Abstract
In this study, a DeoR/GlpR-type transcription factor was investigated for its potential role as a global regulator of sugar metabolism in haloarchaea, using Haloferax volcanii as a model organism. Common to a number of haloarchaea and Gram-positive bacterial species, the encoding glpR gene was chromosomally linked with genes of sugar metabolism. In H. volcanii, glpR was cotranscribed with the downstream phosphofructokinase (PFK; pfkB) gene, and the transcript levels of this glpR-pfkB operon were 10- to 20-fold higher when cells were grown on fructose or glucose than when they were grown on glycerol alone. GlpR was required for repression on glycerol based on significant increases in the levels of PFK (pfkB) transcript and enzyme activity detected upon deletion of glpR from the genome. Deletion of glpR also resulted in significant increases in both the activity and the transcript (kdgK1) levels of 2-keto-3-deoxy-d-gluconate kinase (KDGK), a key enzyme of haloarchaeal glucose metabolism, when cells were grown on glycerol, compared to the levels obtained for media with glucose. Promoter fusions to a β-galactosidase bgaH reporter revealed that transcription of glpR-pfkB and kdgK1 was modulated by carbon source and GlpR, consistent with quantitative reverse transcription-PCR (qRT-PCR) and enzyme activity assays. The results presented here provide genetic and biochemical evidence that GlpR controls both fructose and glucose metabolic enzymes through transcriptional repression of the glpR-pfkB operon and kdgK1 during growth on glycerol.
The archaeal basal transcriptional machinery closely resembles the eucaryal RNA polymerase II (RNAP II) apparatus. Along with a multisubunit RNAP (46), archaea encode two basal transcription factors, TATA-binding protein (TBP) and transcription factor B (TFB), which are homologs of the eucaryal TBP and general transcription factor TFIIB, respectively (5, 39). Although archaeal transcriptional components are fundamentally eukaryote-like in nature (35), the majority of candidate transcriptional regulators are homologous to bacterial activators and repressors (2, 26). Only a few archaeal candidate regulators resemble eukaryotic gene-specific transcription factors, one of the best characterized of which is GvpE, an activator of gas vesicle biosynthesis in haloarchaea which resembles the eukaryotic basic leucine zipper proteins (25, 31). While bioinformatics predicts many candidate archaeal regulators, only a limited number have been characterized at the molecular level, most of which are from hyperthermophiles (4, 13, 23, 24, 27, 42). Molecular data pertaining to haloarchaeal transcriptional regulation, specifically regulators of carbon utilization, are severely limited. Only a few global regulators, namely, transcription factors (8, 11, 36), have been implicated in regulating carbon utilization in haloarchaea. Specifically, in Halobacterium salinarium, pairs of general transcription factors TBP and TFB control gene clusters (8, 11), and transcription factor TrmB regulates diverse metabolic pathways in response to nutrient limitation (36). To our knowledge, no transcriptional regulators of sugar metabolism in the model haloarchaeon Haloferax volcanii have been characterized to date, although homologs of transcription factors can be predicted based on primary sequence. Furthermore, DeoR/GlpR-type regulators in archaea have yet to be characterized.
In this study, biochemical and genetic approaches were used to characterize GlpR, a putative transcriptional regulator of glycerol (Gly) and/or sugar metabolism in H. volcanii. Here, we present evidence that GlpR not only is autoregulatory but also regulates transcription of the downstream phosphofructokinase (PFK) gene pfkB as well as the distant chromosomal 2-keto-3-deoxy-d-gluconate kinase (KDGK) gene kdgK1. Taken together, our results provide the first example of a DeoR/GlpR-type repressor protein that controls key enzymes of sugar metabolism in haloarchaea and allow valuable insight into haloarchaeal metabolic transcriptional regulation.
MATERIALS AND METHODS
Materials.
Biochemicals and enzymes used for PFK and KDGK activity analysis were purchased from Sigma-Aldrich (St. Louis, MO). the other organic and inorganic analytical-grade chemicals were from Fisher Scientific (Atlanta, GA) and Bio-Rad (Hercules, CA). Desalted oligonucleotides were from Integrated DNA Technologies (Coralville, IA). 2′-Deoxyuridine-5′-triphosphate coupled by an 11-atom spacer to digoxigenin (DIG) (DIG-11-dUTP), alkaline phosphatase-conjugated antibody raised against DIG, disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl) phenyl phosphate (CSPD), and the other DIG-related biochemicals were from Roche Molecular Biochemicals (Indianapolis, IN). Positively charged membranes for Southern hybridization were from Ambion (Austin, TX). Phusion and Taq DNA polymerases, restriction enzymes, T4 polynucleotide kinase, and T4 DNA ligase were from New England Biolabs (Ipswich, MA). Molecular biology-grade agarose used for the separation of DNA for Southern blotting and routine analysis was from Bio-Rad. Hi-Lo DNA standards were from Minnesota Molecular, Inc. (Minneapolis, MN).
Strains, media, and plasmids.
Strains, oligonucleotide primers used for cloning, and plasmids are summarized in Table 1 and Table S1 in the supplemental material. Escherichia coli Top10 was used for routine recombinant DNA experiments. H. volcanii strains were transformed (7) using plasmid DNA isolated from E. coli GM2163. Liquid cultures were aerated at 200 rpm. E. coli strains were grown at 37°C in Luria-Bertani medium supplemented with 100 mg per liter ampicillin as needed. H. volcanii strains were grown at 42°C in various media, including Casamino Acids (CA), yeast extract-peptone-Casamino Acids (YPC), YPC supplemented with glucose (Glu) or fructose (Fru), and minimal medium (MM) supplemented with glycerol (Gly), Glu, Fru, and various combinations of these carbon sources. Medium formulae were according to The Halohandbook (10), with the following exception: Glu, Fru, and/or Gly was included at 20 mM each where indicated. Media were supplemented as needed with novobiocin (0.1 μg·ml−1), 5-fluoroorotic acid (5-FOA) (50 μg·ml−1), and uracil (10 and 50 μg·ml−1 for growth in the presence and absence of 5-FOA, respectively). Uracil and 5-FOA were solubilized in 100% (vol/vol) dimethyl sulfoxide (DMSO) at 50 mg·ml−1 prior to addition to the growth medium.
TABLE 1.
List of strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| Top10 | F−recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA relA1 | Invitrogen |
| GM2163 | F−ara-14 leuB6 fhuA31 lacY1 tsx78 glnV44 galK2 galT22 mcrA dcm-6 hisG4 rfbD1 rpsL136 dam13::Tn9 xylA5 mtl-1 thi-1 mcrB1 hsdR2 | New England Biolabs |
| H. volcanii | ||
| DS70 | Wild-type isolate DS2 cured of plasmid pHV2 | 32 |
| H26 | DS70 pyrE2 | 1 |
| KS8 | H26 glpR (devoid of GlpR) | This study |
| KS4 | H26 glpK (devoid of GlpK) | 38 |
| KS10 | H26 glpK glpR (devoid of GlpR and GlpK) | This study |
| Plasmids | ||
| pTA102 | Apr Nvr; pGB70 containing H. alicantei bgaH derived from pMLH32 | 9 |
| pTA131 | Apr; pBluescript II containing Pfdx-pyrE2 | 1 |
| pJAM202 | Apr Nvr; pBAP5010 containing P2rrnA-psmB-his6; β-His6 expressed in H. volcanii | 21 |
| pJAM202c | Apr Nvr; control plasmid derived from pJAM202 | 45 |
| pJAM809 | Apr Nvr; pJAM202 containing P2rrnA-hvo1862-strepII (KpnI site inserted upstream of StrepII coding sequence) | 19 |
| pJAM2676 | Apr; pTA131 containing glpR with ∼700 bp of genomic DNA flanking 5′ and 3′ of the glpR coding region | This study |
| pJAM2677 | Apr; pJAM2676-derived glpR suicide plasmid | This study |
| pJAM2678 | Apr Nvr; pJAM202-derived plasmid containing P2rrnA-bgaH from pTA102 | This study |
| pJAM2682 | Apr Nvr; pJAM809 containing P2rrnA-glpR-strepII | This study |
| pJAM2689 | Apr Nvr; pJAM2678 containing PglpR-pfkB-188 bp-bgaH | This study |
| pJAM2702 | Apr Nvr; pJAM2678 containing PkdgK2-232 bp-bgaH | This study |
| pJAM2703 | Apr Nvr; pJAM2678 containing PHVO_A0327-kdgK2-122 bp-bgaH | This study |
| pJAM2705 | Apr Nvr; pJAM2678 containing PkdgK1-89 bp-bgaH | This study |
| pJAM2706 | Apr Nvr; pJAM2678 containing PkdgK1-524 bp-bgaH | This study |
| pJAM2714 | Apr Nvr; pJAM2678-derived plasmid devoid of P2rrnA from pTA102 and the Shine-Dalgarno site | This study |
| pJAM2715 | Apr Nvr; pJAM2678-derived plasmid devoid of P2rrnA from pTA102 | This study |
Apr, ampicillin resistance; Nvr, novobiocin resistance.
For enzyme activity and RNA extraction experiments, cells were freshly inoculated from −80°C glycerol stocks onto appropriate agar-based media on plates. Cells were twice subcultured and used as an inoculum for final analysis under various conditions as described below. Each subculture was inoculated to a final optical density at 600 nm (OD600) of 0.03 to 0.04. For PFK and KDGK enzyme activity assays and RNA preparation, cells were grown in 25 ml of medium in 250-ml flasks. For β-galactosidase activity measurements, cells were grown in 3 ml of medium in 13- by 100-cm culture tubes. Cell growth was monitored by an increase in OD600 (where 1 OD600 unit equals approximately 1 × 109 CFU·ml−1 for all strains used in this study). All experiments were performed at least in triplicate.
DNA isolation and analysis.
DNA was separated by electrophoresis using 0.8% or 2% (wt/vol) agarose gels in 1× TAE electrophoresis buffer (40 mM Tris-acetate, 2 mM EDTA, pH 8.5). Gels were stained with ethidium bromide at 0.5 μg·ml−1 and photographed with a mini-visionary imaging system (Fotodyne, Hartland, WI). The sizes of the DNA fragments were estimated using Hi-Lo DNA molecular weight markers. Plasmid DNA was isolated from E. coli strains by use of a QIAprep spin miniprep kit (Qiagen, Valencia, CA). PCR products were purified by MinElute (Qiagen) prior to modification by restriction enzymes (BamHI, HindIII, KpnI, NdeI, BlpI, or XbaI) or T4 DNA polynucleotide kinase. For rapid PCR screening, template DNA was extracted from H. volcanii mutant and parent strains and recombinant E. coli Top10 as described previously (45). For Southern blotting, H. volcanii genomic DNA was isolated from 5-ml cultures by DNA spooling (10), subjected to restriction enzyme digestion using MluI and HpaI, transferred to a nylon membrane, hybridized with a DIG-labeled probe specific for a 700-bp region flanking the 5′ end of the target coding region (primers listed in Table S1 in the supplemental material), and detected by CSPD-mediated chemiluminescence as described previously (45).
PCRs.
High-fidelity double-stranded DNA used for the construction of plasmids listed in Table 1 was generated by PCR using Phusion DNA polymerase. Taq DNA polymerase was used for screening of mutant strains and for the generation of the DIG-labeled double-stranded DNA probes that were used for Southern blotting. All PCRs were performed according to the instructions of the suppliers, with the following modifications: 3% (vol/vol) DMSO was included as needed and 0.1 mM deoxyribonucleoside triphosphate mix was added to the standard DIG-labeling reaction mixture, which included 1× DIG deoxyribonucleoside triphosphate (catalog no. 1277065; Roche). Primer pairs and template DNA used for the PCRs are outlined in Table S1 in the supplemental material. PCR was performed using an iCycler or GeneCycler (Bio-Rad Laboratories), and products were analyzed by DNA electrophoresis.
Chromosomal knockout of glpR.
The open reading frame (ORF) (HVO_1501; glpR) encoding a putative DeoR/GlpR repressor protein was targeted for markerless deletion from the chromosome of H. volcanii H26 using the pyrE2-based “pop-in/pop-out” method (1, 6). Colonies were screened for the absence of a 750-bp PCR product by use of internal primers specific for the coding region of the gene (the Negative-Forward and Negative-Reverse primers) and confirmed to be mutant strains by Southern blotting and PCR with the primer pair comprising Confirm-Forward and Confirm-Reverse, which anneal 100 bp upstream and downstream of the genomic region cloned into suicide plasmid pJAM2677. The products of the latter PCR were sequenced to further confirm mutant strain fidelity (as described below). The primers used for the cloning and screening of mutant colonies are provided in Table S1 in the supplemental material.
PFK and KDGK activity assays.
Exponential-growth-phase cells were harvested by centrifugation (20 min, 4,300 × g, 4°C), washed once with 20 ml buffer A (100 mM Tris-HCl at pH 7.5, 2 M NaCl), resuspended in 1 ml of buffer A containing 1 mM phenylmethylsulfonyl fluoride (PMSF), and lysed by sonication (4 × 20 s at 140 W). Debris was removed by centrifugation (10 min, 12,000 × g, 4°C). Protein concentration was estimated using the Bradford assay, with bovine serum albumin as a standard. PFK and KDGK activity assays were carried out as previously detailed (20), with the following exceptions: all enzyme activities were carried out aerobically at 37°C in a 96-well microplate reader filled with 0.1-ml assay mixtures. It was ensured that in coupled enzymatic assays, the auxiliary enzymes were not rate-limiting. Background change in absorbance for reaction mixtures containing no substrate was subtracted from the value for reactions in which substrate was included to yield the overall change in absorbance. Reaction mixtures containing boiled enzyme and no NADH were also included as controls. All experiments were performed in biological triplicate, and the means ± standard deviations (SD) of the results were calculated. One unit (U) of enzyme activity is defined as 1 μmol substrate consumed or product formed per min.
1-Phosphofructokinase (EC.2.7.1.56) specific activity was determined at 37°C by measuring the ATP-dependent formation of fructose-1,6-bisphosphate (FBP) from fructose-1-phosphate (F1P), which was coupled to the oxidation of NADH via FBP aldolase, triosephosphate isomerase (TIM), and glycerol-3-phosphate dehydrogenase (G3PDH). The assay mixture contained 100 mM Tris-HCl at pH 8.5 with 30 mM MgCl2, 1 M KCl, 10 mM F1P sodium salt, 2 mM ATP, 0.3 mM NADH, 0.54 U FBP-aldolase, 2 U TIM, 0.34 U G3PDH, and cell extract (1 to 3 μg protein).
2-Keto-3-deoxy-d-gluconate kinase (EC 2.7.1.45) specific activity was determined at 37°C by measuring the ATP- and gluconate-dependent formation of pyruvate, which was coupled to the oxidation of NADH via lactate dehydrogenase (LDH). The assay mixture contained 100 mM Tris-HCl at pH 8.5 with 1 M KCl, 10 mM MgCl2, 2 mM ATP, 0.3 mM NADH, 10 mM sodium gluconate, 11 U LDH, and cell extract (5 μg protein).
RNA purification and analysis.
Total RNA was isolated from H. volcanii parent H26 and glpR mutant KS8 strains (exponential phase; OD600, 0.3 to 0.5) using RNeasy RNA purification columns (Qiagen). RNA was treated with amplification-grade DNase I according to the supplier's recommendations (Sigma-Aldrich), with the following modifications: 3 U enzyme was added per 1 μg RNA, and the mixture was incubated for 45 min at room temperature. The integrity of the RNA was determined by agarose gel electrophoresis. RNA concentration was determined by A260 measurement using a Bio-Rad SmartSpec 3000 instrument.
Reverse transcription-PCRs (RT-PCRs) and quantitative RT-PCRs (qRT-PCRs) were performed using H. volcanii total RNA as a template (0.1 μg), appropriate primers (see Table S1 in the supplemental material), iQ SYBR green supermix (Bio-Rad), and an iCycler (Bio-Rad). RNA was reverse transcribed into cDNA using iSCRIPT (Bio-Rad) according to the manufacturer's instructions. After cDNA synthesis (25°C for 5 min, 42°C for 30 min, and 85°C for 5 min), qRT-PCR mixtures were preheated to 95°C (4 min), followed by 40 amplification cycles consisting of denaturation (95°C, 30 s), annealing (temperatures are listed in Table S1 in the supplemental material; 1 min), and elongation (72°C, 17 s). For RT-PCR, the reaction mixture was preheated to 95°C (4 min), followed by 35 amplification cycles consisting of denaturation (95°C, 30 s), annealing (55°C, 1 min), and elongation (72°C, 19 s), after which a final extension was performed at 72°C (10 min). For each primer pair, negative and positive controls were included to exclude genomic DNA contamination and confirm primer pair function, respectively. For the controls, the reactions were identical, with the following exceptions: the sample was maintained on ice during the reverse transcription step for the negative control, and H. volcanii genomic DNA prepared as previously described (29) was used as a template for the positive control. The products from the (q)RT-PCRs were sequenced as described below to confirm specificity.
Transcriptional reporter construction and assay.
A plasmid-based reporter system in which the promoter regions of glpR-pfkB (HVO_1501-HVO_1500), kdgK1 (HVO_0549), and kdgK2 (HVO_A0328) were fused to the Haloferax alicantei-derived bgaH open reading frame encoding β-galactosidase was used to analyze transcription (9). The bgaH gene was amplified from pTA102 (primers are provided in Table S1 in the supplemental material) and cloned into pJAM202 using NdeI and BlpI, which fused the bgaH gene downstream of the strong rRNA P2 promoter of Halobacterium cutirubrum to generate pJAM2678. The regions upstream of glpR-pfkB, kdgK1, and kdgK2 were amplified from H. volcanii genomic DS70 DNA using the primers listed in Table S1 in the supplemental material and fused with bgaH using XbaI and NdeI to generate pJAM2689 (188-bp glpR-pfkB promoter region), pJAM2705 (89-bp kdgK1 promoter region), pJAM2706 (524-bp kdgK1 promoter region), pJAM2702 (232-bp kdgK2 promoter region), and pJAM2703 (122-bp HVO_A0327-kdgK2 promoter region) (where base pair number represents the region upstream of the translational start codon of the first gene listed for each construct). Promoters were predicted upstream of glpR-pfkB, kdgK1, and kdgK2 according to the methods described by Schneider et al. (37). Plasmid controls pJAM2714 and pJAM2715 were constructed using pJAM2678 by removal of the H. cutirubrum rRNA P2 promoter and the Shine-Dalgarno site (using XbaI and NdeI) and through removal of only the rRNA P2 promoter (using XbaI and BamHI), respectively.
The promoter activity of each construct was assessed quantitatively by assaying β-galactosidase activity as previously described (18). Briefly, cells were grown in 3 ml of appropriate media and harvested at exponential growth (0.3 to 0.5 OD600 units) by centrifugation (15 min, 6,000 × g, 4°C). Cell pellets were washed once in buffer B (50 mM Tris-HCl at pH 7.2 with 2.5 M NaCl, 10 μM MnCl2), resuspended in 300 μl of bgaH buffer (buffer B with 0.1% [wt/vol] β-mercaptoethanol), and lysed using 150 μl of 2% Triton X-100. Debris was removed by centrifugation (10 min, 6,000 × g, 4°C), and the protein concentration in the cell extract was estimated using the Bradford assay (as above). Cells with promoter fusion constructs were assayed at 25°C for β-galactosidase specific activity by measuring the increase in absorbance at 405 nm due to the liberation of o-nitrophenol from o-nitrophenyl-β-d-galactopyranoside (ONPG). The assay mixture (100 μl) contained 20 μl of cell lysate (3 to 5 μg protein), 70 μl of bgaH buffer, and 2.66 mM ONPG (stock solution with 8 mg·ml−1 in 100 mM potassium phosphate buffer, pH 7.2). Negative controls included cells carrying pJAM2714 and pJAM2715 (the bgaH reporter plasmids devoid of promoter elements). Background values in which no substrate (ONPG) was added to the reaction mixture were subtracted from the value for reaction mixtures containing substrate for each lysate tested. All experiments were performed in biological triplicate, and the means ± standard deviations (SD) of the results were calculated. One unit of β-galactosidase activity is defined as the amount of enzyme catalyzing the hydrolysis of 1 μmol ONPG·min−1 with a molar extinction coefficient for o-nitrophenol of 3,300 M−1·cm−1.
DNA sequencing.
The specificity of all PCR and (q)RT-PCR products, including DNA cloned into plasmids listed in Table 1 and amplified from the knockout region of mutant strains, was confirmed by Sanger automated DNA sequencing using an Applied Biosystems model 3130 genetic analyzer (ICBR Genomics Division, University of Florida).
RESULTS AND DISCUSSION
Identification of GlpR as a putative transcriptional repressor of metabolic enzymes.
We previously demonstrated that H. volcanii exhibits glycerol-mediated catabolite repression of glucose metabolism when grown on minimal media containing both glycerol and glucose (38). In order to provide further insight into the central metabolism of haloarchaea, the H. volcanii genome (17) was searched for ORFs encoding proteins with DNA-binding domains that clustered near sugar metabolic operons. One such ORF (HVO_1501; designated glpR), when searched against the NCBI protein database, clustered with the DeoR/GlpR family of transcriptional regulators of sugar metabolism (COG1349) (see Fig. S1A in the supplemental material). The DeoR/GlpR protein family is widespread among bacteria, and its members often serve as transcriptional repressors (3, 16, 28, 34, 41, 44) or activators (14) of either sugar or nucleoside metabolism. DeoR/GlpR-type repressors are composed of approximately 250 amino acids, possess a helix-turn-helix DNA-binding motif near the amino terminus, and generally bind a sugar phosphate effector molecule of the relevant metabolic pathway in the C-terminal portion of the protein, which can also contain oligomerization domains (43). In H. volcanii as well as some haloarchaea and many Gram-positive bacterial species, glpR clusters on the genome with pfkB, encoding PFK (see Fig. S1B in the supplemental material), a key enzyme of fructose metabolism in haloarchaea such as H. volcanii (12, 20). Dendrogram analysis revealed that H. volcanii GlpR clusters to the DeoR/GlpR family of transcriptional regulators, with closest relationship to uncharacterized proteins of haloarchaea and firmicutes (see Fig. S1B and S1C in the supplemental material). This result suggested that GlpR may be involved in regulating sugar metabolic enzymes at the level of transcription in H. volcanii and was thus targeted for further analyses (see below).
Transcripts encoding GlpR and PFK are under the control of a common promoter and are repressed by glycerol.
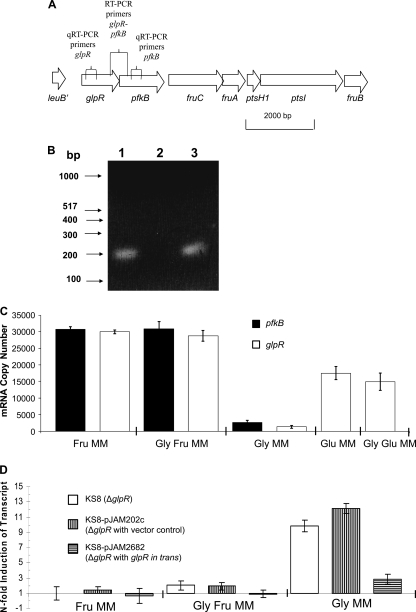
In haloarchaea, PFK is involved in the metabolism of fructose through a modified Embden-Meyerhof-Parnas (EMP) pathway (12). Due to the close proximity of glpR and pfkB on the chromosome (4-bp overlap in coding sequence) (Fig. 1A), we investigated whether these genes were cotranscribed in an operon. RT-PCR was performed using primers designed to amplify a portion of each coding region (Fig. 1A). A single PCR product of expected size (0.2 kb) was detected using synthesized cDNA (Fig. 1B) and was further confirmed by DNA sequencing. No product was detected in the negative-control reaction mixture containing RNA as a template (Fig. 1B). Thus, glpR and pfkB are linked at the level of transcription.
FIG. 1.
Genomic organization and transcript analysis of pfkB encoding phosphofructokinase (PFK) and glpR encoding GlpR, a putative transcriptional repressor of the DeoR/GlpR family in H. volcanii. (A) Schematic representation of glpR and pfkB on the H. volcanii genome and locations of annealing sites (represented as vertical lines) for (q)RT-PCR primers. (B) qRT-PCR reveals that pfkB and glpR are cotranscribed from a common promoter. Total RNA from parent H26 was extracted and reverse transcribed to generate cDNA, which was used as a template for PCR (lane 1). RNA which had not undergone reverse transcription was used as a negative-control template for PCR (lane 2). Genomic DNA was used as a positive-control template for PCR (lane 3). Quick Load DNA markers (100 bp) and molecular sizes are indicated on the left. (C) Absolute quantifications of pfkB- and glpR-specific transcripts reveal increased transcript levels for both genes in the presence of fructose and glucose. Transcript levels were derived from qRT-PCR. (D) Relative quantifications of transcript levels specific for pfkB in glpR mutant KS8, KS8 with a plasmid vector control (pJAM202c), and KS8 with glpR in trans (pJAM2682) compared to the level for parent H26. PFK transcript is upregulated in the absence of fructose (in Gly) in KS8 compared to the level for H26. Transcript levels were derived by qRT-PCR. Calculations are based on the n-fold value for induction of transcript levels in KS8 (and subsequent control and complementary strains) compared to the level for parent H26. Results were normalized to the level for the internal control, ribL. Experiments were performed in triplicate, and the means ± SD were calculated.
Since glpR and pfkB are cotranscribed and PFK activity is largely fructose inducible and to some extent glucose inducible in haloarchaea (20), we investigated whether this regulation was controlled at the level of the pfkB-specific transcript and whether glpR was also induced by sugars. RNA was extracted from parent H26 grown in minimal media supplemented with different carbon sources, including fructose, glucose, and glycerol and various combinations thereof, and was subjected to qRT-PCR using primers specific for the 200-bp coding region of glpR and pfkB (see Table S1 in the supplemental material). Absolute quantification was performed for each transcript. In addition, a transcript specific for the ribosomal protein L10 gene (ribL) was used as an internal control based on previous studies (6a) and our confirmation by qRT-PCR that the n-fold value for induction of transcripts specific for ribL was close to 1.0 when parent H26 was grown in minimal media. With this approach, transcripts were found to be highly upregulated for both glpR (>20-fold ± 7-fold) and pfkB (>10-fold ± 3-fold) in the presence of fructose, regardless of glycerol supplementation, compared to the levels obtained with glycerol alone (Fig. 1C). Transcripts for glpR were also upregulated (>10-fold ± 4-fold) during growth on glucose, regardless of glycerol supplementation, compared to the levels obtained with glycerol alone (Fig. 1C). Thus, our data reveal that glpR and pfkB are cotranscribed from a common promoter and that the transcript levels of this operon are increased by fructose and, to a lesser extent, by glucose. These results are consistent with the observed sugar-dependent alterations in PFK activity for Halococcus saccharolyticus (20), in which the addition of fructose to peptide-rich media stimulated the level of PFK activity and suggest that PFK is regulated at the level of transcription in haloarchaea.
GlpR represses PFK transcription during growth on glycerol.
To analyze the role of GlpR in regulating sugar metabolism, the glpR gene (HVO_1501) was targeted for knockout in H. volcanii. A markerless knockout strategy was used with the H26 ΔpyrE2 parent strain as previously described (1, 6) to generate glpR mutant strain KS8. Gene deletion was confirmed by PCR, Southern blotting, and sequencing analysis (see Fig. S2 in the supplemental material). qRT-PCRs were used to compare the transcript levels of KS8 to those of parent strain H26 on various minimal media. With this approach, a pfkB-specific transcript was found to be significantly upregulated by the glpR knockout (10- to 12-fold) compared to the level for parent H26 when cells were grown in the presence of glycerol as the sole carbon source (Fig. 1D). This was complemented at least in part by providing a copy of glpR in trans, thus ruling out polar effects of the markerless deletion of glpR on the glpR-pfkB operon (Fig. 1D). In contrast to growth on glycerol alone, the glpR knockout had little if any impact on pfkB-specific-transcript levels when cells were grown in the presence of fructose with or without glycerol (Fig. 1D). Transcript levels remained high for pfkB in fructose-containing media for all strains analyzed (Fig. 1C) and were not significantly altered by the glpR mutation (Fig. 1D). Thus, while GlpR was required for the glycerol-mediated repression of pfkB-specific transcripts, GlpR was not needed for the high-levels of the pfkB-specific transcript present when cells were grown on fructose.
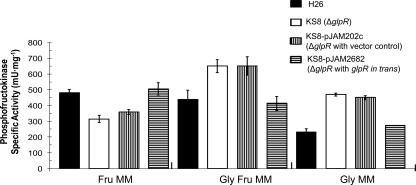
On the basis of the qRT-PCR findings showing that GlpR may serve as a repressor of pfkB-specific transcription in the absence of fructose and presence of glycerol, PFK activity in both parent and glpR-deficient strains was tested to determine if enzyme activity was altered. Consistent with the qRT-PCR results, PFK activity was increased in wild-type cells grown in media supplemented with fructose (versus media with glycerol alone) (Fig. 2). Also consistent with our qRT-PCR results, PFK specific activity was significantly increased by the glpR knockout (2-fold compared to the levels for the parent and glpR-complemented strains) when cells were grown on media with glycerol alone (Fig. 2). Deletion of glpR also resulted in a 1.5-fold increase in PFK activity compared to the levels for the parent and glpR-complemented strains when cells were grown in the presence of both fructose and glycerol (Fig. 2). In contrast to media with glycerol (with or without fructose), PFK activity was decreased during growth on fructose by deletion of glpR (Fig. 2). The reason for the latter finding remains to be determined but does not appear to be at the level of transcription, based on our qRT-PCR results.
FIG. 2.
PFK activity increases when cells are grown on glycerol minimal medium after deletion of glpR. Specific activity of PFK was determined from cell lysates of H. volcanii H26 (parent), KS8 (glpR mutant), KS8-pJAM202c (glpR mutant with vector control), and KS8-pJAM2682 (glpR mutant with glpR in trans) cells grown to log-phase in Fru, Gly-Fru, and Gly as indicated. Experiments were performed in biological triplicate, and the means ± SD were calculated.
The PFK activity of H. volcanii was also measured after growth on peptide-rich YPC medium with or without fructose or glucose. Similar to what was observed for minimal media, PFK activity was reduced when YPC was not supplemented with hexose sugars (see Fig. S3 in the supplemental material). GlpR, however, was not required for this decrease, which is in contrast to the GlpR-dependent reduction in PFK activity observed when sugars were excluded from glycerol minimal medium (Fig. 2; see also Fig. S3 in the supplemental material). It should be noted that the range of PFK activity values determined for the H. volcanii strains under the various conditions (140 to 650 mU·mg−1) was in agreement with PFK activities reported for other haloarchaea (22 mU·mg−1 for H. saccharolyticus [20] and 1,300 mU·mg−1 for Haloarcula vallismortis [33] in peptide media with 25 to 28 mM fructose).
To determine whether the glpR-pfkB operon is regulated by GlpR at the level of transcription, (i) the 188-bp genomic region upstream of the start codon of glpR was fused to the coding region of the H. alicantei bgaH reporter, and (ii) the β-galactosidase activity of this reporter in parent and glpR mutant strains grown on various carbon sources was monitored. With this approach, significant and comparable levels of reporter activity were detected for both parent and glpR mutant strains when cells were grown in the presence of fructose, regardless of glycerol supplementation (Table 2). Under these conditions, the β-galactosidase specific activity measured for the glpR-pfkB promoter was 25 to 34 mU·mg−1, compared to the levels for the negative controls, which lacked promoter elements (less than or equal to 12 mU·mg−1 for all conditions tested) (Table 2). In contrast, when cells were grown on glycerol alone, the promoter activity of the glpR-pfkB operon was reduced in parent strain H26 to levels comparable to that for the vector control, while deletion of glpR increased promoter activity to levels similar to those obtained with growth on fructose (Table 2). Overall, the data obtained with the promoter fusions are consistent with the qRT-PCR and PFK specific activity data and reveal that GlpR is required for transcriptional repression of the glpR-pfkB operon during growth on glycerol minus fructose, possibly by interacting with promoter elements within the 188-bp region upstream of this operon. The results also reveal that the glpR-pfkB promoter is relatively moderate at driving the expression of heterologous genes such as bgaH compared to the strong and constitutive H. cutirubrum rRNA P2 promoter, which reached levels of up to 260 mU·mg−1 of β-galactosidase activity for the reporter fusion (Table 2).
TABLE 2.
Transcription of a bgaH β-galactosidase reporter gene from glpR-pfkB, kdgK1, and kdgK2 promoters in parent and glpR mutant strains grown on various carbon sources
| Minimal medium | Mutation | β-Galactosidase sp act (mU/mg)a for indicated promoter |
||||||
|---|---|---|---|---|---|---|---|---|
| glpR-pfkB (188 bp) | kdgK1 (524 bp) | kdgK1 (89 bp) | HvoA0327-kdgK2 (122 bp) | kdgK2 (232 bp) | P2 rrnA (551 bp) | None (Shine-Dalgarno site only) | ||
| Gly | None | 9.2 ± 2 | 160 ± 3 | 180 ± 3 | 85 ± 0.01 | 14 ± 0.01 | 260 ± 10 | 8.1 ± 0.1 |
| ΔglpR | 32 ± 3 | 270 ± 10 | 250 ± 8 | 99 ± 0.8 | 14 ± 0.01 | 250 ± 1 | 8.2 ± 0.2 | |
| Fru | None | 27 ± 1 | ND | ND | ND | ND | 160 ± 7 | 8.2 ± 0.1 |
| ΔglpR | 25 ± 0.3 | ND | ND | ND | ND | 150 ± 5 | 7.9 ± 0.2 | |
| Gly-Fru | None | 27 ± 0.2 | ND | ND | ND | ND | 190 ± 10 | 7.3 ± 0.9 |
| ΔglpR | 34 ± 0.1 | ND | ND | ND | ND | 180 ± 9 | 8.2 ± 0.2 | |
| Glu | None | ND | 300 ± 4 | 280 ± 0.01 | 190 ± 0.01 | 41 ± 0.1 | 250 ± 7 | 7.3 ± 0.9 |
| ΔglpR | ND | 250 ± 2 | 280 ± 3 | 160 ± 0.01 | 39 ± 0.02 | 250 ± 0.01 | 12.0 ± 0.02 | |
| Gly-Glu | None | ND | 230 ± 5 | 200 ± 0.01 | 150 ± 4 | 33 ± 0.01 | 260 ± 7 | 8.1 ± 0.05 |
| ΔglpR | ND | 300 ± 4 | 280 ± 1 | 160 ± 0.01 | 34 ± 0.01 | 260 ± 0.01 | 8.2 ± 0.2 | |
β-Galactosidase activities were determined from the lysates of cells carrying transcriptional fusion constructs of promoters grown to log phase in minimal media with Fru, Glu, and Gly as indicated. Promoter fusions included the start codon and genomic region (indicated in bp) immediately upstream of each target gene. Note that glpR-pfkB and HvoA0327-kdgK2 are operons most likely transcribed from these promoters. β-Galactosidase activity values increased by glpR mutation are in bold. Experiments were performed in biological triplicate, and the means ± SD were calculated. ND, not determined.
GlpR represses KDGK transcription during growth on glycerol.
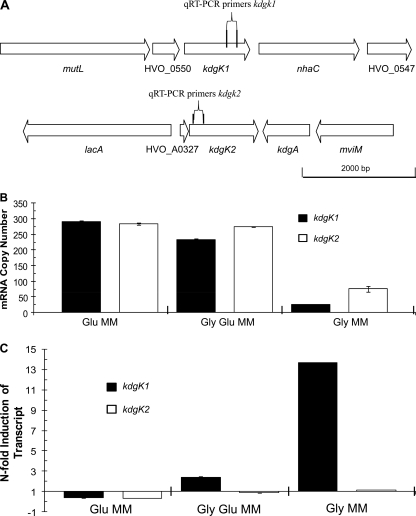
To address whether GlpR is involved in the repression of other sugar catabolic pathways, qRT-PCR primers were designed for analysis of the transcript levels of genes encoding homologs of KDGK, including chromosomally located kdgK1 (HVO_0549) and pHV4-carried kdgK2 (HVO_A0328) (Fig. 3A). In haloarchaea, KDGK is involved in the metabolism of glucose through a modified Entner-Doudoroff pathway (12). In H. volcanii, both kdgK1 and kdgK2 are predicted to encode active KDGK enzymes based on their conservation of active-site residues and their close relationship to KDGKs from other microbes that have been characterized at the biochemical level (see Fig. S4 in the supplemental material). Since KdgK1 and KdgK2 are close homologs (62% identity and 74% similarity in primary amino acid sequence), DNA sequencing was used to confirm qRT-PCR product specificity. On the basis of qRT-PCRs, the transcripts of both kdgK genes were significantly upregulated (4- to 12-fold) when parent strain H26 was grown on media containing glucose (with or without glycerol) compared to the levels obtained with glycerol alone (Fig. 3B). Furthermore, chromosomally located kdgK1 was significantly upregulated in the glpR mutant compared to the level for parent H26 when cells were grown on glycerol minimal medium (14-fold) but was relatively unaltered when cells were grown on media with glucose (i.e., 0.38-fold ± 0.02-fold lower in the presence of glucose alone and 2.4-fold ± 0.14-fold higher on glucose plus glycerol) (Fig. 3C). Unlike kdgK1, the glpR mutation had little if any impact on kdgK2 transcript levels compared to those for parent H26 on all media examined (i.e., Glu MM, Gly-Glu MM, and Gly MM) (Fig. 3C).
FIG. 3.
Genomic organization and transcript analysis of KDGK genes located on the chromosome and megaplasmid pHV4 of H. volcanii. (A) Schematic representations of chromosomal kdgK1 (upper) and related pHV4-carried kdgK2 (lower) genes and their neighbors and locations of annealing sites (represented as vertical lines) for qRT-PCR primers. (B) Absolute quantification of transcripts specific for kdgK1 and kdgK2 reveals increased transcript levels for both genes during growth on glucose, regardless of glycerol supplementation. Transcripts specific for kdgK1 and kdgK2 were upregulated 12-fold and 4-fold, respectively, in the presence of glucose-containing media compared to the levels obtained with glycerol alone. Transcript levels were derived from qRT-PCR as described in Materials and Methods. (C) Relative quantifications of transcript levels specific for kdgK1 and kdgK2. Chromosomal kdgK1 transcripts are upregulated in a glpR mutant (KS8) in the presence of glycerol, regardless of glucose supplementation, compared to the level for parent H26. Transcript levels were derived by qRT-PCR. Calculations are based on the n-fold value for induction of transcripts in the designated minimal media for glpR-deficient cells compared to the level for parent H26. Results were normalized to the n-fold value for induction of the internal control, ribL. Experiments were performed in triplicate, and the means ± SD were calculated.
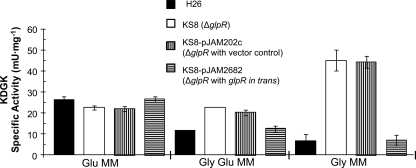
On the basis of the qRT-PCR finding that GlpR may serve as a repressor of kdgK1 transcript levels during growth on glycerol, KDGK enzyme activity in both parent and glpR mutant strains was monitored to determine the role of GlpR at the protein level. Similar to transcript levels, KDGK activity was reduced when cells were grown on glycerol compared to the level obtained with glucose (Fig. 4). Likewise, deletion of glpR resulted in significant increases in KDGK activity on glycerol, with the increase on glycerol and glucose modest (2-fold) compared to the level obtained with glycerol alone (7-fold) (Fig. 4). These levels were reduced to parental levels by providing glpR in trans (Fig. 4). The glpR knockout did not affect KDGK activity when cells were grown with glucose as the sole carbon source (Fig. 4) or on peptide-rich YPC media with or without glucose or fructose (see Fig. S5 in the supplemental material). On the basis of these results, the KDGK activity of H. volcanii was consistent with the qRT-PCR data in which glpR was needed for repression of kdgK1 transcript levels on glycerol. Furthermore, KDGK specific activity (7.1 to 45 mU·mg−1), although higher, was within a reasonable range observed for other haloarchaea (i.e., H. saccharolyticus at 2 to 5 mU·mg−1) (20).
FIG. 4.
KDGK activity is increased by deletion of glpR in cells grown on glycerol minimal medium. KDGK specific activities were determined from cell lysates of log-phase H. volcanii H26 (parent), KS8 (glpR mutant), KS8-pJAM202c (glpR mutant with vector control), and KS8-pJAM2682 (glpR mutant with glpR in trans) grown on Glu, Gly-Glu, and Gly as indicated. Experiments were performed in biological triplicate, and the means ± SD were calculated.
To further investigate the role of GlpR at the level of KDGK transcription, the kdgK1 and kdgK2 promoter regions were fused to a bgaH-based transcriptional reporter (as described above). Reporter fusions of various lengths and start sites were generated to ensure that the entire promoter and potential regulatory elements were included for analysis, and transcription of the kdgK promoter constructs was monitored by an assay of β-galactosidase activity (see Materials and Methods and Table 2 for details). Although the β-galactosidase activities of the kdgK1- and kdgK2-bgaH transcriptional fusions were somewhat high on glycerol alone (Gly MM), these activities increased significantly upon glucose supplementation (Table 2). Furthermore, the β-galactosidase activities of both kdgK1-bgaH fusions were higher in the glpR mutant than in the parent when the cells were grown on glycerol alone (Table 2). In contrast to kdgK1, deletion of glpR did not affect transcription of the kdgK2-promoter fusions as measured using the reporter constructs (Table 2). Thus, GlpR appears to reduce KDGK activity and kdgK1 transcript levels through transcriptional repression of kdgK1 when cells are grown on glycerol. It should be noted that both the 89- and the 524-bp kdgK1 and 122-bp HVO_A0327-kdgK2 promoters are relatively robust at driving expression of the heterologous bgaH reporter. β-Galactosidase activity for the kdgK promoters was greater than that of the H. cutirubrum rRNA P2 promoter (Table 2), used routinely for high-level production of proteins in H. volcanii (22, 40).
While our data reveal that GlpR is required for the transcriptional repression of kdgK1 and the reduction of KDGK enzyme activity when the cells are grown on glycerol, it remains to be determined why the levels of β-galactosidase activity are relatively high for both kdgK1-bgaH reporter fusions on glycerol compared to the nearly baseline level of transcripts detected for kdgK1 by qRT-PCR during growth on this same medium. Posttranscriptional mechanisms and/or insufficient levels of GlpR, needed to repress transcription of the kdgK1-bgaH reporter fusion on the multicopy plasmids pJAM2705 and pJAM2706, may explain these findings.
GlpR and diauxic growth.
The data presented here indicate that GlpR represses transcription of the glpR-pfkB operon and kdgK1 in the presence of glycerol and that GlpR is no longer an active repressor of these operons when cells are grown on media with fructose or glucose. It is unclear, however, whether GlpR and/or these operons are control points for the diauxie previously observed during growth on glucose and glycerol, in which glucose metabolism is repressed by glycerol (38) (see Fig. S6 in the supplemental material). To directly examine the role of GlpR in this phenomenon and determine whether glycerol represses fructose metabolism, the glpR deletion was introduced into a previously described glycerol kinase (glpK) mutant strain, KS4 (38), for phenotypic analysis. Since both glpR and glpK deletions are in separate operons and markerless, this double mutant strain was readily generated. Parent and glpK mutant strains were analyzed as a replicate of previous experiments (38) along with single glpR and double glpR glpK mutant strains for growth and carbon utilization on minimal media, including fructose, glucose, and/or glycerol. Interestingly, unlike the glycerol-mediated catabolite repression of glucose metabolism observed for H. volcanii cells (38), glycerol and fructose are coutilized when provided at equimolar concentrations (see Fig. S7 in the supplemental material). Furthermore, GlpR did not appear to regulate the diauxic growth of H. volcanii on glycerol and glucose, based on the finding that the glpR mutation did not alleviate this catabolite repression (see Fig. S6 in the supplemental material).
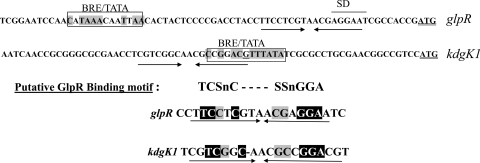
The promoter regions for kdgK1 and glpR-pfkB include a putative GlpR binding motif.
Based on the finding that GlpR likely controls pfkB and kdgK1 transcription within the 188-bp and 89-bp region upstream of the translational start codon for each gene, respectively, these regions were analyzed to determine putative promoter elements and the GlpR binding motif(s). Shine-Dalgarno sites and promoter elements, including the TFB responsive element (BRE) and the TATA box, were predicted based on consensus sequences (15). Next, inverted repeats located near these elements were aligned to identify the potential GlpR binding site(s). With this approach, an inverted hexameric repeat, TCSnCn(3-4)SSnGGA (where S is G or C and n is any nucleotide), which overlaps the putative kdgK1 promoter and is downstream of the putative glpR-pfkB promoter, was identified (Fig. 5). This motif was not found within the kdgK2 promoter region, consistent with our findings that GlpR regulates both kdgK1 and pfkB but not kdgK2. Future investigation is expected to provide insight as to whether GlpR binds this motif and represses transcription from the kdgK1 and glpR-pfkB promoters when cells are grown on glycerol.
FIG. 5.
Genomic regions upstream of glpR-pfkB and kdgK1 include a conserved inverted repeat that may serve in GlpR binding and transcriptional repression. Regions upstream of glpR-pfkB and kdgK1 were analyzed for potential GlpR binding sites, such as inverted repeats near putative Shine Dalgarno (SD) sites and promoter elements, including the TFB responsive element (BRE) and the TATA box (the consensus sequences were CRnAAT for BRE and TTTAWA for the TATA box, where W is A or T, R is A or G, and n is any nucleotide base). Residues matching the consensus BRE/TATA box sequence are boxed and highlighted in gray. SD sites are indicated by a line above the DNA sequence, and the translational start codon is double underlined. An inverted hexameric repeat with a consensus sequence, TCSnCn(3-4)SSnGGA (where S is C or G and n is any nucleotide base), was found common to the glpR-pfkB and kdgK1 promoter regions. Inverted repeats are indicated, with arrows displaying directionality.
Conclusions.
In this study, we demonstrate that the DeoR/GlpR-type GlpR protein represses the transcription of genes encoding both fructose and glucose metabolic enzymes when H. volcanii cells are grown on glycerol minimal medium. The transcript levels and activities of key enzymes of fructose and glucose metabolism, PFK (pfkB) and KDGK (kdgK1), were reduced in the presence of glycerol alone compared to the levels obtained with fructose or glucose. Analysis of the transcriptional fusions of the pfkB and kdgK1 promoters in a glpR mutant strain revealed that GlpR was required for transcriptional repression of these genes during growth on glycerol. In contrast, transcription from the kdgK2 promoter, although reduced by glycerol, was not significantly altered by the glpR mutation, suggesting that an additional regulator is used for kdgK2. The glpR and pfkB genes were cotranscribed under a common promoter based on RT-PCR analysis, suggesting that GlpR also serves as an autoregulator, and transcription of the glpR-pfkB operon was reduced during growth on glycerol alone compared to that obtained with growth on fructose or glucose. The results presented here provide the first genetic and biochemical evidence of a DeoR/GlpR-type transcriptional repressor protein in haloarchaea. Future biochemical characterization of GlpR and its effector is expected to provide further insight into the transcriptional regulation of sugar metabolism in H. volcanii as well as other microorganisms with similar gene organizations. Further analysis will also provide new insight into how DeoR/GlpR family members can interact with and regulate archaeal basal-transcriptional machineries composed of eukaryote-like RNAP and TBP and TFB proteins.
Supplementary Material
Acknowledgments
We thank T. Allers for providing parent strain H26, plasmid pTA131, and plasmid pTA102. We also thank Savita Shanker at the UF Genomics Core of UF ICBR for DNA sequencing.
This work was funded in part by NIH R01 GM057498 and DOE DE-FG02-05ER15650 to J.A.M.-F.
Footnotes
Published ahead of print on 8 October 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allers, T., H. P. Ngo, M. Mevarech, and R. G. Lloyd. 2004. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70:943-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 1999. DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 27:4658-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrière, C., M. Veiga-da-Cunha, N. Pons, E. Guédon, S. A. van Hijum, J. Kok, O. P. Kuipers, D. S. Ehrlich, and P. Renault. 2005. Fructose utilization in Lactococcus lactis as a model for low-GC gram-positive bacteria: its regulator, signal, and DNA-binding site. J. Bacteriol. 187:3752-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, S. D. 2005. Archaeal transcriptional regulation—variation on a bacterial theme? Trends Microbiol. 13:262-265. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S. D., and S. P. Jackson. 2001. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 4:208-213. [DOI] [PubMed] [Google Scholar]
- 6.Bitan-Banin, G., R. Ortenberg, and M. Mevarech. 2003. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185:772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Brenneis, M., O. Hering, C. Lange, and J. Soppa. 2007. Experimental characterization of cis-acting elements important for translation and transcription in halophilic archaea. PLoS Genet. 3:e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cline, S. W., W. L. Lam, R. L. Charlebois, L. C. Schalkwyk, and W. F. Doolittle. 1989. Transformation methods for halophilic archaebacteria. Can. J. Microbiol. 35:148-152. [DOI] [PubMed] [Google Scholar]
- 8.Coker, J. A., and S. DasSarma. 2007. Genetic and transcriptomic analysis of transcription factor genes in the model halophilic Archaeon: coordinate action of TbpD and TfbA. BMC Genet. 8:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delmas, S., L. Shunburne, H. P. Ngo, and T. Allers. 2009. Mre11-Rad50 promotes rapid repair of DNA damage in the polyploid archaeon Haloferax volcanii by restraining homologous recombination. PLoS Genet. 5:e1000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyall-Smith, M. 2008. The Halohandbook: protocols for halobacterial genetics. http://www.haloarchaea.com/resources/halohandbook_2008_v7.pdf.
- 11.Facciotti, M. T., D. J. Reiss, M. Pan, A. Kaur, M. Vuthoori, R. Bonneau, P. Shannon, A. Srivastava, S. M. Donohoe, L. E. Hood, and N. S. Baliga. 2007. General transcription factor specified global gene regulation in archaea. Proc. Natl. Acad. Sci. U. S. A. 104:4630-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falb, M., K. Müller, L. Königsmaier, T. Oberwinkler, P. Horn, S. von Gronau, O. Gonzalez, F. Pfeiffer, E. Bornberg-Bauer, and D. Oesterhelt. 2008. Metabolism of halophilic archaea. Extremophiles 12:177-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiorentino, G., R. Ronca, R. Cannio, M. Rossi, and S. Bartolucci. 2007. MarR-like transcriptional regulator involved in detoxification of aromatic compounds in Sulfolobus solfataricus. J. Bacteriol. 189:7351-7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaurivaud, P., F. Laigret, M. Garnier, and J. M. Bové. 2001. Characterization of FruR as a putative activator of the fructose operon of Spiroplasma citri. FEMS Microbiol. Lett. 198:73-78. [DOI] [PubMed] [Google Scholar]
- 15.Gregor, D., and F. Pfeifer. 2005. In vivo analyses of constitutive and regulated promoters in halophilic archaea. Microbiology 151:25-33. [DOI] [PubMed] [Google Scholar]
- 16.Haghjoo, E., and J. E. Galán. 2007. Identification of a transcriptional regulator that controls intracellular gene expression in Salmonella typhi. Mol. Microbiol. 64:1549-1561. [DOI] [PubMed] [Google Scholar]
- 17.Hartman, A. L., C. Norais, J. H. Badger, S. Delmas, S. Haldenby, R. Madupu, J. Robinson, H. Khouri, Q. Ren, T. M. Lowe, J. Maupin-Furlow, M. Pohlschroder, C. Daniels, F. Pfeiffer, T. Allers, and J. A. Eisen. 2010. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One 5:e9605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes, M. L., and M. L. Dyall-Smith. 2000. Sequence and expression of a halobacterial β-galactosidase gene. Mol. Microbiol. 36:114-122. [DOI] [PubMed] [Google Scholar]
- 19.Humbard, M. A., G. Zhou, and J. A. Maupin-Furlow. 2009. The N-terminal penultimate residue of 20S proteasome alpha1 influences its N(alpha) acetylation and protein levels as well as growth rate and stress responses of Haloferax volcanii. J. Bacteriol. 191:3794-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnsen, U., M. Selig, K. B. Xavier, H. Santos, and P. Schönheit. 2001. Different glycolytic pathways for glucose and fructose in the halophilic archaeon Halococcus saccharolyticus. Arch. Microbiol. 175:52-61. [DOI] [PubMed] [Google Scholar]
- 21.Kaczowka, S. J., and J. A. Maupin-Furlow. 2003. Subunit topology of two 20S proteasomes from Haloferax volcanii. J. Bacteriol. 185:165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaczowka, S. J., C. J. Reuter, L. A. Talarico, and J. A. Maupin-Furlow. 2005. Recombinant production of Zymomonas mobilis pyruvate decarboxylase in the haloarchaeon Haloferax volcanii. Archaea 1:327-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanai, T., J. Akerboom, S. Takedomi, H. J. van de Werken, F. Blombach, J. van der Oost, T. Murakami, H. Atomi, and T. Imanaka. 2007. A global transcriptional regulator in Thermococcus kodakaraensis controls the expression levels of both glycolytic and gluconeogenic enzyme-encoding genes. J. Biol. Chem. 282:33659-33670. [DOI] [PubMed] [Google Scholar]
- 24.Keese, A. M., G. J. Schut, M. Ouhammouch, M. W. Adams, and M. Thomm. 2010. Genome-wide identification of targets for the archaeal heat shock regulator phr by cell-free transcription of genomic DNA. J. Bacteriol. 192:1292-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krüger, K., T. Hermann, V. Armbruster, and F. Pfeifer. 1998. The transcriptional activator GvpE for the halobacterial gas vesicle genes resembles a basic region leucine-zipper regulatory protein. J. Mol. Biol. 279:761-771. [DOI] [PubMed] [Google Scholar]
- 26.Kyrpides, N. C., and C. A. Ouzounis. 1999. Transcription in archaea. Proc. Natl. Acad. Sci. U. S. A. 96:8545-8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S. J., A. Engelmann, R. Horlacher, Q. Qu, G. Vierke, C. Hebbeln, M. Thomm, and W. Boos. 2003. TrmB, a sugar-specific transcriptional regulator of the trehalose/maltose ABC transporter from the hyperthermophilic archaeon Thermococcus litoralis. J. Biol. Chem. 278:983-990. [DOI] [PubMed] [Google Scholar]
- 28.Munch-Petersen, A., and N. Jensen. 1990. Analysis of the regulatory region of the Escherichia coli nupG gene, encoding a nucleoside-transport protein. Eur. J. Biochem. 190:547-551. [DOI] [PubMed] [Google Scholar]
- 29.Ng, W. L., C. F. Yang, J. T. Halladay, P. Arora, and S. DasSarma. 1995. Archaea: a laboratory manual, p. 179-184. Cold Spring Harbor Laboratory Press, New York, NY.
- 30.Reference deleted.
- 31.Offner, S., and F. Pfeifer. 1995. Complementation studies with the gas vesicle-encoding p-vac region of Halobacterium salinarium PHH1 reveal a regulatory role for the p-gvpDE genes. Mol. Microbiol. 16:9-19. [DOI] [PubMed] [Google Scholar]
- 32.Oren, A. 1994. Enzyme diversity in halophilic archaea. Microbiologia 10:217-228. [PubMed] [Google Scholar]
- 33.Rangaswamy, V., and W. Altekar. 1994. Characterization of 1-phosphofructokinase from halophilic archaebacterium Haloarcula vallismortis. Biochim. Biophys. Acta 1201:106-112. [DOI] [PubMed] [Google Scholar]
- 34.Ray, W. K., and T. J. Larson. 2004. Application of AgaR repressor and dominant repressor variants for verification of a gene cluster involved in N-acetylgalactosamine metabolism in Escherichia coli K-12. Mol. Microbiol. 51:813-826. [DOI] [PubMed] [Google Scholar]
- 35.Reeve, J. N., K. Sandman, and C. J. Daniels. 1997. Archaeal histones, nucleosomes, and transcription initiation. Cell 89:999-1002. [DOI] [PubMed] [Google Scholar]
- 36.Schmid, A. K., D. J. Reiss, M. Pan, T. Koide, and N. S. Baliga. 2009. A single transcription factor regulates evolutionarily diverse but functionally linked metabolic pathways in response to nutrient availability. Mol. Syst. Biol. 5:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider, K. L., K. S. Pollard, R. Baertsch, A. Pohl, and T. M. Lowe. 2006. The UCSC archaeal genome browser. Nucleic Acids Res. 34:D407-D410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherwood, K. E., D. J. Cano, and J. A. Maupin-Furlow. 2009. Glycerol-mediated repression of glucose metabolism and glycerol kinase as the sole route of glycerol catabolism in the haloarchaeon Haloferax volcanii. J. Bacteriol. 191:4307-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soppa, J. 1999. Transcription initiation in Archaea: facts, factors and future aspects. Mol. Microbiol. 31:1295-1305. [DOI] [PubMed] [Google Scholar]
- 40.Uthandi, S., B. Saad, M. A. Humbard, and J. A. Maupin-Furlow. 2010. LccA, an archaeal laccase secreted as a highly stable glycoprotein into the extracellular medium by Haloferax volcanii. Appl. Environ. Microbiol. 76:733-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weissenborn, D. L., N. Wittekindt, and T. J. Larson. 1992. Structure and regulation of the glpFK operon encoding glycerol diffusion facilitator and glycerol kinase of Escherichia coli K-12. J. Biol. Chem. 267:6122-6131. [PubMed] [Google Scholar]
- 42.Xie, Y., and J. N. Reeve. 2005. Regulation of tryptophan operon expression in the archaeon Methanothermobacter thermautotrophicus. J. Bacteriol. 187:6419-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng, G., S. Ye, and T. J. Larson. 1996. Repressor for the sn-glycerol 3-phosphate regulon of Escherichia coli K-12: primary structure and identification of the DNA-binding domain. J. Bacteriol. 178:7080-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng, X., and H. H. Saxild. 1999. Identification and characterization of a DeoR-specific operator sequence essential for induction of dra-nupC-pdp operon expression in Bacillus subtilis. J. Bacteriol. 181:1719-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, G., D. Kowalczyk, M. A. Humbard, S. Rohatgi, and J. A. Maupin-Furlow. 2008. Proteasomal components required for cell growth and stress responses in the haloarchaeon Haloferax volcanii. J. Bacteriol. 190:8096-8105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zillig, W., K. O. Stetter, and D. Janekovic. 1979. DNA-dependent RNA polymerase from the archaebacterium Sulfolobus acidocaldarius. Eur. J. Biochem. 96:597-604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.