Abstract
We show that phage lysogenization, lysogens, and prophage induction are all targeted by CRISPR. The results demonstrate that genomic DNA is not immune to the CRISPR system, that the CRISPR system does not require noncytoplasmic elements, and that the system protects from phages entering and exiting the lysogenic cycle.
The CRISPR system was recently identified as an adaptive defense mechanism against bacteriophages and extrachromosomal elements. The function of the CRISPR system in antiviral defense was demonstrated experimentally for the first time in 2007 by Barrangou et al. for Streptococcus thermophilus (3). CRISPR protection from lytic proliferation of phages and horizontal gene transfer has since been reported for different bacterial species (5, 11, 17). However, studies of the CRISPR response to temperate phages are scarce. It has been shown that Streptococcus pyogenes strains harboring a CRISPR system contain few or no prophages, whereas strains lacking a CRISPR system are polylysogens. It has also been shown that strains harboring spacers against prophages are free of these prophages (4, 7, 9, 12). Another study has shown that the presence of two spacers against a phage of Pseudomonas aeruginosa, DMS3, inhibits biofilm formation and swarming motility of the lysogen in a CRISPR-dependent manner (20). This phenomenon shown for P. aeruginosa demonstrates a function of the CRISPR system in controlling group behavior responses of a lysogen by an uncharacterized mechanism. Nevertheless, experimental evidence of CRISPR protection from lysogenization and lysogens, which may explain the mutually exclusive relationship between CRISPR spacers and prophages, has not yet been demonstrated.
The CRISPR activity demonstrated thus far against plasmid transfer or lytic growth of phages involves passage of the alien DNA through membranes. It is still unknown whether this membrane passage is essential for the activity of the CRISPR system, which perhaps requires involvement of a membrane or periplasmic protein(s). A mechanism requiring such proteins should be excluded if the CRISPR system were shown to act on prophages already present in the cytoplasm. Showing CRISPR activity against an integrated prophage will also address whether the bacterial genome is protected from the CRISPR system. Recent findings show that small parts of the CRISPR system—CRISPR arrays—are protected by virtue of their flanking repeats, at least in Staphylococcus epidermidis (13). Activity of the CRISPR system against a prophage integrated in the genome would suggest that the genomic DNA is not recognized as “self” due to unique modifications, as is the case with protection from restriction endonucleases (14). In this short report, several remarkable aspects of CRISPR-mediated protection are revealed. (i) Lysogenization is prevented by the CRISPR system. (ii) Integrated prophage is targeted by CRISPR, resulting in cellular death. (iii) The CRISPR system can rescue bacteria from prophage induction. The results also demonstrate that the CRISPR system is active in the cytoplasm without requiring noncytoplasmic elements and that DNA immunity is not conferred by its integration into the genome per se.
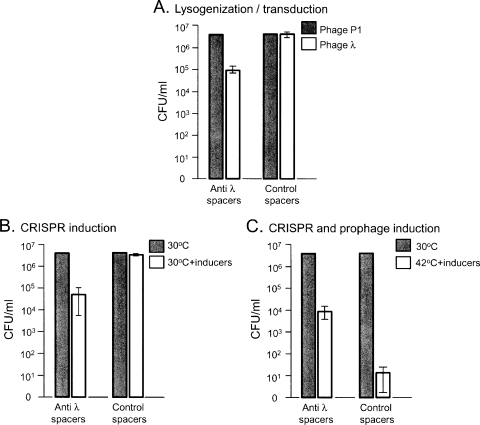
Escherichia coli and its temperate phage λ have been well characterized, and the activity of the CRISPR system against a lytic variant of phage λ (λvir), but not against a lysogenic variant, has been recently demonstrated (5). We therefore examined the effectiveness of the CRISPR system in preventing lysogenization of the host. We used an hns mutant strain from the Keio collection (2), since the hns product, H-NS, is a repressor of the CRISPR system in E. coli (16), and we wanted to examine an active system, as has been recently described (15). The hns mutant strain also harbored a T7-RNA polymerase in its genome, under an arabinose-induced promoter to allow efficient expression from T7 promoters. Plasmids pWUR478 and pWUR477, harboring, respectively, spacers with homology to λ genes J, O, R, and E, here termed “anti-λ spacers,” or control spacers without homology to λ (described in reference 5; see also Table S1 in the supplemental material), both expressed from a T7 promoter, were individually introduced into this host. The two strains harboring anti-λ or control spacers were grown with 0.2% arabinose and 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), incubated at 30°C with λcI857tel, a λ phage which confers tellurite resistance upon lysogenization, and then plated on LB agar supplemented with tellurite, arabinose, and IPTG to determine lysogenization efficiency. As shown in Fig. 1A, the strain harboring the anti-λ spacers lysogenized ∼100-fold less efficiently than the strain carrying the control spacers, suggesting that the CRISPR system attacks the phage early enough in the infection stage that the lysogenic growth cycle could be prevented. The two strains were capable of receiving phage DNA with no homology to the λ spacers at comparable efficiencies, as determined by a control experiment using P1 phage transduction, indicating that the observed reduction in lysogenization is a direct consequence of specific CRISPR activity. Although maybe not surprising, this is the first demonstration of CRISPR protection from a temperate phage entering the lysogenic cycle. This demonstration further supports the notion that the CRISPR system does not use an RNA interference (RNAi) mechanism to exert its effect, as is probably the case in archaea (8), but rather targets the DNA, as elegantly shown for Staphylococcus epidermidis (11), because none of the genes targeted by the anti-λ spacers are known to be required for lysogenization.
FIG. 1.
CRISPR effects on different stages in phage λ growth. (A) Lysogenization. An hns mutant strain with anti-λ spacers or control spacers was incubated at 30°C with the λcI857tel phage at a multiplicity of infection of 1 and then inoculated on LB agar supplemented with 2.5 μg/ml tellurite to determine lysogenization efficiency. As a control for phage infection, we incubated the cells with a lysate of phage P1 grown on Tetr cells and then inoculated them on LB agar supplemented with 10 μg/ml tetracycline. CFU were counted after 18 h of incubation at 30°C. (B) CRISPR induction in lysogens. λcI857kan E. coli BL21AI lysogens transformed with plasmids encoding the CRISPR system genes cas3 and casABCDE and a plasmid encoding a CRISPR array harboring anti-λ or control spacers were inoculated on LB plates supplemented with 100 μg/ml ampicillin, 50 μg/ml streptomycin, and 35 μg/ml chloramphenicol, with or without inducers (0.2% l-arabinose and 1 mM IPTG). CFU were counted after 18 h of incubation at 30°C. (C) CRISPR and prophage induction. λcI857kan E. coli BL21AI lysogens transformed as described for panel B were inoculated and incubated on LB plates as in panel B at 30°C without inducers or at 42°C with inducers. CFU were counted after 18 h of incubation. The control P1 transduction for panel A and the nontreated samples for panels B and C were normalized to 5 × 106 CFU/ml, and the treated samples were calculated accordingly. Bars in all panels represent averages ± standard deviations from three independent experiments.
We speculated that activation of the CRISPR system against a prophage integrated in the genome would cause catastrophic DNA breaks, resulting in cell death. To examine the fate of an established lysogen upon CRISPR activation, we used an inducible CRISPR system, which can be activated following lysogenization. λcI857kan was used to lysogenize E. coli BL21AI, a strain which does not code for any of the known CRISPR system genes. Lysogens were then transformed with plasmids encoding the CRISPR system genes cas3 and casABCDE and a plasmid encoding the anti-λ spacers (pWUR478) or control spacers (pWUR477), as described in reference 5 and in Table S1 in the supplemental material, except that the kanamycin resistance cassette of pWUR397 was replaced with an ampicillin resistance cassette (pWUR397A) to allow appropriate selection for λcI857kan, encoding a kanamycin resistance cassette. The casABCDE, cas3, and CRISPR arrays are controlled by a T7 promoter and a lacI operator and are fully expressed when IPTG is added and when transcription of T7 RNA polymerase is induced by l-arabinose. While establishing the system, we observed that transformation of the lysogen with the plasmid encoding the anti-λ spacers is much less efficient than transformation with the control plasmid. We also observed that plasmid loss is markedly high in cells harboring spacers against the lysogen, but not with the control spacers (see Table S1 in the supplemental material). This result indicates that even low CRISPR activity, due to leakage of its expression, is toxic to cells harboring a lysogen against which the spacers are targeted. Mutants neutralizing the CRISPR system may evolve easily due to this toxicity. We therefore verified our results by carrying out the experiments described in the following paragraph by using 10 individual colonies picked directly from the transformation plate and minimizing the growth cycles of these transformants. To examine its effect against the integrated λ prophage, the CRISPR system was induced by inoculating cells on LB agar plates containing 0.2% l-arabinose and 1 mM IPTG and supplemented with antibiotics to maintain the plasmids. Figure 1B shows that whereas CRISPR induction had no effect on the growth of bacteria harboring the control spacers, it killed over 98% of the bacteria harboring the anti-λ spacers. We speculate that bacterial death due to induction of the CRISPR system is a result of neutralization of the prophage DNA, most likely by cleavage of the corresponding target DNA, which eventually leads to genomic DNA destruction. Direct evidence for DNA cleavage has still not been provided and is the subject of current studies. An alternative explanation is that the CRISPR system affects the level of cI repressor in the cell, leading to induction of the prophage. We ruled out the latter possibility experimentally by monitoring phage production in the culture supernatants. Cultures grown as described above, harboring control or anti-λ spacers, were induced with arabinose and IPTG and aerated for 5 h at 30°C or at 42°C as a control. Supernatants were then collected from both cultures. Burst sizes of less than 1 PFU/cell were measured for either the induced or noninduced supernatants grown at 30°C, whereas burst sizes of ∼100 PFU/cell were measured for the cultures grown at 42°C. These results indicate that the observed cell death of the induced culture harboring anti-λ spacers at 30°C is due to the CRISPR activity and not to prophages entering the lytic cycle. Our demonstration that lysogenic DNA is not protected from attack by the CRISPR system indicates, at least for E. coli, that genomic DNA enjoys no inherent protection from CRISPR activity and that the main protective mechanism must be in the adaptation step (the step in which spacers are acquired). The adaptation step, in some bacterial species, selects for spacers with a corresponding unique adjacent motif in the target DNA, by an unknown mechanism (6). Our results emphasize that the adaptation step must also discriminate between environmental DNA samples and bacterial genomic DNA samples, in order to prevent self targeting. Unraveling the mechanism of the adaptation step will shed light on the discrimination of acquisition of self versus alien DNA.
The suicidal activity of the CRISPR system in this case represents altruistic behavior that has also been exemplified in several abortive infection mechanisms in which a cell “commits suicide” in order to stop replication of an infecting phage (10). From an evolutionary standpoint, such activity is beneficial, as it protects the entire bacterial population, which carries a genetic load similar to that of the “suicidal” bacterium, from phage infection. The activity reported for the CRISPR system in preventing biofilm formation and swarming motility of a P. aeruginosa lysogen is also explained, by a similar rationale, as a defensive activity of an individual bacterium from the spread of lysogens (20). The lysogenized bacterium, in this case, sacrifices valuable resources by reducing its contact with the bacterial population in order to prevent viral spread.
Our results fit very well with a recent study showing that self-targeting spacers having homologies to genomic sequences, be they prophages or other chromosomal elements, are not evolutionarily conserved (18). The study suggests that “accidental” insertions of spacers targeting genomic sequences are deleterious to the organism harboring them (18). We provide experimental evidence for this study by demonstrating that spacers against a prophage are lethal to the bacteria, and unless the CRISPR system is rendered inactive (e.g., by plasmid loss), the cells are killed. CRISPR activity against an endogenous gene has been recently reported also for Pelobacter carbinolicus (1).
Observing that the CRISPR system acts against the prophage, we speculated that in certain circumstances it can protect bacteria from prophage induction. Prophage λcI857kan can be induced at 42°C due to a temperature-sensitive cI variant. We examined simultaneous induction of the prophage and CRISPR system by inducing transcription of the latter using arabinose and IPTG, while simultaneously shifting the temperature to 42°C. Despite the induction of these two lethal pathways, namely, the CRISPR system's catastrophic activity on a lysogen and prophage induction, the survival frequency of cells harboring anti-λ spacers was over 500-fold higher than that for bacteria harboring control spacers (Fig. 1C). Although induction of the CRISPR system during prophage induction protected only a small fraction of the bacteria in our system, the survivors provide a proof of principle for the system's ability to rescue bacteria from prophage induction and cure the cells of the prophage. To test whether the survivors lost the integrated prophage, which carries a kanamycin resistance gene, we inoculated them on kanamycin plates. All of the examined survivors (40 of 40) were sensitive to kanamycin, indicating that these cells had cleared the phage from their genome without lethal damage to the chromosome. We believe that phage excision from the genome is not synchronous with the induction of the CRISPR system under our experimental conditions. Therefore, while some cells are at the exact time point at which they can be saved by the CRISPR system acting on an excised phage, most cells either still harbor the prophage and are thus killed by genomic breakdown by the CRISPR system or are at advanced stages of the lytic cycle during which the CRISPR system cannot adequately act. We performed timing experiments to try to induce the CRISPR system at the exact time point at which all phages are excised from the genome but could not significantly raise the survival frequencies. Perhaps in “real-life” situations, in which prophage induction is sensed by cellular regulators, the response is more timely and efficient. One possible regulator that may synchronize the response is H-NS, which has been recently shown to repress the E. coli CRISPR system under LB broth growth conditions (16). Some promoters of the system (casD, cas2) are controlled by a stress response sigma factor, σ32, which might provide additional regulation for CRISPR activation exactly when required, while keeping the system dormant before prophage induction (19). In addition, the molar ratio of the Cas proteins may play an important role, and other, still-unknown regulatory or effector elements of CRISPR activity may allow the activity to take place only during prophage induction. Nevertheless, the increased overall CRISPR-dependent survival demonstrates that the system may rescue bacteria, under certain circumstances, from prophage induction by clearing the induced phage.
Our experiments simulate a possible scenario in which the acquisition of a spacer against a phage occurs after lysogenization by that phage. We show two possible outcomes, both beneficial to the bacteria from an evolutionary standpoint. The first is that the bacteria are killed along with the lysogens and therefore stop propagating it. The second is that the CRISPR system eliminates the prophage while keeping the host alive. As a whole, our results demonstrate the diversity with which the CRISPR system exerts its functions against lysogenization, lysogens, and prophage induction, in addition to its reported activity against lytic phages and extrachromosomal DNA invaders.
Supplementary Material
Acknowledgments
We thank John van der Oost for providing the plasmids encoding the CRISPR system and Lynn C. Thomason for providing the λcI857kan phage. We also thank Nir Osherov for critically reading the manuscript and Camille Vainstein for professional language editing of the manuscript.
This research was supported by the Israel Science Foundation (grant 611/10), the Binational Science Foundation (grant 2009218), the German-Israel Foundation (grant 2061/2009), and Marie Curie International Reintegration grants PIRG-GA-2009-256340 and GA-2010-266717.
Footnotes
Published ahead of print on 1 October 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aklujkar, M., and D. R. Lovley. 2010. Interference with histidyl-tRNA synthetase by a CRISPR spacer sequence as a factor in the evolution of Pelobacter carbinolicus. BMC Evol. Biol. 10:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709-1712. [DOI] [PubMed] [Google Scholar]
- 4.Bolotin, A., B. Quinquis, A. Sorokin, and S. D. Ehrlich. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151:2551-2561. [DOI] [PubMed] [Google Scholar]
- 5.Brouns, S. J., M. M. Jore, M. Lundgren, E. R. Westra, R. J. Slijkhuis, A. P. Snijders, M. J. Dickman, K. S. Makarova, E. V. Koonin, and J. van der Oost. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deveau, H., R. Barrangou, J. E. Garneau, J. Labonte, C. Fremaux, P. Boyaval, D. A. Romero, P. Horvath, and S. Moineau. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190:1390-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grissa, I., G. Vergnaud, and C. Pourcel. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hale, C. R., P. Zhao, S. Olson, M. O. Duff, B. R. Graveley, L. Wells, R. M. Terns, and M. P. Terns. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvath, P., and R. Barrangou. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167-170. [DOI] [PubMed] [Google Scholar]
- 10.Labrie, S. J., J. E. Samson, and S. Moineau. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317-327. [DOI] [PubMed] [Google Scholar]
- 11.Marraffini, L. A., and E. J. Sontheimer. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marraffini, L. A., and E. J. Sontheimer. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11:181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marraffini, L. A., and E. J. Sontheimer. 2010. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463:568-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClelland, M. 1981. The effect of sequence specific DNA methylation on restriction endonuclease cleavage. Nucleic Acids Res. 9:5859-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pougach, K., E. Semenova, E. Bogdanova, K. A. Datsenko, M. Djordjevic, B. L. Wanner, and K. Severinov. 2010. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol. 77:1367-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pul, U., R. Wurm, Z. Arslan, R. Geissen, N. Hofmann, and R. Wagner. 2010. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol. 75:1495-1512. [DOI] [PubMed] [Google Scholar]
- 17.Sorek, R., V. Kunin, and P. Hugenholtz. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6:181-186. [DOI] [PubMed] [Google Scholar]
- 18.Stern, A., L. Keren, O. Wurtzel, G. Amitai, and R. Sorek. 2010. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 26:335-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wade, J. T., D. C. Roa, D. C. Grainger, D. Hurd, S. J. Busby, K. Struhl, and E. Nudler. 2006. Extensive functional overlap between sigma factors in Escherichia coli. Nat. Struct. Mol. Biol. 13:806-814. [DOI] [PubMed] [Google Scholar]
- 20.Zegans, M. E., J. C. Wagner, K. C. Cady, D. M. Murphy, J. H. Hammond, and G. A. O'Toole. 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J. Bacteriol. 191:210-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.