaryote taxonomy (animals, fungi, plants and protists), thus reflecting a widely accepted view, which is otherwise not supported in most other molecular databases.
GOBASE VERSUS OTHER MITOCHONDRIAL DATABASES
In the past five years, several other mitochondrial databases have emerged, but their aim, data content and functionality are very different from GOBASE. MitoDat (9), MITOMAP (10) and MitOP (11) and AMmtDB (12) specialize in the study of human diseases and disorders that are associated with mitochondrial mutations and dysfunctionalities, and in population-associated variations of mtDNA in humans and animals. MitBASE (13) is taxonomically broader, and in this sense shares more common ground with GOBASE than the other mitochondrial databases, but the focus of MitBASE is on data compilation, with a special emphasis on mtDNA variants and RNA editing in plants, while the focus of GOBASE is on query capability for comparative genomics studies.
In summary, GOBASE is unique in that it integrates most diverse data types related to mitochondria from all eukaryotes (it will soon also include data from chloroplasts and model eubacteria, as described below) and provides carefully validated and completed sequence annotations, in conjunction with complex search capabilities for data retrieval. In its current form, GOBASE can be regarded as one of the most accurate sources of mitochondrial sequence information.
DATA VALIDATION
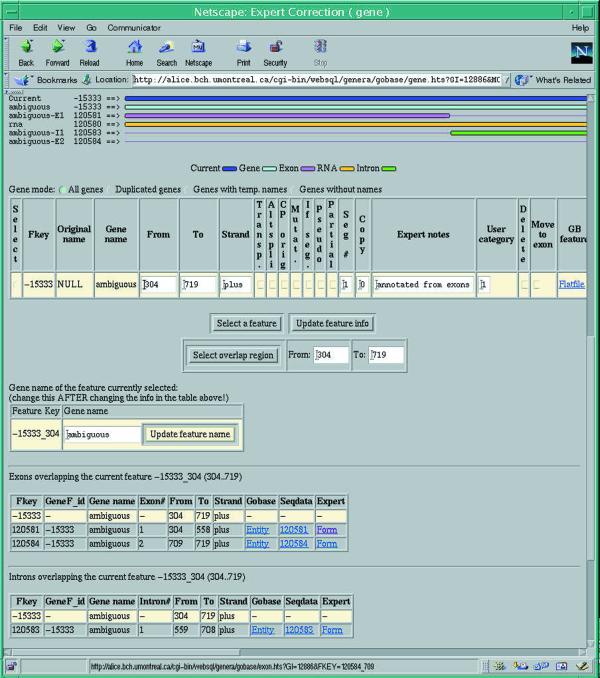
As public sequence repositories such as GenBank are archival in nature, data validation is performed only to a limited degree, and consequently, the quality and completeness of annotations vary strongly among records. As a result, a sizable fraction of the sequence information in the public databases is either difficult to spot (missing annotations about source, nature and type of genetic features) or even misleading due to incorrect annotations. The elimination of these limitations is the raison-d’être of GOBASE. However, with currently about 84 000 (DNA, RNA and protein) sequence records stored in GOBASE, expert data validation cannot be achieved by case-to-case visual inspection of all records, but requires substantial assistance from software tools. Therefore, we have developed a number of SQL procedures to extract potentially inconsistent and erroneous sequence features (overlapping genes; introns without downstream exons, over/under-sized genes, etc.). Detection of other, more subtle errors and omissions, like mis-identified and overlooked (especially tRNA) genes, is currently tackled by gene identification programs, executed in batch mode by UNIX scripts (note that due to their unorthodox structure, mitochondrial tRNAs are often difficult to identify with currently available search programs). A listing of problematic records is then presented to the experts on a correction web page (Fig. 1), which allows inspection of all available and newly detected features, and direct modification of data values in the database back-end. It should be noted that we now provide GOBASE users with a description of the nature and rationale for such corrections, via hyperlinks (‘Expert notes’) on the result pages.
Figure 1.
GOBASE’s web-based expert correction form. Potential erroneous data are presented to the expert under the following categories: missing gene name; overlapping genes; duplicated genes; etc. The expert form permits graphical display of the genetic elements of a particular sequence (upper part of the page) and access to related entities, such as INTRON, EXON (lower part of the page). The expert selects a certain feature (‘Fkey’) with the ‘Select’ button, modifies its attributes such as location (‘From’, ‘To’, ‘Strand’), trans-splicing (‘Transp.’), completeness (‘Partial’), gene name (‘Gene name’; in a separate box, in the middle of the page), adds an expert note (‘Expert notes’) for display to the public, or deletes the feature (‘Delete’). After entering the new values, they are submitted by clicking on ‘Update feature info’ or ‘Update feature name’ to the database back-end for modification of the corresponding tables.
THE CURRENCY MANAGER
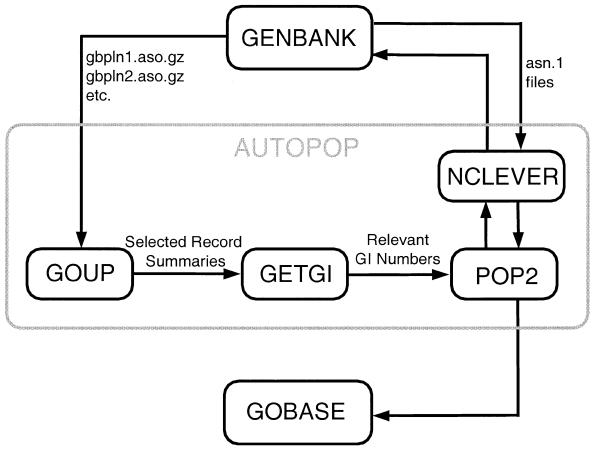
Data currency crucially determines the value of any, but especially the exponentially growing molecular sequence database. To keep up with the rapid expansion of public sequence archives, we have developed a suite of programs to retrieve data from GenBank and to populate GOBASE tables with a minimum of human intervention. Our goal is to update the sequence and taxonomic data in GOBASE at least once per month. The data actualization process currently involves the following three consecutive steps: (i) identification of updated records and new entries related to mitochondrial-encoded sequences in GenBank’s cumulative update files; (ii) gene and product name standardization; and (iii) population of GOBASE tables and transfer of previously made expert corrections. To carry out these steps, we have enhanced our original currency manager (now dubbed AUTOPOP), with regard to automation. As illustrated in Figure 2, AUTOPOP coordinates the execution of three specialized helper utilities (GOUP, GETGI and POP2) that scan the GenBank release for relevant records, parse and extract new or updated entries of interest, and populate GOBASE tables. For Genbank release no.118 (1 SEP 2000), it took ∼5 days to complete the population process, involving execution of close to 20 SQL procedures that fill GOBASE’s primary tables. AUTOPOP is not only designed to replace tedious manual operations, but it is less error-prone than the previous, mostly manually executed procedures, because it verifies proper completion at each step, and requests human (database manager and/or biological expert) interventions when problems occur.
Figure 2.
Flowchart of the software tool AUTOPOP that synchronizes data in GOBASE with new GenBank releases. AUTOPOP begins with a call to GOUP, which downloads from NCBI’s ftp server, the released divisional sequence files (in ASN.1 format). Once downloaded, these sequence files are decompressed and decoded by asntool (NCBI Toolkit), and summaries are produced of the individual records. These summaries contain submission and modification dates, GI numbers and the information whether or not the keyword ‘mitoch*’ or ‘kinetoplast’ is present in the record’s datafields ‘descr.source.genome’, ‘descr.modif’ or ‘descr.title’ of the ASN.1 record. The summaries are then passed on to GETGI, which extracts the GI numbers of records that are already in GOBASE, but have been updated, and of those that are not in GOBASE, but contain the keyword ‘mitoch*’ or ‘kinetoplast’. The extracted GI numbers are then used by POP2 as inputs to NCLEVER (14) to download the corresponding ASN.1 records via NCBI’s network retrieval system. To avoid duplications, records that have been treated (cached) in previous runs are skipped. Finally, the retrieved ASN.1 records are used to populate GOBASE tables.
FUTURE DEVELOPMENTS
We will continue to improve the mitochondrial data sets already present in GOBASE, by including more complete protist organismal information (hyperlinks pointing to external databases), genetic maps, secondary structures of tRNAs (run-time generated), ribosomal RNAs, RNase P RNAs, and group I and II introns.
To transform GOBASE from a mitochondrial into a comprehensive comparative organelle database, we are currently adding chloroplast data to the database. We expect to release this version 5 of GOBASE at the beginning of the year 2001. In a subsequent step, we will also include in the database complete genomes of α-proteobacteria and cyanobacteria that are close relatives of the mitochondrial and chloroplast ancestors.
Finally, we plan to build a sequence analysis suite (‘workbench’) into GOBASE, to allow immediate analysis on data sets compiled through GOBASE queries. The workbench will support basic sequence analysis functions such as identification of reading frames, sequence similarities and motifs; multiple sequence alignments; phylogenetic and protein structure analyses; detection of organellar tRNAs and introns; gene order analysis; generation of genetic maps; and sequence feature annotation (as part of our goal to offer online confidential data preparation, for submissions to public sequence repositories).
Acknowledgments
ACKNOWLEDGEMENTS
We wish to acknowledge Tim G. Littlejohn (Australian National Genomics Information Service, ANGIS, Sidney, NS, AU) for his valuable contributions during the design phase of GOBASE, Pierre Rioux (Incyte Genomics Inc., Palo Alto, CA, USA) and Nicolas Brossard (Wysdom, Montreal, PQ, Canada) for their contributions to the database and web interface development and Maria Korab-Laskowska (National Center for Biotechnology Information, Bethesda, MD, USA) for her role in database design and management. We thank Alexander Nip for substantial technical support and assistance in the preparation of the manuscript, and M. W. Gray and M. Schnare (Dalhousie University, Halifax, NS, Canada) for designing the secondary structure models for ribosomal RNAs available through GOBASE. This research is supported by the Medical Research Council Canada (MRC)/Canadian Institutes of Health Research (CIHR) (current grant no. GOP-15331), an IMASCO fellowship and a Canadian National scholarship of the Canadian Institute of Advanced Research (CIAR) to B.F.L. and G.B., and a generous equipment grant from SUN Microsystems (Palo Alto, CA, USA).
References
- 1.Lang B.F., Gray,M.W. and Burger,G. (1999) Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet., 33, 351–397. [DOI] [PubMed] [Google Scholar]
- 2.Lang B.F., Seif,E., Gray,M.W., O’Kelly,C.J. and Burger,G. (1999) A comparative genomics approach to the evolution of eukaryotes and their mitochondria. J. Eukaryot. Microbiol., 46, 320–326. [DOI] [PubMed] [Google Scholar]
- 3.Paquin B., Laforest,M.J., Forget,L., Roewer,I., Wang,Z., Longcore,J. and Lang,B.F. (1997) The fungal mitochondrial genome project: evolution of fungal mitochondrial genomes and their gene expression. Curr. Genet., 31, 380–395. [DOI] [PubMed] [Google Scholar]
- 4.Boore J.L. (1999) Animal mitochondrial genomes. Nucleic Acids Res., 27, 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korab-Laskowska M., Rioux,P., Brossard,N., Littlejohn,T.G., Gray,M., Lang,B.F. and Burger,G. (1998) The organelle genome database project (GOBASE). Nucleic Acids Res., 26, 138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2000) GenBank. Nucleic Acids Res., 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tateno Y., Miyazaki,S., Ota,M., Sugawara,H. and Gojobori,T. (2000) DNA data bank of Japan (DDBJ) in collaboration with mass sequencing teams. Nucleic Acids Res., 28, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker W., van den Broek,A., Camon,E., Hingamp,P., Sterk,P., Stoesser,G. and Tuli,M.A. (2000) The EMBL nucleotide sequence database. Nucleic Acids Res., 28, 19–23. Updated article in this issue: Nucleic Acids Res. (2001), 29, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemkin P.F., Chipperfield,M., Merril,C. and Zullo,S. (1996) A World Wide Web (WWW) server database engine for an organelle database, MitoDat. Electrophoresis, 17, 566–572. [DOI] [PubMed] [Google Scholar]
- 10.Kogelnik A.M., Lott,M.T., Brown,M.D., Navathe,S.B. and Wallace,D.C. (1997) MITOMAP: an update on the status of the human mitochondrial genome database. Nucleic Acids Res., 25, 196–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scharfe C., Zaccaria,P., Hoertnagel,K., Jaksch,M., Klopstock,T., Dembowski,M., Lill,R., Prokisch,H., Gerbitz,K.D., Neupert,W., Mewes,H.W. and Meitinger,T. (2000) MITOP, the mitochondrial proteome database: 2000 update. Nucleic Acids Res., 28, 155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanave C., Liuni,S., Licciulli,F. and Attimonelli,M. (2000) Update of AMmtDB: a database of multi-aligned metazoa mitochondrial DNA sequences. Nucleic Acids Res., 28, 153–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Attimonelli M., Altamura,N., Benne,R., Brennicke,A., Cooper,J.M., D’Elia,D., Montalvo,A., Pinto,B., De Robertis,M., Golik,P., Knoop,V., Lanave,C., Lazowska,J., Licciulli,F., Malladi,B.S., Memeo,F., Monnerot,M., Pasimeni,R., Pilbout,S., Schapira,A.H., Sloof,P. and Saccone,C. (2000) MitBASE: a comprehensive and integrated mitochondrial DNA database. The present status. Nucleic Acids Res., 28, 148–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rioux P., Gilbert,W.A. and Littlejohn,T.G. (1994) A portable search engine and browser for the Entrez database. J. Comp. Biol., 1, 293–295. [DOI] [PubMed] [Google Scholar]