Abstract
Plasmids pRAS3.1 and pRAS3.2 are natural variants of the IncQ-2 plasmid family, that except for two differences, have identical plasmid backbones. Plasmid pRAS3.1 has four 22-bp iterons in its oriV region, while pRAS3.2 has only three 6-bp repeats and pRAS3.1 has five 6-bp repeats in the promoter region of the mobB-mobA/repB genes and pRAS3.2 has only four. In previous work, we showed that the overall effect of these differences was that when the plasmid was in an Escherichia coli host, the copy numbers of pRAS3.1 and pRAS3.2 were approximately 41 and 30, respectively. As pRAS3.1 and pRAS3.2 are likely to have arisen from the same ancestor, we addressed the question of whether one of the variants had an evolutionary advantage over the other. By constructing a set of identical plasmids with the number of 22-bp iterons varying from three to seven, it was found that plasmids with four or five iterons displaced plasmids with three iterons even though they had lower copy numbers. Furthermore, the metabolic load that the plasmids placed on E. coli host cells compared with plasmid-free cells increased with copy number from 10.9% at a copy number of 59 to 2.6% at a copy number of 15. Plasmid pRAS3.1 with four 22-bp iterons was able to displace pRAS3.2 with three iterons when both were coresident in the same host. However, the lower-copy-number pRAS3.2 placed 2.8% less of a metabolic burden on an E. coli host population, and therefore, pRAS3.2 has a competitive advantage over pRAS3.1 at the population level, as pRAS3.2-containing cells would be expected to outgrow pRAS3.1-containing cells.
Plasmids of the IncQ family are characterized by their relatively small size (∼6 to 15 kb), their ability to replicate in a very wide range of bacterial host cells, and by being readily mobilizable by certain self-transmissible plasmids, in particular the broad-host-range IncP plasmids such as RP4 or RK2 (22). IncQ family plasmids have been subdivided into the IncQ-1 and IncQ-2 groups depending on whether their mobilization genes are of the three-gene IncQ type or the five-gene IncP type. Both subgroups of IncQ family plasmids have similar replicons that consist of three genes that encode a helicase (repA), a primase (repB), a DNA-binding initiator protein (repC), and an oriV region. The oriV region typically contains three complete, identical 22-bp iterons (or 20-bp iterons with 2-bp spacers) that serve as the binding site for RepC proteins (18, 22). Although some IncQ family plasmids may possess more than three copies of the iterons, the additional iterons are either partial copies, have sequence variations, or lack the spacer regions (22). It has been shown that even a single-base-pair replacement in a single iteron could result in a nonfunctional iteron-containing region and the inability of the IncQ-1 plasmid RSF1010 to replicate (19). Furthermore, in IncQ plasmids, the iterons have been shown to serve as the primary incompatibility determinants with cloned iterons on their own being able to displace the plasmid from which they were derived in the absence of selection. This displacement is a result of competition for RepC binding, and the rate at which the IncQ-like plasmid was displaced was dependent on the number of iterons cloned in trans (16).
Plasmids pRAS3.1 and pRA3.2 are almost identical IncQ plasmids that were isolated from the fish pathogen Aeromonas salmonicida (pRAS3.2) or atypical A. salmonicida (pRAS3.1 and pRAS3.2) in Norway (12). The mobilization and replication genes of pRAS3 plasmids are similar in order and sequence to the mobilization and replication genes of both pTF-FC2 (7) and pTC-F14 (9), with greater sequence identity to pTF-FC2 than to pTC-F14 (17). Together these plasmids constitute the only known members of the IncQ-2 subgroup of IncQ plasmids. The plasmid backbones of pRAS3.1 and pRAS3.2 differ in only two features. One of these differences is in the promoter region of the mobB-mobA/repB genes where pRAS3.2 has four 6-bp GCGGGG repeats, while pRAS3.1 has a fifth identical 6-bp repeat (Fig. 1). The additional 6-bp repeat was shown to increase the level of transcription of the mobB-mobA/repB genes by ∼2-fold, and the plasmid copy numbers (PCNs) of two plasmids that were identical except that one plasmid had an additional 6-bp repeat increased by a corresponding ∼2-fold. The second difference was in the oriV region, where pRAS3.2 has three copies of a perfectly conserved 22-bp iteron typical of the IncQ family plasmids, while pRAS3.1 is unusual in that it has an additional, identical, fourth 22-bp iteron. Comparison of pRAS3 derivative plasmids indicated that the effect of an additional 22-bp iteron was to reduce the copy number of the plasmid by 26 to 30%, provided the number of 6-bp repeats was the same for both plasmids.
FIG. 1.
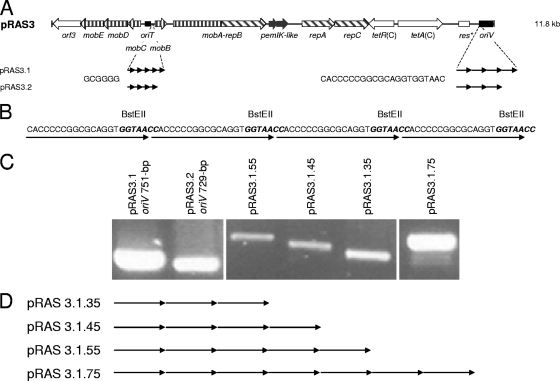
(A) Genetic map of pRAS3 plasmids with the differences in the plasmid backbones of pRAS3.1 and pRAS3.2 shown below the map. (B) Nucleotide sequence of the four 22-bp repeats of pRAS3.1 indicating the location of the unique BstEII site used to construct plasmids with different numbers of iterons. (C) Agarose gel showing oriV regions of pRAS3 containing different numbers of iterons. (D) Diagram showing derivatives of pRAS3.1 with different numbers of iterons.
The overall effect of pRAS3.1 in having a PCN-increasing additional 6-bp repeat in the promoter region of the mobB-mobA/repB genes but a PCN-decreasing additional 22-bp iteron compared with pRAS3.2 was that the PCN of pRAS3.1 was 41 while that of pRAS3.2 was 30 copies per chromosome. This raised the question as to whether one of the two natural, nearly identical pRAS3 plasmids has a competitive advantage over the other. For example, did one variant have a stability advantage over the other, would one plasmid be able to displace the other when both were present in the same host cell, and how did the plasmids compare with respect to the metabolic load placed on the host? Were there any observable advantages or disadvantages to having three or four 22-bp iterons besides their effect on copy number? This is a report on the investigation into these questions carried out in Escherichia coli host cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Escherichia coli strains, cloning vectors, and plasmid constructs are shown in Table 1. Cultures of E. coli were grown in either Luria-Bertani (LB) broth or on Luria agar (LA) plates. The growth medium was supplemented with the following antibiotics as required at the indicated concentrations: ampicillin, 100 μg·ml−1; chloramphenicol, 20 μg·ml−1; kanamycin, 30 μg·ml−1; nalidixic acid, 35 μg·ml−1; streptomycin, 35 μg·ml−1; and tetracycline, 10 μg·ml−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristics and/or description | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | φ80dlacZΔM15 endA1 recA1 gyrA96 thi-1 hsdR17 (rK− mK+) relA1 supE44 deoR Δ(lacZYA-argF)U196 | Promega Corp., Madison, WI |
| JM109 | endA1 gyrA96 hsdR17(rK− mK+) mcrB+recAl relA1 supE44 thi-1 Δ(lac-proAB) F′ [traD36 proAB lacIqZΔM15] | 26 |
| Plasmid vectors | ||
| pACYC177 | Apr Kmr; p15A replicon; cloning vector | 6 |
| pBAD28 | Apr Cmr; arabinose-inducible expression vector; pACYC184 replicon | 11 |
| pBR322 | Apr Tcr; ColE1 replicon; cloning vector | 3 |
| pOU82 | Apr; lacZYA; R1 replicon | 10 |
| Plasmid constructs | ||
| pACYC177ΔApr | Kmr Amps; 683-bp deletion of a BamHI-ScaI fragment from pACYC177 | This study |
| pACYC177-TA(Apr) | Apr; 731-bp PCR fragment containing pRAS3.1 pemIK-like genes (nucleotide positions 8819 to 8088) cloned into the XhoI-BamHI sites of pACYC177 | This study |
| pACYC177-TA(Kmr) | Kmr; 731-bp PCR fragment containing pRAS3.1 pemIK-like genes (nucleotide positions 8819 to 8088) cloned into the BamHI-ScaI sites of pACYC177 | This study |
| pBAD28-repC | Apr Cmr; 1,015-bp PCR fragment containing the pRAS3.1 repC (nucleotide positions 6903 to 5889) cloned behind the PBAD promoter | 17 |
| pOU82-TA | Apr; 731-bp PCR fragment containing pRAS3.1 pemIK-like genes (nucleotide positions 8819 to 8088) cloned into pOU82 | 17 |
| pRAS3.1 | Tcr; natural 11,851-bp plasmid isolated from Aeromonas salmonicida subsp. salmonicida with four iterons and five 6-bp repeats | 12 |
| pRAS3.1Km | Kmr; pRAS3.1 with Tcr replaced by Kmr from pSKm2 at the BamHI-EcoRV sites | 17 |
| pRAS3.1.34 | Tcr; pRAS3.1.35 derivative with four 6-bp repeats from pRAS3.2 by exchanging the 2.9-kb HindIII-PvuI region | This study |
| pRAS3.1.35 | Tcr; pRAS3.1 derivative with three iterons obtained by random ligation of short iteron fragments after BstEII digestion | This study |
| pRAS3.1.35Km | Kmr; pRAS3.1.35 with Tcr replaced by Kmr from pSKm2 at the BamHI-EcoRV sites | This study |
| pRAS3.1.44 | Tcr; pRAS3.1 derivative with four 6-bp repeats from pRAS3.2 by exchanging the 2.9-kb HindIII-PvuI region | This study |
| pRAS3.1.55 | Tcr; pRAS3.1 derivative with five iterons obtained by random ligation of short iteron fragments after BstEII digestion | This study |
| pRAS3.1.55Km | Kmr; pRAS3.1.55 with Tcr replaced by Kmr from pSKm2 at the BamHI-EcoRV sites | This study |
| pRAS3.1.74 | Tcr; pRAS3.1.75 derivative with four 6-bp repeats from pRAS3.2 by exchanging the 2.9-kb HindIII-PvuI region | This study |
| pRAS3.1.75 | Tcr; pRAS3.1 derivative with seven iterons obtained by random ligation of short iteron fragments after BstEII digestion | This study |
| pRAS3.1.75Km | Kmr; pRAS3.1.75 with Tcr replaced by Kmr from pSKm2 at the BamHI-EcoRV sites | This study |
| pRAS3.2 | Tcr; natural 11,823-bp plasmid isolated from atypical Aeromonas salmonicida with three iterons and four 6-bp repeats | 12 |
| pRAS3.2Km | Kmr; pRAS3.2 with Tcr replaced by Kmr from pSKm2 at the BamHI-EcoRV sites | 17 |
| pSKM2 | Apr Kmr; plasmid with the 1.45-kb HindIII-SmaI kanamycin resistance cassette from Tn5 cloned into the HindIII-SmaI sites in pBluescript (SK) | Stellenbosch University laboratory collection (D. E. Rawlings) |
General DNA techniques.
Plasmid preparation, restriction endonuclease digestion, gel electrophoresis, and cloning were performed by standard methods (2, 24). Where no suitable restriction sites were present, single-strand DNA primers were designed, and DNA fragments to be cloned were amplified by PCR as described previously (19).
Copy number determinations.
The relative and absolute plasmid copy numbers were determined by real-time quantitative PCR (qPCR) using the primers and standard curves described previously (19). Total genomic DNA was prepared from E. coli DH5α cultures containing the respective plasmids during exponential growth (optical density at 600 nm [OD600] of 0.8) using a QIAamp DNA minikit (Qiagen); the DNA was prepared, placed in 60 μl of elution buffer, and quantified using a NanoDrop spectrophotometer.
Real-time qPCR amplification was performed using a LightCycler (version 2.0) with the LightCycler FastStart DNA master SYBR green I kit (Roche Diagnostics). Four nanograms of total DNA was added to each amplification reaction mixture, and the thermal cycling protocol of Lee et al. (13) was followed except that primer annealing was performed at 56°C for 4 s and DNA extension was performed at 72°C for 15 s.
Relative plasmid copy numbers were determined with pRAS3.1.35 as a reference standard using the relative expression software tool (REST) analysis software tool (21). The absolute plasmid copy numbers of pRAS3.1 and pRAS3.2 were determined using the threshold cycle (CT) values to extrapolate the total amount of plasmid and chromosome present in a sample using LightCycler software (version 3.5) by the calculation method of Lee et al. (13).
Plasmid stability assay.
The plasmids tested for stability were transformed into E. coli DH5α containing a resident pACYC177 vector with or without the pemIK system, plated on selective Luria agar plates containing tetracycline, and incubated overnight (O/N) at 37°C. Single colonies were inoculated into 5-ml portions of LB or M9 medium containing antibiotics selecting for both plasmids and incubated at 30°C for 24 h, whereafter the cultures were diluted 1 × 106 times into fresh medium containing an antibiotic selecting for only pACYC177 at 24-h intervals and incubated at 30°C while shaking for a total of 5 days. Each day serial dilutions of the cultures were plated onto plates containing an antibiotic selecting for only pACYC177 and incubated at 37°C. The percentage plasmid retention was then determined by replica plating 50 colonies from each of the plates onto plates containing antibiotics selecting for both plasmids or an antibiotic selecting for only pACYC177.
Relative fitness assays.
The fitness of plasmid-containing (P+) cultures was measured relative to the fitness of plasmid-free (P−) cultures (15). Aliquots of competent E. coli JM109 cells were split into two samples. One sample was transformed with the plasmid derivatives and plated on selective medium, while the other sample, which was not transformed, was plated on nonselective medium. A single colony from each of the cultures (P+ and P− cultures) was inoculated into 10 ml of Davis minimal (DM) medium supplemented with 125 μl of d-glucose (20%) per liter, 406 μl of magnesium sulfate (1 M) per liter, and 58.8 μl of thiamine (34 mg/ml) per liter and incubated O/N at 37°C (5). The cultures were acclimated to the DM medium for a total of 3 days by serially diluting the cultures 100-fold into fresh medium every 24 h. Once the cultures were acclimated, 700 μl of each culture was inoculated into 70 ml of fresh medium, mixed, and divided into six 10-ml cultures in 50-ml Erlenmeyer flasks and incubated while shaking O/N at 37°C. After 24 h of growth, the P+ monoculture was mixed with an approximately equal number of P− monoculture cells, determined from colony counts on selective and nonselective media, respectively, and the six mixed cultures were inoculated into fresh DM medium at a 100-fold dilution and incubated at 37°C while shaking. As a control for stability, an approximately equal amount of P+ cells from each of the six cultures was inoculated into DM medium and maintained as a monoculture in the absence of selection for the duration of the assay. The cultures were reinoculated into fresh medium daily for a total of 6 days, and each inoculum, including the time zero (T0) inoculum representing the original mixed culture and monocultures, was serially diluted and plated onto selective and nonselective media and incubated at 37°C for colony counts.
The selection rate constant was estimated from a linear regression of the daily log ratio of P+ to P− [ln(ratio P+/P−)] cells, and the relative fitness was calculated from the selection rate constant and expressed as a per generation fitness percentage [1 − (selection rate constant/average number of generations per day) × 100] (14). The number of P− cells in a mixed culture was estimated by subtracting the number of P+ cells from the total number of cells on the plates with nonselective medium. Plasmid stability was monitored by plotting the number of P+ cells counted on the selective plates against the total number of cells counted on the nonselective plates. Additionally, to verify the stability data, 50 colonies of each P+ monoculture were replica plated onto selective and nonselective media on days 0, 3, and 6. The relative fitness data were analyzed using a single-sample t test with a constant of 100% (selection rate constant of 0) for each of the samples, and the statistical significance of comparisons between different plasmid samples were determined by the Fisher least-significant difference (LSD) test using Statistica version 9 (StatSoft Inc., OK) software.
Displacement assays.
Competent E. coli DH5α cells containing a resident plasmid were transformed with a second incoming plasmid and plated on antibiotic-containing media selecting for both plasmids. A single colony was picked and inoculated into LB broth with double antibiotic selection and incubated at 37°C. After growth O/N, the cultures were serially diluted and plated onto nonselective LA plates. The cultures were also diluted 1 × 106 into fresh nonselective LB broth and incubated at 37°C for 24 h before they were serially diluted and spread onto nonselective media. Fifty colonies from each of the day 0 and day 1 nonselective plates were picked and replica plated onto four sets of solid media, two sets containing single antibiotics to test for the presence of the resident or incoming plasmid separately, one set containing two antibiotics to test for the presence of both plasmids, and one set containing no antibiotics. The presence of either or both plasmids was scored and expressed as a percentage of the total number of colonies tested. The same method was used to determine segregation patterns in the presence or absence of additional RepC except that the competent cells also contained either pBAD28-RepC or pBAD28, and selection was provided for the pBAD28 plasmids throughout the duration of the assay. Controls to check for spontaneous loss of the resident plasmids were carried out using the same procedure except that the initial competent E. coli cells containing the resident plasmids were taken through equal cycles of growth without antibiotic selection before testing for retention of the resident plasmid.
RESULTS
Construction of plasmids with increased numbers of iterons.
To assist in understanding why one of the natural pRAS3 plasmids had the three 22-bp iteron structure typical of the IncQ plasmids and the other had four 22-bp iterons, we amplified the differences by constructing pRAS3 plasmids with even greater numbers of iterons. Examination of the sequences of the pRAS3 plasmids indicated that they contained a recognition site for the BstEII restriction endonuclease within each of the 22-bp iterons and that no other BstEII sites existed. Plasmid pRAS3.1 was therefore digested with BstEII, and a concentrated self-ligation reaction was conducted. E. coli DH5α was transformed with the ligation mix, and plasmid DNA was extracted from 80 colonies. Single-strand DNA primers homologous to either side of the oriV region were used to PCR amplify the region (751 bp in the pRAS3.1 control) containing the iterons (17); the primers were pRAS3oriV fwd and pRAS3oriV rev (the forward and reverse primers are indicated by fwd and rev, respectively, at the end of the primer designation). Four pRAS3.1 derivatives that appeared to have amplicons of different sizes were identified, and DNA sequencing of the region revealed that plasmids containing 3, 4, 5, and 7 iterons had been constructed (Fig. 1). The plasmids are numbered pRAS3.1.35, pRAS3.1.45, pRAS3.1.55, and pRAS3.1.75 with the last two digits indicating the number of 22-bp iterons and 6-bp repeats, respectively. Using this terminology, construct pRAS3.1.45 is equivalent to the wild-type pRAS3.1 plasmid. Attempts to construct a plasmid containing a single iteron by digesting pRAS3.1 with BstEII and then separating the small 22-bp fragments prior to self-ligation of the remaining 11 785-bp fragments at the single remaining BstII site were unsuccessful. As no plasmids containing either one or two iterons were identified, plasmids with less than three iterons appeared to be nonviable.
Effect of iteron number on plasmid copy number.
Unlike plasmids pRAS3.1 and pRAS3.2 that differ in the numbers of both the 22-bp iteron and mobB-mobA/repB 6-bp repeat, plasmids pRAS3.1.35, pRAS3.1(0.45), pRAS3.1.55, and pRAS3.1.75 are identical plasmids differing only in the number of 22-bp iterons. The effect of the number of 22-bp iterons on plasmid copy number (PCN) could therefore be determined. The relative copy numbers of plasmids containing three, four, five, or seven 22-bp iterons relative to pRAS3.1.35 were determined using quantitative real-time PCR (Table 2). Plasmid vector pBR322 which has been reported to have a copy number of ∼18 as determined by both absolute and relative quantification of real-time PCR methods (13) was included as a standard from which to calculate the absolute PCN of each pRAS3.1 variant. The natural pRAS3.1 (pRAS3.1.45) plasmid containing four 22-bp iterons had a PCN of ∼41. A reduction in the number of iterons to three resulted in a copy number ∼44% higher (PCN of 59), while increasing the number of iterons to five or seven resulted in a decrease in copy number of ∼32% (PCN of 28) and ∼54% (PCN of 19), respectively.
TABLE 2.
Relative and absolute copy numbers of pRAS3 plasmid derivatives
| Plasmid | Relative PCNa | SD for relative PCN | Confidence value (P) for relative PCN | No. of repeats | Absolute PCN |
|---|---|---|---|---|---|
| pRAS3.1.35 | 1.00 | 0.090 | 0.001 | 4 | 59 |
| pRAS3.1.45 (pRAS3.1) | 0.69 | 0.062 | 0.001 | 6 | 41 |
| pRAS3.1.55 | 0.48 | 0.092 | 0.001 | 6 | 28 |
| pRAS3.1.75 | 0.32 | 0.070 | 0.001 | 8 | 19 |
| pBR322 | 0.30 | 0.007 | 0.001 | 4 | 18 |
PCN, plasmid copy number.
The abilities of pRAS3.1, pRAS3.2, and derivatives to displace each other within a host cell.
As pRAS3.1 and pRAS3.2 both contain tetracycline resistance genes as their only selectable marker, the kanamycin resistance gene from pSKM2 was inserted into the tetAR genes of both plasmids so as to obtain tetracycline- and kanamycin-resistant variants of both plasmids. When plasmids pRAS3.2 and pRAS3.1Km were placed in an E. coli DH5α host, the selecting factor was removed, and the host was grown for ∼20 generations, plasmid pRAS3.1Km displaced plasmid pRAS3.2 in ∼98% of cells (Table 3). Furthermore, the remaining ∼2% of cells retained both plasmids in all cases, indicating that although pRAS3.1 readily displaced pRAS3.2, pRAS3.2 was not able to displace pRAS3.1Km. When the antibiotic resistance genes were switched, an identical rate of displacement of pRAS3.2Km by pRAS3.1 was observed (Table 3). This showed that plasmid displacement was not noticeably affected by the antibiotic resistance markers and that a plasmid containing four 22-bp iterons (pRAS3.1) displaced a plasmid with three 22-bp iterons (pRAS3.2). As the natural plasmids pRAS3.1 and pRAS3.2 differ both in the number of 6-bp repeats of their mobB-mobA/repB promoter regions and in the number of 22-bp oriV-associated iterons, we investigated the effect of the number of 22-bp iterons on plasmid displacement alone by constructing pRAS3.1.35Km. This plasmid has a backbone identical to that of pRAS3.1 except that it has three rather than four 22-bp iterons. After ∼20 generations of growth without selection in E. coli DH5α, the plasmid with four 22-bp iterons (pRAS3.1) displaced the plasmid with three 22-bp iterons (pRAS3.1.35Km) in 79% ± 4% of host cells, while 21% ± 4% of cells retained both plasmids, and no instances of the 3-iteron plasmid displacing the 4-iteron plasmid were observed. The reciprocal experiment with pRAS3.1.35 and pRAS3.1Km gave largely similar results with the 4-iteron plasmid (pRAS3.1Km) displacing the 3-iteron plasmid (pRAS3.1.35) in 87% ± 10% of host cells and with 12% ± 8% of the host cells retaining both plasmids. The displacement of a 3-iteron plasmid by a 4-iteron plasmid occurred in spite of the observation that when the iteron number of pRAS3.1 was increased from 3 to 4, its PCN decreased from ∼59 to ∼41 (17). The displacement of a plasmid containing three 22-bp iterons with a higher PCN by a plasmid with an identical structure but with a lower PCN was unexpected. Therefore, a plasmid with three 22-bp iterons (pRAS3.1.35) and a plasmid with four 22-bp iterons (pRAS3.1Km) were placed into the same cell with antibiotic selection to ensure that both plasmids were retained and the relative copy numbers were determined by the gel method of Park et al. (20). The plasmid with three 22-bp iterons (pRAS3.1.35) was barely detected, whereas the plasmid with four 22-bp iterons was clearly visible (data not shown). Therefore, a plasmid with four iterons was able to inhibit the replication of a plasmid with three iterons in spite of the four-iteron plasmid having a lower PCN when on its own in a host cell.
TABLE 3.
Segregation patterns of coresident pRAS3 plasmids and derivatives in an E. coli host
| pRAS3 plasmid | Coresident plasmida |
% colonies with resistance to the following antibiotic(s)b: |
Direction and strength of segregation biasc | ||||
|---|---|---|---|---|---|---|---|
| pRAS3.2.Km | pRAS3.1.35.Km | pRAS3.1.Km | Tet only | Km only | Tet and Km | ||
| pRAS3.2 | • | 0 ± 0 | 98 ± 3 | 2 ± 3 | 3 iterons and 4 6-bp repeats <<< 4 iterons and 5 6-bp repeats | ||
| pRAS3.1 | • | 98 ± 0 | 0 ± 0 | 2 ± 0 | 4 iterons and 5 6-bp repeats >>> 3 iterons and 4 6-bp repeats | ||
| • | 79 ± 4 | 0 ± 0 | 21 ± 4 | 4 iterons >>> 3 iterons | |||
| • | 23 ± 7 | 20 ± 8 | 57 ± 11 | Tet ≈ Km | |||
| pRAS3.1.35 | • | 0 ± 0 | 87 ± 10 | 12 ± 8 | 3 iterons <<< 4 iterons | ||
| • | 24 ± 3 | 13 ± 1 | 62 ± 3 | Tet > Km | |||
| pRAS3.1.34 | • | 14 ± 0 | 18 ± 14 | 66 ± 14 | 4 6-bp repeats ≈ 5 6-bp repeats | ||
| pRAS3.1.44 | • | 30 ± 6 | 8 ± 6 | 60 ± 17 | 4 6-bp repeats > 5 6-bp repeats | ||
The small black circles indicate the plasmid in the cell in addition to the pRAS3 plasmid used in each experiment. Each plasmid was completely stable on its own for the duration of the assay in the absence of antibiotic.
Tet, tetracycline; Km, kanamycin.
Direction and strength of segregation bias as a result of iteron and/or 6-bp repeat copy number, as well as the influence of the antibiotic resistance genes. Arrowheads indicate the direction of plasmid displacement, and the number of arrowheads is an indication of the strength of displacement. Tet, tetracycline resistance gene; Km, kanamycin resistance gene.
Experiments to determine the effect of an additional 6-bp repeat in the mobB-mobA/repB promoter region on plasmid stability when the number of iterons was kept the same were less clear. In order to obtain such plasmids, a 2.9-kb HindIII-PvuI region containing the five 6-bp repeats in each of the pRAS3.1 derivatives was replaced with the same region from pRAS3.2. In E. coli host cells containing plasmids pRAS3.1.34 and pRAS3.1.35Km, both of which have three 22-bp iterons but four or five 6-bp repeats, respectively, ∼66% ± 14% of the cells contained both plasmids after ∼20 generations, while approximately equal numbers of cells retained only one of the two plasmids (14% ± 0% of the cells or 18% ± 14% of the cells, respectively). When both plasmids had four 22-bp iterons but differed by having four or five 6-bp repeats, pRAS3.1.44 or pRAS3.1Km, respectively, then after ∼20 generations, ∼60% ± 17% of the cells contained both plasmids after ∼20 generations, while 30% ± 6% of the cells retained pRAS3.1.44 with four 6-bp repeats and only 8% ± 6% of the cells retained pRAS3.1Km with five 6-bp repeats. Taken together, the results suggest that even though a greater level of mobB-mobA/repB expression has been shown to occur in a plasmid with five 6-bp repeats compared with plasmids with four 6-bp repeats (17), this appears not to have an effect on plasmid displacement. This observation is not unexpected, given that the oriV regions of the two plasmids are identical and replication proteins are capable of working in trans, and therefore, the replication proteins produced by one plasmid variant would also be available to the other plasmid. Under such circumstances, plasmids containing identical numbers of iterons would be expected to initiate replication equally efficiently, and therefore, the PCNs of coresident plasmids with different numbers of 6-bp repeats might be expected to be approximately equal.
Effect of iteron number on the displacement of related plasmids.
We had shown that a plasmid with four 22-bp iterons displaced a plasmid with three 22-bp iterons, and this raised the question as to how plasmids with five or seven 22-bp iterons would compete with a typical plasmid with three 22-bp iterons. It is known that the binding of RepC monomers to the iterons is an essential step during the initiation of replication and that iteron binding by DNA-binding proteins may be cooperative (4). That is, DNA-binding proteins may be more effectively sequestered by DNA with larger numbers of tandem iterons. Plasmids with four, five, or seven 22-bp iterons, all plasmids containing identical five 6-bp repeats, were allowed to compete for replication space in an E. coli DH5α host containing a coresident plasmid with three 22-bp iterons. Since RepC is an important player in the initiation of plasmid replication, these experiments were repeated with additional RepC DNA-binding protein expressed in trans from the arabinose-inducible PBAD promoter on pBAD28 (Table 4).
TABLE 4.
Effect of iteron number on coresident plasmid segregation patterns in the absence and presence of excess RepC
| pRAS1 plasmid | Coresident plasmidsa |
% colonies with resistance to the following antibiotic(s): |
Direction and strength of segregation biasb | |||
|---|---|---|---|---|---|---|
| pRAS3.1.35.Km + pBAD28 | pRAS3.1.35.Km + pBAD28-RepC | Tet only | Km only | Tet and Km | ||
| pRAS3.1.35.Tet | • | 53 ± 11 | 21 ± 6 | 28 ± 10 | Tet > Km | |
| • | 55 ± 8 | 17 ± 1 | 29 ± 5 | Tet > Km | ||
| pRAS3.1 | • | 91 ± 6 | 1 ± 1 | 7 ± 5 | 4 iterons >>> 3 iterons | |
| • | 88 ± 4 | 7 ± 3 | 6 ± 0 | 4 iterons >>> 3 iterons | ||
| pRAS3.1.55.Tet | • | 79 ± 3 | 6 ± 3 | 14 ± 6 | 5 iterons ≫ 3 iterons | |
| • | 67 ± 2 | 16 ± 2 | 21 ± 1 | 5 iterons ≫ 3 iterons | ||
| pRAS3.1.75.Tet | • | 63 ± 14 | 25 ± 13 | 15 ± 2 | 7 iterons > 3 iterons | |
| • | 63 ± 11 | 26 ± 13 | 13 ± 4 | 7 iterons > 3 iterons | ||
The small black circles indicate the two plasmids in the cell in addition to the pRAS3 plasmid used in each experiment. All plasmids were completely stable in the absence of antibiotics for the duration of the assay.
Direction and strength of segregation bias as a result of iteron number in the absence or presence of additional RepC. Arrowheads indicate the direction of plasmid displacement, and the number of arrowheads is an indication of the strength of displacement.
When two plasmids each with three 22-bp iterons but with different antibiotic markers (pRAS3.1.35 and pRAS3.1.35.Km) were competed, plasmid incompatibility was weak, with the plasmid containing the tetracycline (Tet) marker having a small advantage over the plasmid containing the kanamycin (Km) marker but with a fairly high proportion (28%) of cells retaining both plasmids. The standard deviations were high, supporting the interpretation that the bias in favor of the plasmid with the Tet marker was relatively weak. The pRAS3.1 plasmid, which contained four 22-bp iterons, displaced the pRAS3.1.35Km plasmid, which contained three 22-bp iterons, in 90% of the cells, much like was found in the earlier experiment in which pRAS3.1 displaced pRAS3.2 in 87% of the cells (Table 4). The plasmids with five and seven 22-bp iterons also displaced the plasmid with three 22-bp iterons, although this displacement became progressively weaker as the number of 22-bp iterons increased. Because of the apparent weak bias in favor of plasmids containing the Tet marker, reciprocal experiments to those shown in Table 4 were carried out; in these experiments, the plasmid with three 22-bp iterons was pRAS3.1.35, rather than pRAS3.1.35Km, and the competing plasmids were Km resistant rather than Tet resistant. Similar results were obtained except that the weak displacement of the plasmid with three 22-bp iterons by the plasmid with seven 22-bp iterons had largely disappeared (data not shown). The presence of additional RepC (second row of each data set in Table 4) did not affect plasmid competition but served to confirm the plasmid displacement results obtained without additional RepC.
Stability of pRAS3 plasmids and derivatives.
We wished to determine how the metabolic loads that the pRAS3.1 or pRAS3.2 plasmids placed on a host cell differed. Before doing this, it was important to determine how stable these plasmids were in the absence of selection, as the addition of antibiotics to growth medium would itself affect the metabolic load experienced by a plasmid-containing cell. Furthermore, the pRAS3 plasmids contain a toxin-antitoxin (TA) system and should one plasmid be lost at a higher rate than the other, this would result in a greater proportion of the daughter cells of a host with the less stable plasmid being unable to continue to grow and divide. This inability to grow on the loss of a TA-containing plasmid would, of itself, appear to increase the metabolic load. The stability of plasmids pRAS3.1 and pRAS3.2 when present in E. coli DH5α host cells was tested in LB for approximately 100 generations in the absence of selection. No loss of either plasmid was detected (data not shown). The stability assay was repeated in M9 minimal medium, and again, 100% of cells appeared to have retained the plasmid after 100 generations. Since the apparent plasmid stability might have been due to the effectiveness of the TA system, the TA system was cloned into vector pACYC177 so that it could be placed in a cell together with the pRAS3 plasmids to neutralize any effect the TA system might have on plasmid loss. Functionality of the pACYC177-TA(Kmr) clone was demonstrated by its ability to eliminate the stabilizing effect of the TA system on an unstable pOU82 test plasmid. Plasmids pACYC177-TA(Kmr) or pACYC177ΔAmp were placed in trans with either pOU82-TA or pOU82, and the stability of the test plasmid was determined. When the pRAS3 TA genes were cloned onto the pOU82 test plasmid, the stability of pOU82-TA was 99% after ∼90 generations. Neutralization of the TA genes on pOU82-TA by having pACYC177-TA(Kmr) in trans decreased plasmid stability to 12% after ∼90 generations. This was identical to the high level of instability observed for pOU82 plasmid that lacked the TA system.
The presence of pACYC177-TA(Apr) in the same cell as either pRAS3.1 or pRAS3.2 did not affect plasmid stability, and both these pRAS3 plasmids were 100% stable even when their TA systems were neutralized. To further investigate the stability of the pRAS3 plasmids, we tested the stability of the pRAS3 derivatives with the highest copy number (pRAS3.1.35) and the lowest copy number (pRAS3.1.74) with PCNs of ∼59 and ∼15, respectively. The stability of these plasmids was tested in E. coli DH5α cultures containing plasmid pACYC177ΔKm or pACYC177-TA(Apr) in trans. Both of these plasmids, pRAS3.1.35 and pRAS3.1.74, were found to be 100% stable for ∼90 generations irrespective of whether the TA system was neutralized by having a coresident pACYC177-TA(Apr) or not. The stability assays were carried in LB and M9 minimal medium and at 30 and 37°C, and in none of the experiments was there any evidence of instability. It thus seemed that the pRAS3 plasmids were sufficiently stable for metabolic load experiments to be carried out without selection for plasmid maintenance.
Comparative metabolic loads of pRAS3 plasmids and derivatives.
The relative effects of pRAS3.1 and pRAS3.2 plasmids on host cell fitness was carried out in E. coli JM109 cells and compared with the fitness of an isogenic plasmid-free host as described by Lenski (14) (Table 5). The wild-type plasmid pRAS3.2 had a smaller impact on host cell fitness with a cost of ∼4.7% than pRAS3.1 with the cost being slightly higher at ∼7.5%. This difference in metabolic load of ∼2.8% was small but statistically significant (P < 0.05). We next tested the metabolic burden imposed by derivatives of pRAS3.1 with various PCNs as a result of their having different combinations of 6-bp repeats and 22-bp iterons (Table 5). As may be expected, plasmid pRAS3.1.34 with a PCN of ∼31, which was approximately equal to the PCN of pRAS3.2 (PCN of ∼30), had a similar metabolic load at ∼5.3% (P > 0.05). Plasmid pRAS3.1.35 with the highest PCN of ∼59 also had the highest metabolic load of ∼10.9%, and plasmid pRAS3.1.74 with the lowest PCN (PCN of ∼15) also had the lowest metabolic load at ∼2.6%.
TABLE 5.
Metabolic burden of plasmids on E. coli host cells relative to plasmid-free cells
| Plasmid | Approximate PCN | % relative fitness | SD for relative fitness | No. of cells tested | P valuea |
|---|---|---|---|---|---|
| pRAS3.2 | 30 ± 5 | 95.30 | 1.56 | 6 | 0.001 |
| pRAS3.1.34 | 31 ± 1 | 94.70 | 1.72 | 5 | 0.002 |
| pRAS3.1.35 | 59 | 89.12 | 1.16 | 6 | 0.000 |
| pRAS3.1 | 41 ± 4 | 92.52 | 2.64 | 6 | 0.001 |
| pRAS3.1.74 | 15 ± 1 | 97.36 | 1.07 | 6 | 0.002 |
P value for the metabolic burden of plasmids on E. coli host cells compared to plasmid-free cells.
DISCUSSION
The backbone sequences of the two natural plasmid variants, pRAS3.1 and pRAS3.2, are identical with the exception that pRAS3.1 has a fourth perfectly conserved 22-bp iteron in its oriV region, whereas pRAS3.2 has only three of the 6-bp repeats within the promoter region of the mobB-mobA/repB genes, pRAS3.1 has five of the 6-bp repeats, and pRAS3.2 has four of these repeats. In previous work, we established that the additional 6-bp repeat in the mobB-mobA/repB gene promoter region of pRAS3.1 had resulted in an increase in plasmid copy number (PCN) of about ∼2-fold (from 30 to 59), whereas the additional 22-bp iteron had resulted in a decrease in PCN of pRAS3.1 (from 59 to 41). The two alterations appear to have been compensatory, with one increasing the PCN and the other decreasing the PCN, at least partially. This raised the question of why two natural variants of the pRAS3 plasmids exist and whether either of the resultant plasmids, pRAS3.1 or pRAS3.2, is more fit evolutionarily.
Plasmid fitness can operate at two levels. One level is competition between related plasmids within a host cell. That is, plasmids with identical (or nearly identical) origins of replication are incompatible when both plasmids are present in an individual host cell. If one of the plasmids has a replication advantage, it will tend to displace the other. The second level is competition at a host cell population level. That is, a population of host cells containing one of the plasmids may be more competitive than a population of the identical host cells containing the other plasmid because of differences in metabolic burden that the two plasmids place on the host.
A high PCN would be expected to decrease the chance of plasmid loss on cell division, but too high a PCN would increase the metabolic load on a cell and thereby reduce host cell competitiveness. In contrast, although a low PCN might decrease the metabolic burden on the host, the host population might have reduced competitiveness should plasmids like the pRAS3 plasmids be lost too easily on cell division. The reason for this is that the pRAS3 plasmids have a functional toxin-antitoxin module and these modules function by inhibiting the growth of cells that fail to inherit a plasmid on cell division. Therefore, should the PCN be so low that a high rate of plasmid loss occurs, cells failing to inherit the plasmid would cease to grow, thereby reducing the competitiveness of the population. At the population level, plasmid copy number is likely to be a compromise between having a high PCN to reduce plasmid loss but with an increased metabolic load and a low PCN with an increased chance of plasmid loss but decreased metabolic load.
The 22-bp-iteron deletion and ligation experiments clearly indicated that the minimum number of 22-bp iterons required to support plasmid replication was three. One would have expected the ligation mix to have contained higher numbers of plasmids with one or two iterons, but since no plasmids of that type were isolated, plasmids with less than three iterons appeared to be nonviable. These findings were consistent with studies by Lin and Meyer (16) using plasmid R1162, an IncQ-1 plasmid that has 20-bp iterons with 2-bp spacers. These workers constructed oriV regions containing 0, ½, 2, and 3 iterons on multicopy vectors and found that only plasmids containing 3 iterons were able to replicate in the presence of repBAC genes. We succeeded in isolating pRAS3.1 plasmids with three, four, five, and seven 22-bp iterons and found that the number of iterons had a marked effect on the copy number. The PCN decreased from 59 to 19 as the number of iterons in tandem increased from 3 to 7 (Table 2) or from 30 to 15 with the same increase in iteron numbers but with one less 6-bp repeat (data not shown). A variation in the number of iterons is therefore a highly effective means of selection for plasmids with a suitable PCN that may apply generally to iteron-containing plasmids.
In incompatibility experiments, plasmids with 4 or 5 22-bp iterons strongly displaced plasmids with three iterons in spite of the PCN of plasmids with 5 iterons being half of that of plasmids with 3 iterons (Table 4). Displacement of 3-iteron plasmids by 7-iteron plasmids was not marked. This suggested that plasmids with 4 or 5 iterons were selectively replicated in preference to plasmids with 3 iterons. A possible reason for this is that binding of the RepC replication initiator protein to the 22-bp iterons was enhanced when 4 or 5 iterons were present due to the phenomenon of cooperative binding. Such cooperative binding by the π replication initiator protein to the seven 22-bp iterons of plasmid R6K has been shown (4). This view is supported by the observation that when a 4-iteron plasmid and a 3-iteron plasmid were present in the same cell with antibiotic selection for both plasmids, the presence of the 3-iteron plasmid was barely detected (data not shown). This indicated that whereas the PCN of the 3-iteron plasmid was higher than the 4-iteron plasmid when each was on its own in a host cell, the 4-iteron plasmid inhibited the normal replication of the 3-iteron plasmid when both plasmids were present in the same cell. We speculate that this cooperative binding may favor the initiation of replication from plasmids containing four or five iterons above plasmids containing three iterons in spite of their copy number being lower. In the case of the 7-iteron plasmid, although cooperative binding of RepC to the iterons may be enhanced with respect to 3-iteron plasmids, the much greater difference in copy number eliminates this advantage, and both plasmids were able to compete for replication approximately equally. The provision of additional RepC protein by the placement of a repC gene under the control of a Para promoter on a coresident vector did not noticeably affect the pattern of plasmid displacement. This is either because the additional RepC resulted in inactive RepC dimer formation (23) or because cooperative binding resulted in the distribution of RepC to iterons in the same proportions irrespective of whether the intercellular levels of RepC were elevated.
Next we investigated what the effect of variations in the pRAS3 plasmids was on the fitness of E. coli host cells. One might predict that plasmids with four or five iterons had a double advantage over otherwise identical plasmids with three iterons. First, they were able to displace coresident three-iteron plasmids, and second, they had a lower copy number and therefore would be expected to place a lower metabolic burden on the host. As mentioned earlier, a possible disadvantage of having a lower copy number is that if this resulted in increased plasmid loss on cell division, then the host population may be negatively affected, as the TA system present on the pRAS3 plasmids may inhibit the growth of plasmid-free cells. However, when grown in an E. coli host for ∼90 generations, the loss of pRAS3-derived plasmids was undetectable over a PCN range of 15 to 59 irrespective of whether the TA system was neutralized or the cells were grown in Luria broth or minimal medium. These results are in agreement with a predicted theoretical loss frequency (based on random segregation) of 1 in 1.6 × 104 generations for a plasmid with a copy number of 15 plasmids per chromosome, and even less for plasmids with a higher copy number (25). The high level of TA system-independent stability may be a result of the relatively high PCN of pRAS3 plasmids relative to other IncQ family plasmids which have been reported to have a PCN varying from 10 to 16 (17). The reason for the high PCN of the pRAS3 plasmids is not known. A possible reason is that amino acid sequence alignments show that the RepC proteins of the pRAS3 plasmids are very different from those of the other sequenced IncQ-like plasmids. It is possible that the formation of inactive RepC dimers occurs at a higher concentration in pRAS3 plasmids than in other IncQ family plasmids; however, this requires further investigation.
The contribution of PCN on host fitness was measured by determining the growth rate of a plasmid-containing (P+) E. coli JM109 host relative to that of an isogenic plasmid-free (P−) host (14). There are several advantages in measuring fitness using this method as opposed to directly competing cultures containing the respective plasmids. First, a plasmid-bearing culture is competed against a host which does not have the additional metabolic burden, thus amplifying the growth rate differences and allowing a more accurate estimation in a shorter time frame. Second, it negates the need to have different antibiotic selection genes on each of the competing plasmids for identifying the number of each competing plasmid-containing strain at different stages of a competition experiment. Different markers may have resulted in a different, artificial burden that could have influenced the results. Last, as cultures were competed against the same reference strain, the percentage relative fitness allows for comparisons between any of the samples rather than just between the samples that were paired in an assay. A disadvantage is that the conclusions derived from comparing the fitness between the different plasmid derivatives are less direct.
It was found that metabolic load on an E. coli host cell population was increased as the PCN increased (Table 5) and that of the two natural plasmids, pRAS3.1 placed a small but significantly higher metabolic load on the host than did pRAS3.2. With this information, we speculate on the evolution of pRAS3.1 and pRAS3.2 and why both plasmids have survived. Although one cannot be certain which plasmid originated first or which of the two differences to the backbones of the plasmids occurred first, a likely scenario is the following. Plasmid pRAS3.2, with three 22-bp iterons plus four 6-bp repeats in the promoter of the mobB-mobA/repB genes and a PCN of ∼30, was most likely the ancestral plasmid. It more closely resembles the rest of the IncQ plasmids in the number of 22-bp iterons and has a PCN closer to that of typical IncQ family plasmids (although still considerably higher). It is also interesting that the pRAS3-like plasmid that has been integrated into the chromosome of the obligately intracellular parasite Chlamydia suis has four 6-bp repeats, similar to pRAS3.2, rather than pRAS3.1 (8). Plasmid pRAS3.1 probably arose from pRAS3.2 in two steps, the first being the acquisition of an additional 6-bp repeat which resulted in an approximately doubling of the PCN to ∼59 and eventual random segregation of the two sister plasmids. The additional metabolic burden placed on the host by this high-copy-number intermediate plasmid provided the evolutionary pressure for the selection of a plasmid with a lower copy number. This was achieved by the acquisition of an additional 22-bp iteron with a corresponding reduction in PCN to ∼41. As shown in this study, this plasmid with four 22-bp iterons would be expected to displace the intermediate plasmid with three 22-bp iterons, even though it had a higher PCN. Should an increase in the number of iterons have occurred first with a corresponding reduction in PCN (such a plasmid, pRAS3.1.44, was constructed, and the PCN was found to be ∼23) (17), the plasmid with a lower PCN and with four iterons would still have displaced the three-iteron plasmid as shown in Table 3. However, since this lower-copy-number plasmid is perfectly stable, it seems less likely for there to have been evolutionary pressure for this plasmid to have gained an additional 6-bp repeat, thereby increasing its PCN but placing an increased metabolic load on the host. A different possible reason for the acquisition of an additional 6-bp repeat is that not only is the expression of repB increased (17) but the expression of the mobA and mobB genes is also increased, as they are part of the same operon (Fig. 1A). If the additional 6-bp repeat altered the mobilization frequency, this could affect the horizontal spread of one of the variants and thereby provide a selective advantage. However, when mobilized by the RP4 conjugative system, the mobilization frequency of plasmids with either four or five 6-bp repeats and identical iteron numbers was found to be similar (17). Therefore, a change in mobilization frequency is unlikely to have provided selective pressure for acquisition of an altered number of 6-bp repeats.
We would have liked to perform plasmid copy number, displacement, competition, and metabolic load experiments in the two different Aeromonas hosts for pRAS3.1 and pRAS3.2, but they are no longer available. Our repeated attempts to transform a different natural isolate of Aeromonas salmonicida were unsuccessful. As IncQ plasmids have a broad host range and are highly promiscuous, we carried out our studies in E. coli as a representative of one of the many types of bacteria in which pRAS3 plasmids could occur. However, it is possible that other hosts might have placed somewhat different evolutionary pressures on the evolution of the pRAS3 plasmids.
Our overall finding from this study was that although pRAS3.2 has the advantage that it places a slightly lower metabolic burden on an E. coli host, it has the disadvantage that it is displaced by pRAS3.1 if both plasmids are present in the same cell. Nevertheless, both plasmids appear to have survived in the natural environment, with pRAS3.1 having been isolated from Aeromonas salmonicida in Norway and Scotland and pRAS3.2 having been isolated from atypical A. salmonicida in Norway (12) and A. salmonicida in Japan (1).
Acknowledgments
We thank Trine L'Abée-Lund for the gift of plasmids pRAS3.1 and pRAS3.2.
This work was supported by grants from the National Research Foundation (Pretoria, South Africa).
Footnotes
Published ahead of print on 1 October 2010.
REFERENCES
- 1.Aoki, T., and A. Takahashi. 1986. Tetracycline resistant gene of a non-transferable R-plasmid from fish pathogenic bacteria Aeromonas salmonicida. Bull. Jpn. Soc. Scient. Fish. 52:1913-1917. [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1993. Current protocols in molecular biology. Wiley Interscience, New York, NY.
- 3.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 4.Bowers, L. M., R. Krüger, and M. Filutowicz. 2007. Mechanism of origin activation by monomers of R6K-encoded π protein. J. Mol. Biol. 368:928-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlton, B. C., and B. J. Brown. 1981. Gene mutation, p. 222-242. In P. Gerhardt, R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. B. Phillips (ed.), Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC.
- 6.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorrington, R. A., and D. E. Rawlings. 1989. Identification and sequence of the basic replication region of a broad-host-range plasmid isolated from Thiobacillus ferrooxidans. J. Bacteriol. 171:2735-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dugan, J., D. D. Rockey, L. Jones, and A. A. Andersen. 2004. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob. Agents Chemother. 48:3989-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner, M. N., S. M. Deane, and D. E. Rawlings. 2001. Isolation of a new broad-host-range IncQ-like plasmid, pTC-F14, from the acidophilic bacterium Acidithiobacillus caldus and analysis of the plasmid replicon. J. Bacteriol. 183:3303-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerdes, K., J. E. Larsen, and S. Molin. 1985. Stable inheritance of plasmid R1 requires two different loci. J. Bacteriol. 161:292-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzman, L., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.L'Abée-Lund, T., and H. Sørum. 2002. A global non-conjugative Tet C plasmid, pRAS3, from Aeromonas salmonicida. Plasmid 47:172-181. [DOI] [PubMed] [Google Scholar]
- 13.Lee, C., J. Kim, S. G. Shin, and S. Hwang. 2006. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J. Biotechnol. 123:273-280. [DOI] [PubMed] [Google Scholar]
- 14.Lenski, R. E. 1992. Relative fitness: its estimation and its significance for environmental applications of microorganisms, p. 183-198. In M. A. Levin, R. J. Seidler, and M. Rogul (ed.), Microbial ecology: principles, methods, and applications. McGraw-Hill, New York, NY.
- 15.Lenski, R. E., S. C. Simpson, and T. T. Nguyen. 1994. Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J. Bacteriol. 176:3140-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, L. S., and R. J. Meyer. 1986. Directly repeated, 20-bp sequence of plasmid R1162 DNA is required for replication, expression of incompatibility, and copy-number control. Plasmid 15:35-47. [DOI] [PubMed] [Google Scholar]
- 17.Loftie-Eaton, W., and D. E. Rawlings. 2009. Comparative biology of two natural variants of the IncQ-2 family plasmids, pRAS3.1 and pRAS3.2. J. Bacteriol. 191:6436-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer, R. 2009. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid 62:57-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao, D.-M., H. Sakai, S. Okamoto, K. Tanaka, M. Okuda, Y. Honda, T. Komano, and M. Bagdasarian. 1995. The interaction of RepC initiator with iterons in the replication of the broad host-range plasmid RSF1010. Nucleic Acids Res. 23:3295-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park, K., E. Han, J. Paulsson, and D. K. Chattoraj. 2001. Origin pairing (′handcuffing') as a mode of negative control of P1 plasmid copy number. EMBO J. 20:7323-7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawlings, D. E., and E. Tietze. 2001. Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 65:481-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakai, H., and T. Komano. 1996. DNA replication of IncQ broad-host-range plasmids in gram-negative bacteria. Biosci. Biotechnol. Biochem. 60:377-382. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Williams, D. R., and C. M. Thomas. 1992. Active partitioning of bacterial plasmids. J. Gen. Microbiol. 138:1-16. [DOI] [PubMed] [Google Scholar]
- 26.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]