Abstract
We report here that YajL is associated with ribosomes and interacts with many ribosomal proteins and that a yajL mutant of Escherichia coli displays decreased translation accuracy, as well as increased dissociation of 70S ribosomes into 50S and 30S subunits after oxidative stress.
YajL belongs to the Pyrococcus furiosus protease I (PfpI)/Hsp31/DJ-1 superfamily, which includes chaperones (33, 36), peptidases (26), and the Parkinson disease protein DJ-1 (10, 13). The crystal structures of YajL and DJ-1 are strikingly similar (45, 46), and their backbone structures are essentially identical (0.9-Å Cα [alpha carbon] root mean square deviation [RMSD]). DJ-1 has been reported to function as a weak protease (45), as an oxidative-stress-activated chaperone toward synuclein (39, 47), as an atypical peroxidase that scavenges H2O2 (3, 10), as a stabilizer of the antioxidant transcriptional regulator Nrf2 (12), as an apoptosis inhibitor via its interaction with Daxx (20), and as a transcriptional or translational (13, 42) regulator of gene expression.
We have shown recently that a yajL mutant of Escherichia coli displays a protein aggregation phenotype that is increased by oxidative stress (24). The aggregated proteins were mainly flagellin; the ATP synthase beta subunit, OmpA; peptidoglycan-associated lipoprotein (PAL); and many ribosomal proteins, including S1, S2, S3, S5, S6, S10, S16, L1, L2, L3, L4, L5, L6, L10, L11, and L25. YajL displays chaperone properties in vitro toward citrate synthase and toward S1 and L3, two ribosomal proteins that aggregate in the yajL mutant. Since many ribosomal proteins aggregate in the yajL mutant, we investigated whether or not ribosomes in this mutant are fully functional. We found that the yajL mutant displayed translational defects.
Association of YajL with translating ribosomes.
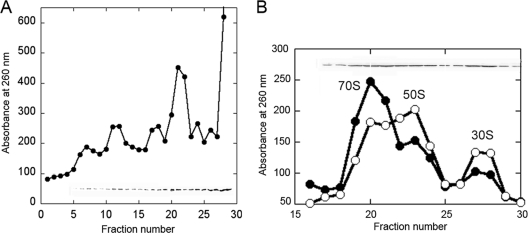
The yajL mutant JW5057 and its parent BW25113 (4) were grown in LB medium at 37°C and lysed as described in reference 9. Polysomes, ribosomes, and ribosomal subunits were immediately separated by centrifugation through sucrose gradients, and proteins were recovered from fractionated gradients. Fifty-two percent of the total amount of YajL from the wild-type strain extract was found in soluble protein fractions at the top of the gradient, but a significant proportion was found in polysomal (21%), 70S (17%), 50S (6%), and 30S (4%) fractions (Fig. 1A), suggesting that YajL is a ribosome-associated protein linked to translating ribosomes.
FIG. 1.
Association of YajL with ribosomes. (A) Lysates from the wild-type strain were centrifuged for 150 min at 36,000 rpm in a SW41 rotor through 15 to 30% sucrose gradients in 10 mM Tris (pH 7.6), 10 mM magnesium acetate, and 50 mM ammonium chloride at 4°C. Ribosomes were detected by their absorbance at 260 nm and YajL by immunoblot analysis (fractions 5 to 29). Representative results are shown for three repeat experiments. (B) Sucrose gradient sedimentation profiles of extracts from the parental strain (filled circles) and from the yajL mutant (empty circles) after they were stressed with 1 mM hydrogen peroxide for 10 min. Ribosomal subunits were centrifuged for 4 h at 36,000 × g in the same conditions as described above. The distribution of YajL in fractions 17 to 30 from the parental strain was detected by immunoblot analysis.
Ribosome profile is altered in the yajL mutant after oxidative stress.
After being stressed with 1 mM hydrogen peroxide for 10 min, both strains displayed increased proportions of free 50S and 30S subunits; however, dissociated subunits were reproducibly higher in the yajL mutant [(50S + 30S)/70S ratio = 65%] than in the parental strain [(50S + 30S)/70S ratio = 40%] (Fig. 1B). After being stressed, a significant proportion of YajL (around 22%) remained associated with the 70S, 50S, and 30S fractions of the parental strain (Fig. 1B, insert).
Specific interactions between YajL and ribosomal proteins.
In order to detect specific interactions between YajL and ribosomal proteins, we prepared the latter by incubating ribosomes overnight in a urea-containing buffer as described by Nomura and colleagues (18) and in the legend to Fig. 2. We incubated 1 volume of ribosomal proteins with 7 volumes of buffer containing 40 mM sodium phosphate (pH 8) and 100 mM NaCl for 10 min at 22°C, either alone or in the presence of purified His-tagged YajL (prepared as described in reference 24) (the final mixture contained 800 μg of ribosomal protein and 80 μg of YajL in 250 μl). Each sample was immediately loaded onto a nickel affinity column (200-μl-bed-volume). After being washed extensively with column buffer (40 mM sodium phosphate [pH 8], 100 mM NaCl, 0.8 M urea, and 10 mM imidazole), the columns were eluted with 50 mM imidazole in the same buffer. The eluate from the column loaded with the YajL-containing sample contained YajL, S2, S10, S19, L1, L5, L13, L14, L27, L28, L32, EF-Tu, DnaJ (a chaperone involved in ribosome biogenesis [2]), RsuA (16S pseudouridine and 516 synthase), TnaA (tryptophanase), and AceE (pyruvate dehydrogenase EI subunit) (Fig. 2). Several of these proteins (S2, S10, L1, and L5) were found in protein aggregates of the yajL mutant (24), suggesting that YajL is important for their biogenesis. Some of them are involved in the early stages of ribosome biogenesis (S19, L1, L5, and L13), whereas others are involved in the late stages (S2, S10, L14, L27, L28, and L32) (21). The main protein retained in the control experiment was Crp (catabolite gene activator), which is known to be retained by metal affinity columns (7). Proteins L13 and L27 and/or L28 were more extensively retained on the YajL affinity column than the others. These proteins are not, however, involved during the same stages of ribosome biogenesis, nor are they close to one another in the ribosomal structure (21), suggesting that their preferential retention likely results from their higher affinity for YajL. Like YajL, the ribosomal biogenesis factor ObgE interacts with numerous ribosomal proteins with diverse affinities (37).
FIG. 2.
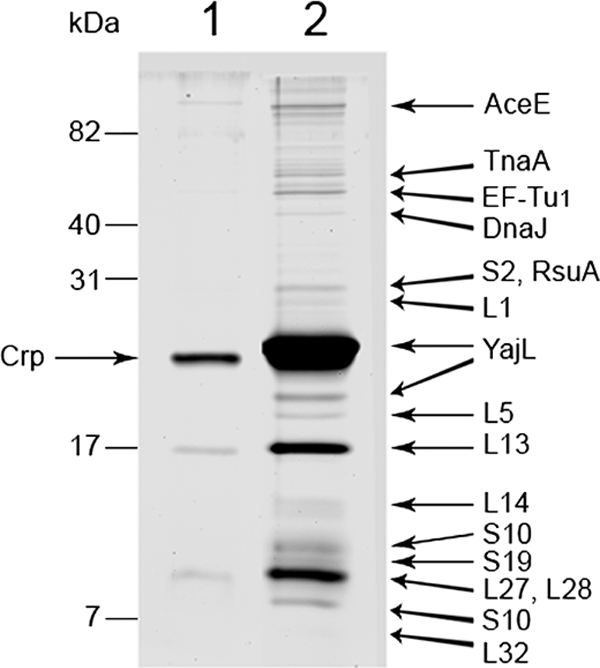
Specific interaction between YajL and ribosomal proteins. Ribosomal proteins from the wild-type strain were prepared by incubating ribosomes (prepared as described in reference 9) overnight in 10 mM Tris (pH 7.6), 6 mM MgCl2, 60 mM NH4Cl, 1 mM 2-mercaptoethanol, 6 M urea, and 1 M LiCl at 0°C as described by Nomura and colleagues (18). The samples were centrifuged at 35,000 × g for 10 min in order to remove the precipitated RNA, and the ribosomal proteins (800 μg) were diluted 8-fold in 250 μl of 40 mM sodium phosphate (pH 8) and 100 mM NaCl in the absence (lane 1) or in the presence (lane 2) of 80 μg of purified His-tagged YajL. Proteins retained on nickel affinity columns (His-Select nickel affinity gel, 200-μl-bed-volume; Sigma-Aldrich) were analyzed by 15% acrylamide SDS-PAGE (silver stained) and identified by matrix-assisted laser desorption ionization-time of flight/time of flight (MALDI-TOF/TOF) mass spectrometry (with an Applied Biosystems 4800 proteomics analyzer).
Increase in −1 and +1 frameshifting in the yajL mutant.
To test translation accuracy, we checked the processivity of the decoding process by measuring the occurrence of plus 1 (+1) or minus 1 (−1) frameshift events and codon-anticodon recognition by measuring the readthrough of stop codons. A series of lacZ plasmids containing either a frameshift-inducing signal or a nonsense mutation in the 5′ end of the lacZ coding region were constructed (6, 34, 43), resulting in β-galactosidase synthesis only when ribosomes shifted reading frames or read a nonsense codon. An in-phase plasmid was used as a control (44).
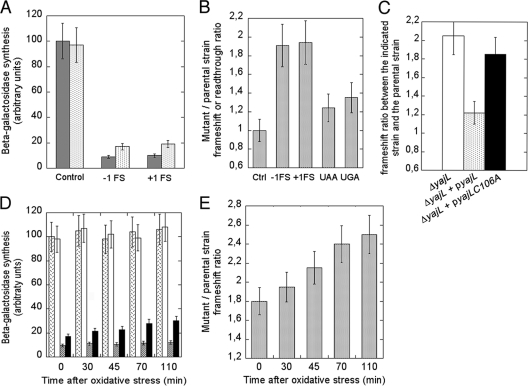
The β-galactosidase activities of the strains transformed with the in-phase or frameshift plasmids were measured in isopropyl-β-d-thiogalactopyranoside (IPTG)-induced wild-type and yajL mutant cells in the exponential phase (44). The in-phase β-galactosidase activities were similar in both strains, whereas the −1- and +1-frameshift-dependent β-galactosidase activities of the parental strain were 9 and 10%, respectively, of the in-phase β-galactosidase activity. By contrast, the −1- and +1-frameshift-dependent β-galactosidase activities of the mutant were 17 and 19%, respectively (Fig. 3A). Thus, the ratios of frameshift-dependent β-galactosidase activities in the mutant to those in the parental strain were 1.8 (−1 frameshift) and 1.9 (+1 frameshift) (Fig. 3B). We measured the −1-frameshift ratio between the mutant and its parental strain after stressing them with 100 μM hydrogen peroxide. This moderate stress did not affect bacterial growth (data not shown), nor did it affect the synthesis of the in-phase β-galactosidase in either strain (Fig. 3D). The −1-frameshift-dependent β-galactosidase activity of the parental strain moderately increased after stress (from 9.5 to 11.5% of the in-phase β-galactosidase activity), whereas that of the yajL mutant increased from 17 to 30% of the in-phase β-galactosidase activity (thus, the −1-frameshift ratio between the mutant and its parental strain rose from 1.8 to 2.6 [Fig. 3E], suggesting that ribosomes in the mutant were more inaccurate after oxidative stress). Since YajL is a ribosome-associated protein, the YajL defect might affect only a fraction of ribosomes, so that a 2-fold increase in frameshift events might be an average between higher values for ribosomes affected by the YajL defect and 1 for unaffected ribosomes.
FIG. 3.
Effect of the yajL mutation on −1 and +1 frameshifting and on UAA and UAG readthroughs. (A) The wild-type (dark gray) and yajL mutant (light gray) strains were transformed with plasmids encoding β-galactosidase expressed with N-terminal frameshifts or nonsense codons or with the wild-type lacZ sequence (control). Bacteria were in the exponential phase of growth during the entire experiment. The β-galactosidase activities of the control strain and of the −1- and +1-frameshift strains were measured as described in reference 44. The values are averages of the results of at least three independent experiments. (B) The β-galactosidase activities of the control, frameshift, and nonsense strains were measured, and the values for β-galactosidase synthesis in the mutant strains relative to those for the parental strains are shown, after being normalized to the ratio obtained with the in-phase plasmid. The miscoding value of the parental strain was normalized to 1. (C) Complementation of the yajL mutant with its wild-type allele but not with the C106A allele. The in-phase and −1-frameshift-dependent β-galactosidase activities of the parental strain, the yajL mutant, and the yajL mutant complemented with plasmids pBAD33-yajL and pBAD33-yajLC106A were measured, and frameshift ratios calculated as described for panel B are shown for the mutant (white), the mutant complemented with the wild-type yajL gene (gray), and the mutant complemented with the C106A yajL gene (black). Bacteria were grown in LB medium containing 0.05% arabinose, 0.5 mM IPTG, and the appropriate antibiotics. (D) Measurement of the −1 frameshift as a function of the time elapsed after the onset of stress with 100 μM hydrogen peroxide. β-galactosidase activities of the wild-type strain and of the yajL mutant expressing either the in-phase β-galactosidase (wild type, light gray; mutant, white) or the −1-frameshift-dependent β-galactosidase (wild type, dark gray; mutant, black) are shown. Bacteria were in the exponential phase of growth and had similar doubling times during the entire experiment. The values are averages of the results of three independent experiments. (E) The −1 frameshift ratios between the yajL mutant and its parental strain were calculated from the results shown in Fig. 3D and described in the legend to Fig. 3B and are shown as functions of the time elapsed after the onset of stress with 100 μM hydrogen peroxide.
Since the yajL deletion removes the last 12 codons of the overlapping panE gene (which codes for ketopantoate reductase), one might suspect that the yajL mutant is also a panE mutant. This is probably not the case, however, since the panE gene encounters a termination codon 13 codons from the beginning of the yajL deletion (4), and since the yajL mutant, in contrast to a panE mutant, is not auxotrophic for pantoate or pantothenate in minimal medium (32) even when the medium contains isoleucine and valine (which repress the synthesis of the acetohydroxy acid isomeroreductase IlvC, which can also catalyze the conversion of ketopantoate to pantoate) (data not shown)). In order to ascertain whether the frameshift defect results from a YajL defect, we complemented the yajL mutant with the wild-type yajL allele and the C106A allele. The wild-type yajL gene was transferred from the pET-21a-yajL expression vector to pBAD33 (as described in reference 1), an expression vector that is under the tight control of the arabinose pBAD promoter, yielding pBAD33-yajL, and the C106A allele was constructed by site-directed mutagenesis with a Stratagene QuikChange kit as described in reference 1, thus yielding pBAD33-yajLC106A. We compared the −1-frameshift values of the yajL mutant with those of the mutants complemented with pBAD33-yajL and pBAD33-yajLC106A. As shown in Fig. 3C, the yajL mutant displayed a −1-frameshift ratio of 2.2 with the parental strain, whereas the mutants complemented with either the wild-type yajL allele or the C106A allele displayed frameshift ratios of 1.2 and 1.8, respectively, suggesting that the frameshift defect results from a YajL defect and that cysteine 106 is important for YajL's function in ribosome biogenesis.
Moderate increases in UAA and UGA readthrough in the yajL mutant.
Levels of β-galactosidase activities dependent on the misreading of UAA and UGA codons as sense codons were 31% (UAA) and 25% (UGA) higher in the yajL mutant than in the parental strain (Fig. 3B). Thus, the yajL mutant displays slightly higher readthrough levels (around 1.3 ± 0.1-fold) than the wild-type strain for UAA and UGA translation termination codons.
Implications.
A substantial fraction of YajL was found in association with polysomes, ribosomes, and ribosomal subunits. Whereas several ribosome-associated proteins specifically interact with the 30S or 50S subunit (RrmJ and Era interact with the 50S subunit [21]) or with a few ribosomal proteins (EF-G interacts with L7/L12 and S12 [27], and trigger factors interact with L23 [23]), YajL, like the yeast peroxiredoxin Tsa1 (41), interacts with polysomes, ribosomes, and ribosomal subunits. The YajL-dependent retention of many ribosomal proteins (S2, S10, S19, L1, L5, L13, L14, L27, L28, and L32) on the nickel affinity column suggests that YajL interacts with many ribosomal proteins and is involved in their biogenesis, possibly as a ribosomal protein-folding chaperone or scaffold protein, and might explain its interaction with the ribosomes and the two ribosomal subunits.
The 2-fold increase in frameshifting of the yajL mutant is similar to that observed in ribosomal mutants such as the U2449, G517, or U2555 mutants (21, 28, 29, 30, 35) or in mutants deficient in the 23S RNA methyltransferase RrmA (17). The yajL mutant displayed only a 1.3-fold increase in the readthrough of stop codons, suggesting that it is less efficient in maintaining the translational reading frame than in avoiding missense errors. Although most mutants deficient in translation accuracy are affected in both reading frame maintenance and missense-error avoidance (5, 6, 28, 35), a few of them display only frame maintenance defects (15). Frameshift errors are more prejudicial than missense errors, since the sequence of amino acids incorporated after the shift is aberrant and codes for an inactive product. The restoration of nearly normal translation accuracy by complementation of the yajL mutant with the wild-type yajL allele but not with the C106A allele suggests that the mutant's translation accuracy defect is linked to the yajL mutation and that cysteine 106 is important for the function of YajL in ribosome biogenesis.
Several YajL-interacting proteins or aggregation-prone proteins of the yajL mutant (S5, L6, L11, and L14) are involved in translational fidelity (22, 38). S5 mutants display a ram phenotype (ribosomal ambiguity mutants) with a severalfold decrease in translation accuracy. L6 defects indirectly affect proofreading and result in decreased translation accuracy (14). L11 mutations result in defects in the binding of translation factors and in decreased translation accuracy (8), and L14 displays functional interactions with proteins L19 and S12 (the most important ribosomal protein for translation accuracy) (25).
To our knowledge, general chaperone mutants have not been reported to display any translational-accuracy defects. DnaK and GroEL are implicated in ribosome biogenesis (2), but translational-accuracy defects have not been reported for dnaK or tig mutants. Many additional factors are involved in ribosome biogenesis, including DEAD-box RNA helicases (CsdA, DbpA, and SrmB) (11, 21), ribosome-dependent GTPases (Era, Der, ObgE, and RsgA) (21, 37), and maturation factors (EryC and RimBCDHMN) (21), but mutants deficient in these factors have not been reported to display any translational-accuracy defect, as opposed to ribosome methyltransferase mutants (17, 21, 44). Interestingly, overexpression of the ribosome biogenesis factor ObgE or Der suppresses the slow growth of the Um2552 methyltransferase rrmJ mutants (40), and the 16S rRNA KsgA methyltransferase suppresses the cold-sensitive phenotype of era mutants (21), suggesting that methyltransferases and maturation factors contribute synergetically to the biogenesis of ribosomes that are thus endowed with optimal translation speed and accuracy.
Although ribosomal defects are not currently reported in Parkinsonism, ribosomal proteins are overrepresented in the proteins associated with soluble alpha-synuclein and/or with DJ-1 in cells treated with the Parkinsonian toxicant rotenone (19). Furthermore, the expression of mitochondrial ribosomal protein S6 has been shown to be altered in the brains of patients with Parkinson's disease (31), suggesting that ribosomal dysfunction might indeed be involved in Parkinsonism. Consequently, it will be of interest to investigate whether translational defects are associated with this disease.
Acknowledgments
We thank H. Mori (Nara Institute of Sciences and Technology, Japan) for the gift of the yajL mutant, M. Chapelle and Cerina Chhuong (Institut Jacques Monod, Paris) for mass spectrometry experiments, and A. Kropfinger for corrections of the English language.
Footnotes
Published ahead of print on 1 October 2010.
REFERENCES
- 1.Abdallah, J., T. Caldas, F. Kthiri, R. Kern, and G. Richarme. 2007. YhbO protects cells against multiple stresses. J. Bacteriol. 189:9140-9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al Refaii, A., and J. H. Alix. 2009. Ribosome biogenesis is temperature-dependent and delayed in Escherichia coli lacking the chaperones DnaK and DnaJ. Mol. Microbiol. 71:748-762. [DOI] [PubMed] [Google Scholar]
- 3.Andres-Mateos, E., C. Perier, L. Zhang, B. Blanchard-Fillion, T. M. Greco, B. Thomas, H. S. Ko, M. Sasaki, H. Ischiropoulos, S. Przedborski, T. M. Dawson, and V. L. Dawson. 2007. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc. Natl. Acad. Sci. U. S. A. 104:14807-14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudin-Baillieu, A., C. Fabret, X. H. Liang, D. Piekna-Przybylska, M. J. Fournier, and J. P. Rousset. 2009. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 37:7665-7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertrand, C., M. F. Prere, R. F. Gesteland, J. F. Atkins, and O. Fayet. 2002. Influence of the stacking potential of the base 3′ of tandem shift codons on −1 ribosomal frameshifting used for gene expression. RNA 8:16-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolanos-Garcia, V. M., and O. R. Davies. 2006. Structural analysis and classification of native proteins from E. coli commonly co-purified by immobilized metal affinity chromatography. Biochim. Biophys. Acta 1760:1304-1313. [DOI] [PubMed] [Google Scholar]
- 8.Bouakaz, L., E. Bouakaz, E. J. Murgola, M. Ehrenberg, and S. Sanyal. 2006. The role of ribosomal protein L11 in class I release factor-mediated translation termination and translational accuracy. J. Biol. Chem. 281:4548-4556. [DOI] [PubMed] [Google Scholar]
- 9.Caldas, T., E. Binet, P. Bouloc, and G. Richarme. 2000. Translational defects of Escherichia coli mutants deficient in the Um2552 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem. Biophys. Res. Commun. 271:714-718. [DOI] [PubMed] [Google Scholar]
- 10.Canet-Avilés, R. M., M. A. Wilson, D. W. Miller, R. Ahmad, C. McLendon, S. Bandyopadhyay, M. J. Baptista, D. Ringe, G. A. Petsko, and M. R. Cookson. 2004. The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. U. S. A. 101:9103-9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charollais, J., M. Dreyfus, and I. Iost. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acid Res. 323:2751-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clements, C. M., R. S. McNally, B. J. Conti, T. W. Mak, and J. P. Ting. 2006. DJ-1, a cancer- and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc. Natl. Acad. Sci. U. S. A. 103:15091-15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cookson, M. R. 2005. The biochemistry of Parkinson's disease. Annu. Rev. Biochem. 74:29-52. [DOI] [PubMed] [Google Scholar]
- 14.Davies, C., D. E. Bussiere, B. L. Golden, S. J. Porter, V. Ramakrishnan, and S. W. White. 1998. Ribosomal proteins S5 and L6: high-resolution crystal structures and roles in protein synthesis and antibiotic resistance. J. Mol. Biol. 279:873-888. [DOI] [PubMed] [Google Scholar]
- 15.Dinman, J. D., and R. B. Wickner. 1995. 5 S rRNA is involved in fidelity of translational reading frame. Genetics 141:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green, R., and H. F. Noller. 1997. Ribosomes and translation. Annu. Rev. Biochem. 66:679-716. [DOI] [PubMed] [Google Scholar]
- 17.Gustafsson, C., and B. C. Persson. 1998. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J. Bacteriol. 180:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Held, W. A., S. Mizushima, and M. Nomura. 1973. Reconstitution of Escherichia coli 30S ribosomal subunits from purified molecular components. J. Biol. Chem. 248:5720-5730. [PubMed] [Google Scholar]
- 19.Jin, J., G. J. Li, J. Davis, D. Zhu, Y. Wang, C. Pan, and J. Zhang. 2007. Identification of novel proteins associated with both alpha-synuclein and DJ-1. Mol. Cell. Proteomics 6:845-859. [DOI] [PubMed] [Google Scholar]
- 20.Junn, E., H. Taniguchi, B. S. Jeong, X. Zhao, G. H. Ichijo, and M. M. Mouradian. 2005. Interaction of DJ-1 with Daxx inhibits apoptosis signal-regulating kinase 1 activity and cell death. Proc. Natl. Acad. Sci. U. S. A. 102:9691-9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaczanowska, M., and M. Ryden-Aulin. 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 71:477-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirthi, N., B. Roy-Chaudhuri, T. Kelley, and G. M. Culver. 2006. A novel single amino acid change in small subunit ribosomal protein S5 has profound effects on translational fidelity. RNA 12:2080-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramer, G., T. Rauch, W. Rist, S. Vorderwülbecke, H. Patzelt, A. Schulze-Specking, N. Ban, E. Deuerling, and B. Bukau. 2002. L23 protein functions as a chaperone docking site on the ribosome. Nature 419:171-173. [DOI] [PubMed] [Google Scholar]
- 24.Kthiri, F., H. T. Le, V. Gautier, T. Caldas, A. Malki, A. Landoulsi, C. Bohn, P. Bouloc, and G. Richarme. 2010. Protein aggregation in a mutant deficient in yajL, the bacterial homolog of the Parkinsonism-associated protein DJ-1. J. Biol. Chem. 285:10328-10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maisnier-Patin, S., W. Paulander, A. Pennhag, and D. I. Andersson. 2007. Compensatory evolution reveals functional interactions between ribosomal proteins S12, L14 and L19. J. Mol. Biol. 366:207-215. [DOI] [PubMed] [Google Scholar]
- 26.Malki, A., T. Caldas, J. Abdallah, R. Kern, V. Eckey, S. J. Kim, S. S. Cha, H. Mori, and G. Richarme. 2005. Peptidase activity of the Escherichia coli Hsp31 chaperone. J. Biol. Chem. 280:14420-14426. [DOI] [PubMed] [Google Scholar]
- 27.Nechifor, R., and K. S. Wilson. 2007. Crosslinking of translation factor EF-G to proteins of the bacterial ribosome before and after translocation. J. Mol. Biol. 368:1412-1425. [DOI] [PubMed] [Google Scholar]
- 28.O'Connor, M., and A. E. Dahlberg. 1993. Mutations at U2555, a tRNA-protected base in 23S rRNA, affect translational fidelity. Proc. Natl. Acad. Sci. U. S. A. 90:9214-9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Connor, M., H. U. Göringer, and A. E. Dahlberg. 1992. A ribosomal ambiguity mutation in the 530 loop of E. coli 16S rRNA. Nucleic Acids Res. 20:4221-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogle, J. M., A. P. Carter, and V. Ramakrishnan. 2003. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 28:259-266. [DOI] [PubMed] [Google Scholar]
- 31.Papapetropoulos, S., J. Ffrench-Mullen, D. McCorquodale, Y. Qin, J. Pablo, and D. C. Mash. 2006. Multiregional gene expression profiling identifies MRPS6 as a possible candidate gene for Parkinson's disease. Gene Expr. 13:205-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Primerano, D. A., and R. O. Burns. 1983. Role of acetohydroxy acid isomeroreductase in biosynthesis of pantothenic acid in Salmonella typhimurium. J. Bacteriol. 153:259-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quigley, P. M., K. Korotkov, F. Baneyx, and W. G. Hol. 2003. The 1.6-Å crystal structure of the class of chaperones represented by Escherichia coli Hsp31 reveals a putative catalytic triad. Proc. Natl. Acad. Sci. U. S. A. 100:3137-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rettberg, C. C., M. F. Prere, R. F. Gesteland, J. F. Atkins, and O. Fayet. 1999. A three-way junction and constituent stem-loops as the stimulator for programmed −1 frameshifting in bacterial insertion sequence IS911. J. Mol. Biol. 286:1365-1378. [DOI] [PubMed] [Google Scholar]
- 35.Rodnina, M. V., and W. Wintermeyer. 2001. Ribosome fidelity: tRNA discrimination, proofreading and induced fit. Trends Biochem. Sci. 26:124-130. [DOI] [PubMed] [Google Scholar]
- 36.Sastry, M. S., K. Korotkov, Y. Brodsky, and F. Baneyx. 2002. Hsp31, the Escherichia coli yedU gene product, is a molecular chaperone whose activity is inhibited by ATP at high temperatures. J. Biol. Chem. 277:46026-46034. [DOI] [PubMed] [Google Scholar]
- 37.Sato, A., G. Kobayashi, H. Hayashi, H. Yoshida, A. Wada, M. Maeda, S. Hiraga, K. Takeyasu, and C. Wada. 2005. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells 10:393-408. [DOI] [PubMed] [Google Scholar]
- 38.Sato, H., K. Ito, and Y. Nakamura. 2006. Ribosomal protein L11 mutations in two functional domains equally affect release factors 1 and 2 activity. Mol. Microbiol. 60:108-120. [DOI] [PubMed] [Google Scholar]
- 39.Shendelman, S., A. Jonason, C. Martinat, T. Leete, and A. Abeliovich. 2004. DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2:e362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan, J., U. Jakob, and J. C. Bardwell. 2002. Overexpression of two GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 184:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trotter, E. W., J. D. Rand, C. M. Vickerstaff, and C. M. Grant. 2008. The yeast Tsa1 peroxiredoxin is a ribosome-associated antioxidant. Biochem. J. 412:73-80. [DOI] [PubMed] [Google Scholar]
- 42.van der Brug, M. P., J. Blackinton, J. Chandran, L. Y. Hao, A. Lal, K. Mazan-Mamczarz, J. Martindale, C. Xie, R. Ahmad, K. J. Thomas, A. Beilina, J. R. Gibbs, J. Ding, A. J. Myers, M. Zhan, H. Cai, N. M. Bonini, M. Gorospe, and M. R. Cookson. 2008. RNA binding activity of the recessive Parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 105:10244-10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss, R. B., D. M. Dunn, A. E. Dahlberg, J. F. Atkins, and R. F. Gesteland. 1988. Reading frame switch caused by base-pair formation between the 3′ end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 7:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Widerak, M., R. Kern, A. Malki, and G. Richarme. 2005. U2552 methylation at the ribosomal A-site is a negative modulator of translational accuracy. Gene 347:109-114. [DOI] [PubMed] [Google Scholar]
- 45.Wilson, M. A., J. L. Collins, Y. Hod, D. Ringe, and G. A. Petsko. 2003. The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson's disease. Proc. Natl. Acad. Sci. U. S. A. 100:9256-9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, M. A., D. Ringe, and G. A. Petsko. 2005. The atomic resolution crystal structure of the YajL (ThiJ) protein from Escherichia coli: a close prokaryotic homologue of the Parkinsonism-associated protein DJ-1. J. Mol. Biol. 353:678-691. [DOI] [PubMed] [Google Scholar]
- 47.Zhou, W., M. Zhu, M. A. Wilson, G. A. Petsko, and A. L. Fink. 2006. The oxidation state of DJ-1 regulates its chaperone activity toward alpha-synuclein. J. Mol. Biol. 356:1036-1048. [DOI] [PubMed] [Google Scholar]