Abstract
Pathogenic strains of the genus Acanthamoeba are causative agents of severe infections, such as fatal encephalitis and a sight-threatening amoebic keratitis. Antimicrobial therapy for these infections is generally empirical, and patient recovery is often problematic, due to the existence of a highly resistant cyst stage in these amoebae. In previous studies, small interfering RNAs (siRNAs) against the catalytic domains of extracellular serine proteases and glycogen phosphorylase from Acanthamoeba were designed and evaluated for future therapeutic use. The silencing of proteases resulted in Acanthamoeba failing to degrade human corneal cells, and silencing of glycogen phosphorylase caused amoebae to be unable to form mature cysts. After the siRNA design and concentration were optimized in order to avoid toxicity problems, cultures of Acanthamoeba were treated with a combination of both siRNAs, and cells were evaluated under an inverted microscope. This siRNA-based treatment dramatically affected the growth rate and cellular survival of the amoebae. These results were observed less than 48 h after the initiation of the treatment. In order to check possible toxic effects of the siRNA combination, three eukaryotic cell lines (HeLa, murine macrophages, and osteosarcoma cells) were treated with the same molecules, and cytotoxicity was examined by measuring lactate dehydrogenase release. The future use of the combination of these siRNAs is proposed as a potential therapeutic approach against pathogenic strains of Acanthamoeba.
Acanthamoeba species are opportunistic agents of disseminated infections (mostly cutaneous and nasopharyngeal infections), a sight-threatening ulceration of the cornea called Acanthamoeba keratitis (AK), and a fatal form of encephalitis known as Acanthamoeba granulomatous encephalitis (AGE) (2, 7, 11, 16).
Current therapeutic measures for Acanthamoeba keratitis rely on topical applications of antimicrobials, including a combination of propamidine isethionate and neomycin or chlorhexidine. However, the length of these treatments makes the process arduous. Furthermore, as the treatments are poorly effective against the cyst stage of the protozoan, residual infection often remains in these cases (3, 7, 17). No fully effective treatment against AGE has been established, although therapeutic measures have been used with apparent effect as an adjunct to surgery (6, 7, 8, 12, 17, 19).
Pathogenic and nonpathogenic Acanthamoeba strains have been isolated from the environment, but the amoebic determinants vital to pathogenesis are poorly understood. Pathogenic Acanthamoeba strains have greater temperature tolerance, growth rates, and adherence properties; more secreted cytotoxic products; and better immune evasion mechanisms than nonpathogenic Acanthamoeba strains (6, 7, 11).
The pathogenesis of Acanthamoeba infection is dependent upon host and amoebic factors, as well as environmental aspects, such as temperature and osmotic pressure. Amoebic factors seemed to be mediated by adhesion to the host cell, followed by the secretion of proteases (mainly serine proteases), phagocytosis, and direct killing of the host cell (7).
Small interfering RNA (siRNA) is the most essential and best-known double-stranded RNA (dsRNA) that is used for RNA interference (RNAi). It follows the exogenous pathway of gene silencing, as it is usually not present in the cell and is incorporated artificially, or it may infect the cell through certain viruses that have RNA as their genetic material (15). Since it can be synthesized artificially, it may be considered for use as a potent therapeutic agent. siRNA is produced from long dsRNA precursors that are further formed by folding of single-stranded RNA (ssRNA) molecules. In certain cases, it is also produced by using primary siRNA as a template and recruiting RNA-dependent RNA polymerase for the synthesis reaction. siRNA is usually long and cannot interact directly with the RNAi machinery to cause gene silencing. As a result, processing of siRNA takes place, which generates 21- to 23-bp-long siRNA that is responsible for causing gene silencing of a homologous target mRNA. Depending on the nature of the loci and biogenesis of the dsRNA precursor, different versions of siRNAs have been identified (18).
RNAi appears promising for silencing gene expression in parasitic pathogens, such as protozoans and helminths, as well as disease vectors, by specific target mRNA interference (9, 10, 18, 20). Analysis of gene functions in pathogens of infectious diseases and their vectors is important for research in drug development, and the silencing effects may be directly employed to control parasite transmission and development.
Since it was first applied in the genus Acanthamoeba (9), a few genes involved in Acanthamoeba pathogenesis and the formation of mature cysts have been silenced using siRNA in order to identify them as possible targets for chemotherapy and gene therapy or as key items in cellular functions (4, 9, 10, 13, 14). In previous studies, extracellular serine proteases were silenced using siRNAs, resulting in the Acanthamoeba organisms not being able to degrade human corneal cells (9). More recently, it was established by using siRNAs that serine proteases also play an important role in trophozoite differentiation and reemergence during excystment (4). Furthermore, the role of glycogen phosphorylase in the formation of mature cysts in Acanthamoeba was also established recently by using siRNA (10). Thus, blocking the entry and exit paths of these amoebae during their pathogenesis process could be achieved by using these siRNA molecules.
The results of the present study suggest that the combination of these two gene-specific siRNAs may be useful as a future therapeutic approach for the treatment of infectious processes caused by Acanthamoeba.
MATERIALS AND METHODS
Culture of Acanthamoeba.
Acanthamoeba castellanii Neff (ATCC 30010, genotype T4), a type strain from the American Type Culture Collection, was used in this study. Three clinical strains (CLC-16, genotype T3; CLC-41.r, genotype T4; and CLC-51.l, genotype T1) previously isolated from contact lens cases were also included in the study (12). These strains were axenically grown in PYG medium (0.75% [wt/vol] proteose peptone, 0.75% [wt/vol] yeast extract, and 1.5% [wt/vol] glucose) containing 40 μg gentamicin ml−1 (Biochrom AG, Cultek, Granollers, Barcelona, Spain) prior to their use in the assays. Amoebae grown in this way were then seeded in 24-well plates at a concentration of 104 cells per well.
Gene-silencing procedure and encystation conditions.
The soaking method, previously and successfully applied to Acanthamoeba gene silencing (9, 10), was used in the study. Regarding the siRNAs, two different molecules that were designed and validated in previous studies (9, 10) were used in this work. These molecules target the catalytic domains of extracellular serine proteases (SP-siRNA) and the glycogen phosphorylase gene in Acanthamoeba (GP-siRNA).
The application of these siRNAs at a maximum concentration of 15 μg ml−1 each (in order to avoid toxicity problems) was carried out in Costar 24-well cell culture clusters (Corning, Madrid, Spain). These experiments were carried out 3 times in duplicate.
Briefly, trophozoites (104 ml−1) of each amoebic strain were seeded in the plates containing PYG medium until they formed an almost confluent monolayer. At this stage, 15 μg ml−1 of SP-siRNA was introduced into the cells using the X-tremeGene siRNA transfection reagent (Roche Diagnostics, Barcelona, Spain). After 24 h of incubation with SP-siRNA, the PYG medium was poured off and immediately replaced with Neff's encystment medium (NEM) in order to simulate a tear-like effect that in the human eye causes the trophozoites physical and osmotic stress (Fig. 1A). Thus, induction of amoebic encystation was triggered, as this procedure is the main exit path of Acanthamoeba against any stressful environmental conditions, resulting in the formation of a highly resistant double-wall mature cyst. Simultaneously, after NEM was added, 15 μl ml−1 of GP-siRNA was added to the cells, using the same siRNA transfection reagent.
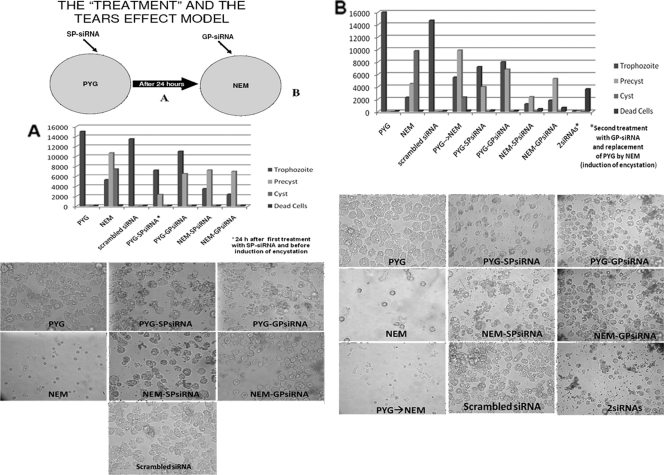
FIG. 1.
The tear model proposed in this study and the treatment scheme undertaken with both siRNAs. Briefly, amoebae were grown in PYG medium and incubated with SP-siRNA (A). After 24 h, the PYG medium was poured off and replaced by NEM (encystation medium). The amoebae under these conditions were then incubated with GP-siRNA (B). The light microscopy (×20 and ×40) images correspond to the assays that were carried out with A. castellanii Neff ATCC 30010; similar results were obtained with the other clinical strains included in this study. Controls are shown, as well as the cell numbers (trophozoites, precysts, and cysts) in each well. 2siRNAs indicates that the amoebae were incubated with both siRNAs. PYG→NEM is a control without the addition of siRNAs but with a change of medium (induction of encystation). Differences between replicate experiments were insignificant.
Controls with only amoebae in PYG medium and NEM alone, and also with one of the siRNAs (SP-siRNA or GP-siRNA only in PYG and NEM) at the same concentrations used in the experiment, were simultaneously developed. A control with scrambled siRNA from a sequence absent from the genus Acanthamoeba and based on the gene encoding the green fluorescent protein was used as previously described (9, 10).
Microscopy.
During the silencing procedure, cells were monitored using a DMIL inverted microscope (Leica, Wetzlar, Germany) at different time intervals in order to check for phenotypic manifestations in the siRNA-treated cells compared to the control cells.
Cell lines.
The following cell lines were used in this study: J774A.1 murine macrophages (ATCC TIB-67), HeLa cells (ATCC CCL-2), and the U-2 OS osteosarcoma cell line (ATCC HTB-96) were purchased from the American Type Culture Collection (ATTC) (LG Promochem, Barcelona, Spain). The three cell lines were maintained as monolayer cultures at 37°C in 5% CO2 in T-75 culture flasks (Corning, Madrid, Spain). Cells were routinely cultured in Dulbecco's modified Eagle medium without phenol red (DMEM) (Sigma, Tres Cantos, Madrid, Spain) supplemented with 10% fetal calf serum, 2 mM glutamine, and 20 μg ml−1 gentamicin (Biochrom AG, Cultek, Granollers, Barcelona, Spain) and subcultured two times per week.
Cytotoxicity assays.
Cytotoxicity assays were performed using a cytotoxicity detection kit based on the release of lactate dehydrogenase enzyme (LDH) (Roche Diagnostics, Sant Cugat del Vallès, Barcelona, Spain) as previously described (1, 9). Briefly, the three cell lines were grown to monolayers in 6-well plates. Next, the cells were incubated with SP-siRNA, GP-siRNA, or both siRNAs at the same concentrations and under the same conditions that were used for the treatment of the Acanthamoeba strains. The cell lines were then incubated in fetal calf serum-free DMEM at 37°C in 5% CO2. The cell monolayers were observed periodically for cytopathic effects for up to 48 h. At the end of this incubation period, cytopathic effects were assessed visually after hematoxylin staining. In addition, the supernatants were collected and cytotoxicity was determined by measuring LDH release following the manufacturer's instructions. Control values were obtained from each cell line incubated alone in fetal calf serum-free DMEM without phenol red. Total LDH release was determined from each cell line treated with 2% Triton X-100 (Sigma, Tres Cantos, Madrid, Spain). These experiments were carried out 3 times in duplicate. Cytotoxicity values between 0% and 10% were considered not cytotoxic, and cytotoxicity values between 10 and 25% were considered low, whereas cytotoxicity values from 25% to 40% were considered moderate levels. Values higher than 40% and up to 100% were considered high cytotoxicity levels.
Sensitivity and viability assays.
Acanthamoeba cell viability was determined by trypan blue staining (trypan blue solution, 0.4%, liquid, sterile filtered, and cell culture tested; Sigma, Tres Cantos, Madrid, Spain). Quantification of cell viability was performed at 0, 24, 48, 72, and 96 h using a hemocytometer.
To test the resistance of Acanthamoeba precysts and cysts, a 5-min treatment with a nonionic detergent (SDS; 0.5% [wt/vol] final concentration) was used as previously described (10).
RESULTS
The addition of SP-siRNA caused the cytoplasm of amoebae to become full of vesicles, and their growth rate was lower than that of the control cells. After 24 h of incubation with the SP-siRNA, it was observed that the SP-siRNA-treated amoebae lowered their division rate in comparison to the control cells (Fig. 1A). At this stage, the PYG medium was removed and replaced by NEM (see Materials and Methods) (Fig. 1A), and GP-siRNA was added to the previously SP-siRNA-treated amoebae. The cells were then monitored for the next 24 h. These double-siRNA-treated cells (SP-siRNA at 0 h plus GP-siRNA after 24 h) were unable to start the formation of a mature cyst stage, and no immature cysts were detected in the wells until 24 h after the induction of osmotic stress (replacement of PYG medium by NEM). Interestingly, GP-siRNA-treated amoebae in NEM seemed unable to start the encystation process and also detached from the culture flask. Similar cell behavior was observed in the control well treated only with SP-siRNA and kept in NEM. A very low growth rate in the control amoebae in PYG medium treated with SP-siRNA was also observed at this stage (Fig. 1B).
At 48 h after the addition of SP-siRNA, amoebae treated with the combination of SP-siRNA and GP-siRNA were all detached from the bottom of the flask. Moreover, most of the cells were dead (they failed to exclude trypan blue), and the remaining amoebae (trophozoites and/or rounded cells) did not survive a 5-min treatment with 0.5% SDS and underwent cell lysis. Thus, the combination of both siRNAs eliminated the amoebae in 48 h, causing cell lysis in most of the amoebae. The rest of the amoebae left in the well were dead, as they also failed to exclude trypan blue (Fig. 1B).
The number of cells of the control amoebae in PYG medium was much higher than that of the amoebae treated with SP-siRNA, also in PYG medium, at this stage. Moreover, amoebae incubated with SP-siRNA or GP-siRNA alone were not able to form mature cysts in NEM, and only trophozoites or rounded cells were observed in these wells. These rounded cells were viable but did not resist a 5-min treatment with 0.5% SDS.
The control with scrambled siRNA and the control with only amoebae (PYG changed to NEM) grew at a normal rate and underwent a normal encystment process when NEM was added (Fig. 2A and B). It was possible to observe mature cysts (with both cyst layers) at 24 to 48 h postinduction of encystation in these wells. Controls consisting of amoebae alone and amoebae treated with siRNA-GP alone in PYG medium showed a confluent monolayer of trophozoites (Fig. 1B).
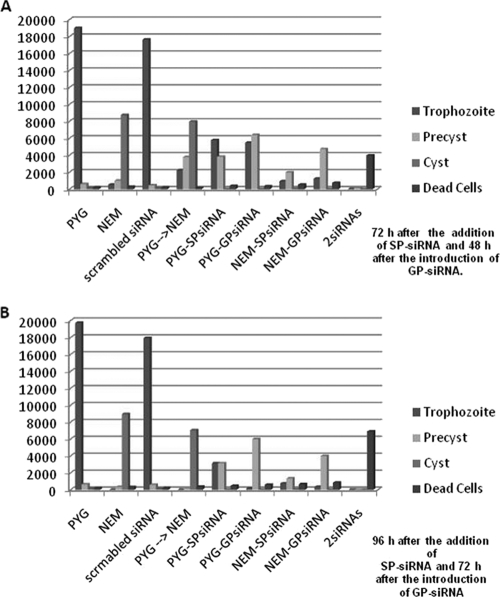
FIG. 2.
Numbers of cells at 72 h (A) and 96 h (B). 2siRNAs indicates that the amoebae were incubated with both siRNAs. PYG→NEM is a control without the addition of siRNAs but with a change of medium (induction of encystation). Differences between replicate experiments were insignificant.
For the cytotoxicity assays, the same pattern of siRNA treatment was used. First, SP-siRNA was added to the cell cultures, and after 24 h, GP-siRNA was also introduced into the cell cultures. At 24 h, all three tested cell lines showed cytotoxicity levels between 10 and 30% when treated with either SP-siRNA or GP-siRNA (Table 1). At 48 h, cells treated with the combination of both siRNAs exhibited cytotoxicity levels around 37% (36.91% ± 3.56) (Table 1), whereas the cytotoxicity levels in the cells incubated with SP- or GP-siRNA alone for 48 h were between 18 and 38% and 24 and 38%, respectively (Table 1).
TABLE 1.
Percentages of cytotoxicity in the three tested eukaryotic cell lines caused by SP-siRNA and GP-siRNA alone and in combination
| Time (h) | Cell line | % Cytotoxicity |
||
|---|---|---|---|---|
| SP-siRNA | GP-siRNA | 2siRNAa | ||
| 24 | Macrophages | 13.23 ± 2.05 | 27.9 ± 2.2 | 34.14 ± 1.38 |
| HeLa | 11.73 ± 2.35 | 30.7 ± 4.11 | 30.9 ± 6.04 | |
| U20S | 23.02 ± 1.9 | 21.2 ± 5.97 | 21.13 ± 1.24 | |
| 48 | Macrophages | 20.14 ± 5.88 | 29.1 ± 4.07 | 37.18 ± 5.96 |
| HeLa | 18.87 ± 3.34 | 38 ± 2.87 | 35.69 ± 2.24 | |
| U20S | 38.83 ± 4.98 | 24.8 ± 3.73 | 36.91 ± 3.56 | |
2siRNA, SP-siRNA and GP-siRNA in combination.
DISCUSSION
A powerful and effective application of siRNAs as a therapeutic tool against the genus Acanthamoeba is presented in this study. Combinations of two different siRNAs (SP-siRNA and GP-siRNA) specific to Acanthamoeba genes and previously developed by our group were used in this work. Extracellular serine proteases are directly involved in the pathogenicity of Acanthamoeba; moreover, larger quantities of extracellular serine proteases are secreted by pathogenic strains than by nonpathogenic strains (5, 7, 9).
SP-siRNAs were added to different pathogenic strains of Acanthamoeba in our previous work (9). In that case, Acanthamoeba failed to secrete large amounts of extracellular serine proteases and mRNAs were also lower, and thus, cytotoxic effects on human corneal cells in vitro were negligible compared to those of controls consisting of Acanthamoeba cells not treated with the SP-siRNAs. Also, it was demonstrated by real-time PCR that the silencing phenomenon was stable up to 96 h after introduction of the SP-siRNA. Moreover, in a recent study, the same SP-siRNAs were used against trophozoites of Acanthamoeba belonging to the T4 genotype, demonstrating that these siRNAs were able to block the encystment and excystment processes in this strain of Acanthamoeba (4). On the other hand, using siRNA-based methods, our group recently reported that glycogen phosphorylase is required for cyst wall assembly, mainly for the formation of the cell wall inner layer. This observation was proven by treating amoebae with specific GP-siRNA and simultaneously inducing encystation by changing PYG medium to NEM. Acanthamoeba cells treated with these siRNAs were able to form only single-layered immature cysts up to 96 h after treatment, and they were degraded in less than 5 min when treated with 0.5% SDS. In comparison, control cells free of GP-siRNA formed mature cysts 24 h after the induction of encystation. At the molecular level, no glycogen phosphorylase mRNAs or protein or activity of the enzyme was detected up to 96 h after treatment with GP-siRNA, confirming the gene-silencing phenomenon (10).
In our experiments in this work and 24 h after the addition of SP-siRNA, it was clear that the SP-siRNA-treated amoebae lowered their division rate in comparison to the control cells (Fig. 1 and 2). At this stage, the PYG medium was removed and replaced by NEM, and also, GP-siRNA was added to the previously SP-siRNA-treated amoebae. The cells were then monitored for the next 24 h. The double-treated cells (SP-siRNA at 0 h plus GP-siRNA after 24 h) did not form mature double-wall cysts, and no signs of initial stages of encystment were observed (Fig. 1B). This observation supports our previous molecular data regarding the status of both extracellular serine protease and glycogen phosphorylase genes at this stage. Interestingly, Acanthamoeba cells were not able to start encystation, which is in accordance with the previous report of a major role of serine proteases in the encystment and excystment processes in Acanthamoeba (4).
Control amoebae treated with GP-siRNA only in NEM were not able to initiate the encystation process but detached from the culture flask, a very early sign of encystment (Fig. 1 and 2). As mentioned above, silencing of glycogen phosphorylase resulted in the formation of single-layered immature cysts but not double-wall mature cysts (10). Similar behavior was observed in the control with SP-siRNA in NEM (Fig. 1 and 2), again supporting the previous observations by our group and recently by Dudley et al. (4). Amoebae cultured in PYG medium and treated with SP-siRNA alone showed a very low growth rate compared to the control nontreated cells in PYG medium. Furthermore, no changes were observed in amoebae treated with GP-siRNA in PYG medium (Fig. 1 and 2).
At 48 h after the introduction of SP-siRNA and 24 h after the addition of GP-siRNA, amoebae treated with this combination of siRNAs all detached from the bottom of the flask, following our previous observations (10). However, most of the cells were dead, and the remaining amoebae (trophozoites and/or rounded cells or single-layered immature cysts) did not survive a 5-min treatment with 0.5% SDS (a test for the sensitivity of cysts) and underwent cell lysis (Fig. 1 and 2). Therefore, the results obtained with the combination of both siRNAs as a therapeutic tool demonstrated that these molecules at a concentration of 15 μg ml−1 are able to eliminate clinical strains of Acanthamoeba in 48 h.
Another interesting observation at this stage was that amoebae incubated with SP-siRNA or GP-siRNA in NEM were not able to form mature cysts, and only trophozoites or rounded cells were observed in these wells, supporting the previous reports mentioned above (4, 9, 10).
Amoebae treated with GP-siRNA alone in PYG medium showed a confluent monolayer of trophozoites, and thus, glycogen phosphorylase does not seem to be involved in any key processes of Acanthamoeba at the trophozoite stage (Fig. 1 and 2). This observation supports our previous data, as no expression of glycogen phosphorylase was detectable in actively growing and stationary trophozoites (10).
In the cytotoxicity assays, SP-siRNA or GP-siRNA alone caused very low cytotoxicity to the three cell lines after 24 h of incubation (Table 1). The combination of both siRNAs also caused low cytotoxicity after 24 h. However, after 48 h, siRNAs alone or in combination caused moderate rates of cytotoxicity (between 20 and 37%) (Table 1). Therefore, this observation should be considered in order to further develop this therapeutic approach. It may be possible to develop liposomal formulations that include these siRNAs or another vehicle of administration for a future siRNA-based amoebicidal agent, which could decrease the cytotoxicity levels caused by these molecules, as well as to develop in vivo models to evaluate this combination of siRNAs at this level.
Overall, the combination of SP-siRNAs and GP-siRNAs against the strains of Acanthamoeba included in this study appears to be a very powerful and effective approach for the establishment and development of more effective treatments for Acanthamoeba infections.
Acknowledgments
This research was funded by project PI081815 (Búsqueda de Marcadores Moleculares de Patogenicidad en Acanthamoeba sp.) from the Fondo de Investigaciones Sanitarias (FIS) and RICET (project no. RD06/0021/0005 of the program Redes Temáticas de Investigación Cooperativa, FIS), Spanish Ministry of Health, Madrid, Spain. J.L.-M. was funded by a postdoctoral grant from the Fundación Canaria Manuel Morales, La Palma, Canary Islands. C.M.M.-N. was funded by the grant Becas Cajacanarias de Postgraduados Convocatoria 2009. A.L.-A. was funded by the grant Ayudas del Programa de Formación de Personal Investigador, para la Realización de Tesis Doctorales from the Agencia Canaria de Investigación, Innovación y Sociedad de la Información from the Canary Islands government.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We declare no conflict of interest.
Footnotes
Published ahead of print on 20 September 2010.
REFERENCES
- 1.Alsam, S., K. S. Kim, M. Stins, A. O. Rivas, J. Sissons, and N. A. Khan. 2003. Acanthamoeba interactions with human brain microvascular endothelial cells. Microb. Pathog. 35:235-241. [DOI] [PubMed] [Google Scholar]
- 2.Clarke, D. W., and Y. Niederkorn. 2006. The pathophysiology of Acanthamoeba keratitis. Trends. Parasitol. 22:175-180. [DOI] [PubMed] [Google Scholar]
- 3.Dart, J. K., V. P. Saw, and S. Kilvington. 2009. Acanthamoeba keratitis: diagnosis and treatment update. Am. J. Ophthalmol. 148:487-499. [DOI] [PubMed] [Google Scholar]
- 4.Dudley, R., S. Alsam, and N. A. Khan. 2008. The role of proteases in the differentiation of Acanthamoeba castellanii. FEMS Microbiol. Lett. 286:9-15. [DOI] [PubMed] [Google Scholar]
- 5.Khan, N. A., E. L. Jarroll, N. Panjwani, Z. Cao, and T. A. Paget. 2000. Proteases as markers for differentiation of pathogenic and nonpathogenic species of Acanthamoeba. J. Clin. Microbiol. 38:2858-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan, N. A. 2003. Pathogenesis of Acanthamoeba infections. Microb. Pathog. 34:277-285. [DOI] [PubMed] [Google Scholar]
- 7.Khan, N. A. 2006. Acanthamoeba: biology and increasing importance in human health. FEMS Microbiol. Rev. 30:564-595. [DOI] [PubMed] [Google Scholar]
- 8.Lackner, P., R. Beer, G. Broessner, R. Helbok, B. Pfausler, C. Brenneis, H. Auer, J. Walochnik, and E. Schmutzhard. 2010. Acute granulomatous Acanthamoeba encephalitis in an immunocompetent patient. Neurocrit. Care 12:91-94. [DOI] [PubMed] [Google Scholar]
- 9.Lorenzo-Morales, J., A. Ortega-Rivas, P. Foronda, N. Abreu-Acosta, D. Ballart, E. Martínez, and B. Valladares. 2005. RNA interference (RNAi) for the silencing of extracellular serine proteases genes in Acanthamoeba: molecular analysis and effect on pathogenicity. Mol. Biochem. Parasitol. 144:10-16. [DOI] [PubMed] [Google Scholar]
- 10.Lorenzo-Morales, J., J. Kliescikova, E. Martinez-Carretero, L. M. De Pablos, B. Profotova, E. Nohynkova, A. Osuna, and B. Valladares. 2008. Glycogen phosphorylase in Acanthamoeba spp.: determining the role of the enzyme during the encystment process using RNA interference. Eukaryot. Cell 7:509-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marciano-Cabral, F., and G. Cabral. 2003. Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 16:273-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martín-Navarro, C. M., J. Lorenzo-Morales, M. G. Cabrera-Serra, F. Rancel, N. M. Coronado-Álvarez, J. E. Piñero, and B. Valladares. 2008. The potential pathogenicity of chlorhexidine-sensitive Acanthamoeba strains isolated from contact lens cases from asymptomatic individuals in Tenerife, Canary Islands, Spain. J. Med. Microbiol. 57:1399-1404. [DOI] [PubMed] [Google Scholar]
- 13.Moon, E. K., D. I. Chung, Y. C. Hong, and H. H. Kong. 2009. Autophagy protein 8 mediating autophagosome in encysting Acanthamoeba. Mol. Biochem. Parasitol. 168:43-48. [DOI] [PubMed] [Google Scholar]
- 14.Moon, E. K., D. I. Chung, Y. C. Hong, and H. H. Kong. 2008. Characterization of a serine proteinase mediating encystation of Acanthamoeba. Eukaryot. Cell 7:1513-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pederson, T. 2004. RNA interference and mRNA silencing, 2004: how far will they reach? Mol. Biol. Cell 15:407-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuster, F. L., and G. S. Visvesvara. 2004. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int. J. Parasitol. 34:1001-1027. [DOI] [PubMed] [Google Scholar]
- 17.Schuster, F. L., and G. S. Visvesvara. 2004. Opportunistic amoebae: challenges in prophylaxis and treatment. Drug Resist. Updat. 7:41-51. [DOI] [PubMed] [Google Scholar]
- 18.Shrey, K., A. Suchit, M. Nishant, and R. Vibha. 2009. RNA interference: emerging diagnostics and therapeutics tool. Biochem. Biophys. Res. Commun. 386:273-277. [DOI] [PubMed] [Google Scholar]
- 19.Singhal, T., A. Bajpai, V. Kalra, S. K. Kabra, J. C. Samantaray, G. Satpathy, and A. K. Gupta. 2001. Successful treatment of Acanthamoeba meningitis with combination oral antimicrobials. Pediatr. Infect. Dis. J. 20:623-627. [DOI] [PubMed] [Google Scholar]
- 20.Ullu, E., C. Tschudi, and T. Chakraborty. 2004. RNA interference in protozoan parasites. Cell. Microbiol. 6:509-519. [DOI] [PubMed] [Google Scholar]