Abstract
Terpenoid phenols, including carvacrol, are components of oregano and other plant essential oils that exhibit potent antifungal activity against a wide range of pathogens, including Candida albicans, Staphylococcus aureus, and Pseudomonas aeruginosa. To gain a mechanistic view of the cellular response to terpenoid phenols, we used Saccharomyces cerevisiae as a model organism and monitored temporal changes in metabolic activity, cytosolic and vacuolar pH, and Ca2+ transients. Using a panel of related compounds, we observed dose-dependent Ca2+ bursts that correlated with antifungal efficacy. Changes in pH were long lasting and followed the Ca2+ transients. A vma mutant lacking functional vacuolar H+-ATPase (V-ATPase) and defective in ion homeostasis was hypersensitive to carvacrol toxicity, consistent with a role for ionic disruptions in mediating cell death. Genomic profiling within 15 min of exposure revealed a robust transcriptional response to carvacrol, closely resembling that of calcium stress. Genes involved in alternate metabolic and energy pathways, stress response, autophagy, and drug efflux were prominently upregulated, whereas repressed genes mediated ribosome biogenesis and RNA metabolism. These responses were strongly reminiscent of the effects of rapamycin, the inhibitor of the TOR pathway of nutrient sensing. The results point to the activation of specific signaling pathways downstream of cellular interaction with carvacrol rather than a nonspecific lesion of membranes, as has been previously proposed.
While the medicinal properties of herbs have been recognized since ancient times, there has been a resurgence of interest in the antimicrobial properties of botanical extracts. Essential oils have been amply documented to kill a wide range of pathogenic fungi and bacteria, such as Candida albicans, Staphylococcus aureus, and Pseudomonas aeruginosa, including their drug-resistant variants (6, 10, 21, 22). Of the herbal extracts tested, essential oils derived from the genus Oreganum were among the most effective, with an in vitro MIC of 500 ppm against C. albicans (27). Major components of oregano extract, which include the terpenoid phenols carvacrol, thymol, and eugenol, have potent antifungal activity of their own (4, 23, 24). Terpenoid phenols have been shown to be efficacious not only on planktonic cells but also on biofilms of Candida albicans that are resistant to many antifungal drugs. Carvacrol demonstrated the strongest antifungal activity against Candida albicans biofilms, with a MIC of <0.03% (9). Furthermore, carvacrol was shown to be effective regardless of the maturity of the biofilm. The terpenoid phenols tested were able to inhibit biofilms of several strains of Candida, including C. albicans, C. glabrata, and C. parapsilosis. In addition to their antimycotic, antibacterial, insecticidal, and bioherbicidal properties, essential oils are also well known for their antioxidant characteristics and are used to inhibit lipid peroxidation in preventing food spoilage or as chemoprotective agents in the treatment of various diseases, including cancer (1, 26).
Although there is abundant evidence for the antifungal efficacy of essential oils and their constituents, there has been relatively little work on the mechanism of killing. A better understanding of the cellular basis of the action of these antifungal agents would improve their therapeutic potential by guiding combination therapy with other established drugs and lead to safer and more innovative treatments. Baker's yeast offers a sophisticated toolkit of experimental approaches. We establish Saccharomyces cerevisiae as a model organism for exploring the effect of terpenoid phenols at a cellular and molecular level. We used compartment-specific cellular probes to follow temporal changes in metabolic activity, Ca2+, and pH as a function of toxicity. Genome-wide profiling of the transcriptional changes to carvacrol revealed large and rapid metabolic, biosynthetic, and stress responses that provide molecular insight into the mechanism of action of essential oils.
MATERIALS AND METHODS
Essential oils and phenolic compounds.
Medicinal-grade oregano oil was purchased from a local health store. Carvacrol, thymol, eugenol, vanillin, guaicol, p-cymene, and γ-terpinene were purchased from Sigma-Aldrich and were at 98% purity. Carvacrol was in liquid form (density, 0.976 g/cm3). The thymol, a white powder, was made into a 10% stock solution in ethanol. Each compound was made into 10%, 1%, and 0.1% stocks by serial dilutions in ethanol and stored at room temperature.
Yeast toxicity assays.
S. cerevisiae strain BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) was grown overnight in a shaking incubator in synthetic complete (SC) medium as specified by Sherman (25). Absorbance was measured at 600 nm after 1:10 dilution in water. Cells (optical density [OD] of 0.025) were added to 1 ml SC medium in each well of a 24-well plate. Finally, the phenolic compounds to be tested were added, in triplicate, to the specified final concentrations. The plates were stored overnight in a 30°C incubator. The plates were gently vortexed to resuspend cells before measurement of absorbance at 600 nm. Halo formation was assessed after soaking sterile filter paper in dilutions of essential oil and overlaying a lawn of freshly plated S. cerevisiae on SC medium. Plates were incubated at 30°C for 1 to 2 days. Mineral oil was used for dilution and as a control.
FUN-1 fluorescence.
The fluorescence indicator FUN-1 (Invitrogen) was used to monitor the loss of metabolic activity according to the method of Millard et al. (19), as previously described (20). Cells emitting green fluorescence were considered metabolically inactive (19). To load the cells with dye, the optical density at 600 nm (OD600) of the yeast culture was measured, and the cells were collected by centrifugation and resuspended in 100 μl of SC media and 2 μl of FUN-1 dye. The tubes were vortexed, wrapped in foil, and then incubated at 30°C for an hour. The cells were washed twice with 2% glucose, and the cell pellet resuspended in 2 ml of 2% glucose per OD unit. To 50 μl of cells, 5 μl of mineral oil or oregano oil was added. After 15 min, the cells were observed under a fluorescence microscope. To quantify these results, 5, 10, and 25 μl of each compound was added, in triplicate, to the cells in a black 96-well microtiter plate. The final volume was 200 μl. Fluorescence (emission 575 nm) was measured for 2 h with a BMG Fluostar Optima plate reader. The averages of the results of the triplicate experiments were graphed against time.
Ca2+-dependent aequorin luminescence.
BY4742 transformed with plasmid pEVP11-Aeq-89 expressing aequorin was grown overnight in SC medium (20). One OD600 unit of cells per microcentrifuge tube was spun down. The cell pellets were resuspended in 1 ml of 2% glucose, spun, and decanted again. Fifty microliters of SC medium was added into each tube. After vortexing to mix cells, 6 μl of coelenterazine (12.5 mg/ml in ethanol, stored at −20°C; Invitrogen) was added into each tube. The cells were incubated for 2 h in a 30°C incubator to allow reconstitution of aequorin with coelenterazine. Finally, the cells were spun down and transferred into 2 ml of SC. After another vortexing, 150 μl was placed in each well of a white 96-well microtiter plate. In addition to this, 150 μl of cells was also put into another microtiter plate to read the OD. The drug was added (0.0125, 0.25, 0.5, and 0.1%) as described in the Fig. 1 legend, and luminescence was measured on a Fluostar Optima microplate reader. Each dose of drug was done in duplicate, and the experiment was repeated at least two times. Luminescence values were collected every second, normalized to the OD, and averaged.
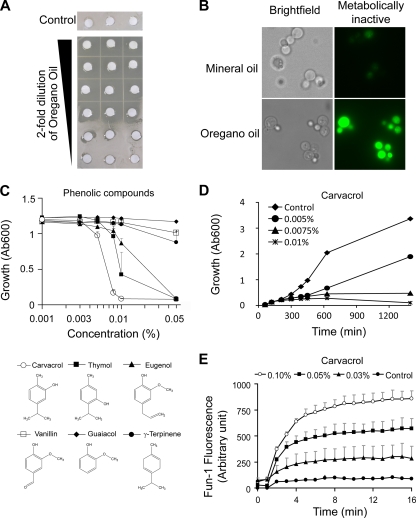
FIG. 1.
Antifungal activities of oregano oil and component terpenoid phenols. (A) Filter disks were soaked in serial dilutions of oregano oil and placed on a lawn of S. cerevisiae. Clear areas or halos were observed up to a dilution of 1:8 and revealed inhibition of yeast growth. Controls show growth in the absence of essential oil. (B) FUN-1 fluorescence (green) following treatment of yeast with oregano oil (described in Materials and Methods) but not with mineral oil indicates loss of metabolic activity. (C) Dose-dependent effects on yeast growth of a panel of terpenoid phenols reveal structure-activity relationships. Inhibition of yeast growth followed the order carvacrol ≥ thymol > eugenol ≫ γ-terpinene, vanillin, and guaicol. (D) Time course of growth inhibition in the presence of 0.005%, 0.0075%, and 0.01% carvacrol. (E) Dose-dependent loss of metabolic activity in response to carvacrol was monitored using FUN-1 green fluorescence. Data represent averages of the results of triplicates. Error bars show standard deviations. Ab600, absorbance at 600 nm.
pH measurements.
Cytoplasmic pH was measured using pHluorin, essentially as described previously (8). Briefly, BY4742 was transformed with plasmid pZR4.1 (33), grown to mid-logarithmic phase, and transferred to clear-bottom 96-well black plates. Carvacrol was injected at the concentration indicated in Fig. 2E, and fluorescence (dual excitation at 410 and 485 nm and emission at 520 nm) was recorded in a BMG Fluostar Optima plate reader. Calibrations were done in buffers of known pH as described previously (8). Vacuolar pH was measured after loading with BCECF AM (Invitrogen), a pH-sensitive fluorophore that localizes preferentially in the vacuole (3). Cells were loaded with BCECF AM and transferred to a 96-well plate. Fluorescence (dual excitation at 450 and 485 nm and emission at 520 nm) was measured in response to carvacrol and calibrated as previously described (3).
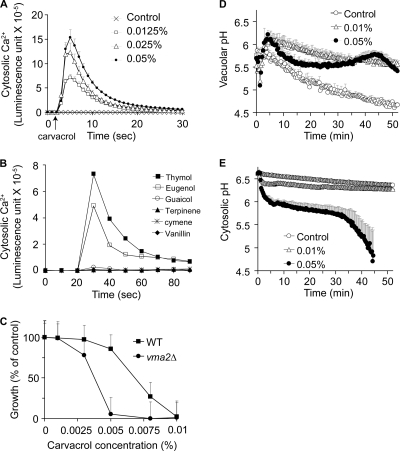
FIG. 2.
Disruption of ion homeostasis correlates with antifungal activity of carvacrol and terpenoid phenols. (A) Cytosolic Ca2+ was monitored by aequorin-coelenterazine luminescence as described in Materials and Methods. The addition of carvacrol (arrow) at the indicated doses elicited an immediate rise in cytosolic Ca2+, followed by rapid decay to baseline. The control response is in the presence of an equivalent volume of solvent (ethanol). (B) Ca2+-induced luminescence in response to the panel of terpenoid phenols (0.05%; added at 20 s) varied in magnitude and was proportional to the ability to inhibit yeast growth. (C) Hypersensitivity to carvacrol in a vma2Δ null mutant, lacking vacuolar H+-ATPase (V-ATPase) activity, relative to the growth of the isogenic wild type. Growth was monitored by absorbance at 600 nm in SC medium as described in Materials and Methods. (D and E) Measurement of vacuolar pH (D) or cytosolic pH (E) in response to indicated concentrations of carvacrol, using compartment-specific pH-sensitive fluorescent probes as described in Materials and Methods. Data are the averages of the results of triplicates. Error bars show standard deviations.
DNA microarray.
An early-log-phase culture of S. cerevisiae BY4742 (OD600 of 0.1) was treated with carvacrol at 0.005% and 0.01% for 15 min. RNA was isolated from control and carvacrol-treated cells as described previously (32). The integrity of the RNA samples was confirmed by polyacrylamide gel electrophoresis. cDNA synthesis, labeling and hybridization, image scanning, and processing were conducted at the Johns Hopkins Microarray Core Facility as described previously (32). Microarray data were imported to Partek GS software for normalization and analysis. The data set was then imported to Gene Cluster 3.0 for hierarchical clustering analyses, along with previously published DNA microarray data for these genes in response to thymol (7), CaCl2, (31), rapamycin (16), amiodarone (32), nitrogen depletion, growth in yeast extract-peptone-dextrose (YPD) (14), diauxic shift (11), and four classes of antifungals (caspofungin, ketoconazole, 5-fluorocytosine, and amphotericin) (2), downloaded from the publisher's website or requested from the authors. The results were displayed with Java Tree View software and edited in Adobe Photoshop (Adobe Systems, Inc.). Geneset enrichment analyses were performed on the server at Munich Information Center for Protein Sequences (MIPS) Functional Catalogue (http://mips.gsf.de/proj/funcatDB/search_main_frame.html).
RESULTS AND DISCUSSION
Efficacy of oregano oils and phenolic derivatives against S. cerevisiae.
A first step in our study was to determine whether the susceptibility of S. cerevisiae to terpenoid phenols recapitulated the results of published studies in various pathogenic fungi. The strength of oregano oil was analyzed by looking at the dose dependence of halo formation on a lawn of yeast. Oregano oil effectively prevented yeast growth up to a dilution of 1:8 (Fig. 1A), and no colonies appeared even after prolonged incubation, consistent with potent fungicidal activity. By comparison, another essential oil with antifungal activity, tea tree (Melaleuca) oil, only worked well at full strength, and small colonies appeared around the filters after 2 weeks (data not shown), suggesting a more fungistatic mechanism of action. As a control, we showed that mineral oil had no effect on yeast growth. In metabolically active cells, the vital stain FUN-1 is converted to a red intravacuolar spindle-like structure, whereas the loss of metabolic activity is associated with bright greenish-yellow fluorescence (19). In yeast cells treated briefly (15 min) with oregano oil but not in yeast cells treated with mineral oil, FUN-1 green fluorescence was indicative of a loss of metabolic activity (Fig. 1B).
Next, we evaluated the relative efficacies of purified components of oregano oil and related compounds. The monoterpenoid phenol carvacrol and its structural isomer thymol together constitute 72 to 83% of extracts from Origanum vulgarum species, although they vary reciprocally as the predominant components in distinct chemotypes (23). Out of a panel of structurally related phenolic compounds (Fig. 1C, bottom), carvacrol was found to be the most potent in inhibiting yeast growth, with a MIC of 0.008% (or 79.8 μg/ml), which was 1,500 times more effective than oregano oil (Fig. 1A). Thymol was only slightly less efficacious, whereas eugenol, a major phenolic component of clove oil (Eugenia sp.), was significantly less effective than carvacrol in the yeast growth assay (Fig. 1C). In contrast, the monoterpene hydrocarbon γ-terpinene, which is the biosynthetic precursor of carvacrol and also a component of oregano oil, was ineffective as a fungicide in the same concentration range as the terpenoid phenols. Other phenolic compounds, vanillin and guaiacol, were also ineffective as fungicides (Fig. 1C). These results parallel the work of Tampieri et al. (27), showing that out of a panel of terpenoid phenols, carvacrol was the most effective in killing C. albicans at a MIC of 100 ppm. Eugenol was slightly less active (MIC = 250 ppm) than carvacrol, and methyl eugenol was even weaker (MIC = 1,000 ppm). Unlike carvacrol, which has a free hydroxyl group, eugenol has a methylated hydroxyl group, and methyl eugenol has two methyl groups. Similarly, hydroxymethyl derivatives of carvacrol, thymol, and eugenol had significantly lower antifungal activities relative to those of the parent compounds (18), although interestingly, the derivatives had superior free radical-scavenging activities and protective effects as antioxidants. This suggests that the antifungal activity depends on the structure and makeup of the terpenoid phenols, specifically, the presence of a free hydroxyl group and an aromatic ring.
Carvacrol exerted dose-dependent inhibition on the yeast growth rate, with complete inhibition of growth at 0.01%, as shown by the results in Fig. 1D. Quantification of FUN-1 fluorescence showed that carvacrol elicited dose-dependent loss of metabolic activity in yeast (Fig. 1E) with more potency than the parent essential oil mixture (not shown), confirming that it was the major active ingredient of oregano oil.
Carvacrol disrupts ion homeostasis in yeast.
We have previously shown that an unrelated membrane-active compound, amiodarone, elicits cytosolic Ca2+ bursts and downstream Ca2+-related stress responses in yeast (15, 20, 33). Therefore, we examined the effect of carvacrol on cytosolic Ca2+ levels in yeast expressing the protein aequorin, after reconstitution with its cofactor coelenterazine. The aequorin-coelenterazine complex emits light upon binding to Ca2+, and the luminescence intensity quantitatively correlates to the Ca2+ concentration. Upon the addition of carvacrol to final concentrations ranging from 0.0125 to 0.05%, we observed immediate dose-dependent Ca2+ elevations, followed by a decrease to baseline within 1 to 2 min (Fig. 2A). These characteristic spikes have been described before (15, 20, 33) and are consistent with rapid influx of Ca2+ from the extracellular medium and from the vacuole and other intracellular stores, followed by sequestration into stores or efflux from cells and concomitant desensitization of channels. Similar Ca2+ bursts were observed with the structural isomer thymol, whereas eugenol showed smaller amplitudes of burst, and the remaining compounds tested (vanillin, guaiacol, γ-terpinene, and p-cymene) failed to elicit any change in luminescence (Fig. 2B). Overall, the ability of phenolic compounds to elicit Ca2+ bursts correlated well with their antifungal activity (Fig. 1C).
Cytosolic Ca2+ levels are tightly controlled within a narrow range that is compatible with cellular viability by an array of ion pumps and transporters. To distinguish whether the Ca2+ burst was directly in the pathway leading to cell death or a mere bystander effect, we evaluated carvacrol toxicity in a yeast vma2Δ mutant lacking a functional vacuolar H+ pump. In the absence of vacuolar acidification, which provides the driving force for H+-coupled Ca2+ exchangers, the clearance of cytosolic Ca2+ is severely impaired (12, 15, 33). We show that vma2Δ mutants are clearly more sensitive to growth inhibition by carvacrol than the isogenic wild type, consistent with toxicity of the Ca2+ burst (Fig. 2C).
Since the vacuolar H+ pump is also critical for pH homeostasis (17), we monitored the effect of carvacrol on both vacuolar and cytosolic pH. Cells were loaded with the acetoxymethyl derivative of the pH-sensitive fluorescent dye, BCECF, which has been shown to stably accumulate in yeast vacuoles (3). The addition of carvacrol elicited a 0.5-unit increase in the vacuolar pH and persistent alkalinization (Fig. 2D), suggesting a loss of protons out of the vacuolar lumen. Concurrently, carvacrol was able to induce immediate acidification of the yeast cytosol (Fig. 2E), which was monitored by the pH-dependent fluorescence of the green fluorescent protein derivative pHluorin (8). Acidification was dose dependent: whereas 0.01% carvacrol elicited a modest drop in pH, 0.05% carvacrol resulted in an immediate drop in pH of ∼0.5 pH unit, followed by a precipitous decrease beginning around 30 min after exposure to the phenolic compound. This second phase of cytosolic acidification induced by 0.05% carvacrol correlated with a reciprocal increase in vacuolar pH (Fig. 2D) and was well downstream of the Ca2+ burst (Fig. 2A). We conclude that carvacrol disrupts both Ca2+ and H+ homeostasis in yeast and that these disruptions likely lead to loss of cell viability.
Transcriptional profiling in carvacrol resembles the Ca2+ stress response.
As an independent approach toward elucidating the antifungal mechanism of terpenoid phenols, we analyzed the transcriptional response to carvacrol in yeast. Because disruption of Ca2+ and H+ homeostasis and loss of metabolic activity occur within minutes of carvacrol treatment, we reasoned that 15 min following drug exposure would be the ideal time to capture the transcriptional effect of carvacrol. Exponentially growing cells were treated for 15 min with 0.005% and 0.01% carvacrol. These concentrations were shown to cause about half-maximal and maximal inhibition of growth rates relative to the growth of the control (Fig. 1D).
As summarized in Table 1, the number of gene transcripts showing 2-fold or greater upregulation increased from 492 to 800 in 0.005% and 0.01% carvacrol, respectively, with 91 genes showing robust (≥2-fold) dose-dependent increases at higher carvacrol concentrations. A smaller number of gene transcripts were downregulated with exposure to carvacrol (Table 1).
TABLE 1.
Summary of differential regulation of genes in response to carvacrol
| Concn (%) of carvacrol | No. of genes that werea: |
|
|---|---|---|
| Upregulated | Downregulated | |
| 0.005 | 492 | 430 |
| 0.01 | 800 | 603 |
| Ratio (0.01% vs 0.005%) | 91 | 7 |
Results are shown for genes that were differentially regulated ≥2-fold.
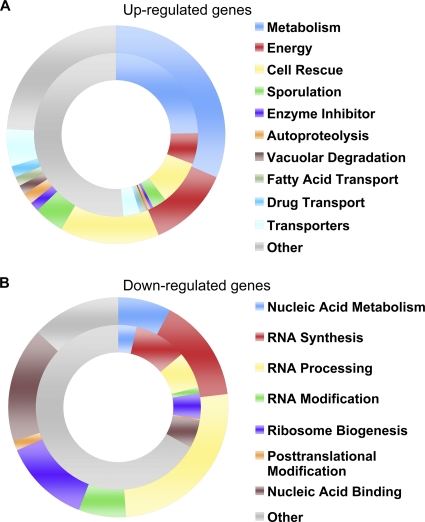
The functional categories of genes, according to the MIPS classification, that were differentially regulated by at least 2-fold in response to 0.01% carvacrol are shown in Fig. 3. The inner circle depicts the functional distribution of the yeast proteome, whereas the outer circle represents categories that were significantly overrepresented in the carvacrol data sets (P ≤ 5 × 10−3). Prominent among the upregulated genes (Fig. 3A) were functions associated with alternative metabolic or energy-handling pathways, including glucogen and trehalose biosynthesis, polyamine degradation, and fatty acid transport and oxidation. Next in abundance were pathways associated with stress response/signaling and cell rescue, including sporulation, oxygen and free radical detoxificaton, heat shock proteins and chaperones, autophagy, and vacuolar degradation mechanisms (Fig. 3A and Table 2). A robust induction of drug efflux mechanisms was observed, including many members of the ABC drug transporter family, such as SNQ2, YOR1, PDR5, PDR10, and PDR15 (Table 2). In contrast to this broad array of cellular functions affected by carvacrol, the pathways represented by repressed (Fig. 3B) genes were overwhelmingly associated with nucleic acid metabolism and RNA synthesis, processing, and modification. In dividing cells, most of the gene transcription (>80% of nucleotides used) is dedicated to synthesis of ribosomes and tRNA (29). Carvacrol rapidly shut down these pathways, consistent with cessation of growth (Table 2 and Fig. 1D).
FIG. 3.
Functional distribution of genes differentially regulated by carvacrol. Genes that showed transcriptional alteration by ≥2-fold in response to 0.01% carvacrol were categorized according to the MIPS classification system (see Materials and Methods). The inner circle shows the functional categorization of the entire yeast genome, whereas the outer ring includes categories that were significantly overrepresented in the carvacrol data set (P ≤ 5 × 10−3). Major functional categories represented in the carvacrol data set are listed on the right.
TABLE 2.
Representative examples of genes that are differentially regulated in response to 0.01% carvacrol
| Response, category | Genesa |
|---|---|
| Upregulated | |
| Drug/ABC transporters | SNQ2, PDR15, YOR1, VMR1, PDR11, PXA2, STE6, PDR18, PDR5, PDR10, PXA1 |
| Autophagy | ATG1, ATG2, ATG3, ATG4, ATG5, ATG7, ATG8, ATG9, ATG14, ATG15, ATG16, ATG20, ATG22, PBN1, COX20, SNX4, LAP4, YPS1, STE13 |
| Heat shock proteins and chaperones | SSA1, SSA3, SSA4, HSP26, HSP42, HSP78, HSP82, HSP104, SSE2, MSS2, ECM10, MDJ1, FMO1, MPD1 |
| Oxidative stress response | FRT2, PRX1, UGA2, GRX1, SNQ2, CTA1, TSA2, CTT1, GTT1, GPX1, SRX1, MCR1, FMP46, GAD1, GRE2, OXR1, GRE1 |
| Energy reserve metabolism | TPS1, TPS2, GLC3, GSY1, GSY2, GSC2, PGM2 |
| Downregulated | |
| rRNA processing | RPF2, UTP11, UTP13, UTP20, UTP23, RRP8, TSR1, TSR2, NOP4, ESF1, RRP1, RRP12 |
| Ribosome biogenesis | RRP7, DBP6, RLI1, RSA4, RRB1, KRI1, NOP1, NOP7, NOP15, MAK16, MAK21 |
| tRNA synthesis and processing | RPB5, LHP1, RPC52, BCD1, RPC10, RPC17, RET1, RPO40, RPO26, SEN34, POP8, POP6, SRM1 |
| Pyrimidine metabolism | URA7, FUI1, FUR4, PRS3, DCD1, PPR1, URK1 |
Only genes whose transcription was altered at least 2-fold are shown. Categories shown have P values of <0.0005.
Yeast transcriptional profiles have been documented in response to a wide array of antifungal drugs (5-fluorocytosine, amphotericin B, caspofungin, and ketoconozole), metabolic conditions (diauxic shift, nitrogen depletion, rapamycin treatment, and YPD) and agents known to induce ionic stress (amiodarone and Ca2+). Clustering analysis of microarray data revealed that the transcriptional response to carvacrol most closely resembled the Ca2+ stress response (Fig. 4A). We had previously observed that the transcriptional response to the antifungal agent amiodarone also clusters closely with Ca2+ stress, consistent with the ability of amiodarone to evoke bursts of cellular Ca2+. Here, we show a similarity in overall gene regulation by carvacrol and amiodarone (Fig. 4A). These observations corroborate our hypothesis that carvacrol elicits Ca2+-mediated cell death. Interestingly, the carvacrol response also closely resembles the effect of rapamycin, an inhibitor of the TOR signaling pathway that controls cell growth in response to nutrients and stress by regulating mRNA transcription and stability, protein translation, ribosome biogenesis and autophagy, and nutrient transport. To examine this further, we evaluated the overlap in transcriptional profiles in response to carvacrol, rapamycin, and Ca2+ stress (Fig. 4B and C). As seen in the Venn depictions, the overall transcriptional response to carvacrol was 58% identical to that in response to rapamycin and 63% identical to that in response to Ca2+ stress. Of the genes upregulated by carvacrol, a third (166) were also induced by the other two conditions and chiefly included genes involved in metabolic pathways of carbohydrate, protein, or energy. An even greater overlap existed among downregulated genes, wherein 60% of genes repressed by carvacrol (257) were common to downregulation by Ca2+ stress and rapamycin. Shared genes belonged largely to categories of RNA metabolism (60.7%) or ribosome biogenesis (55.6%). As these transcriptional responses are characteristic of inhibition of the TOR pathway, these findings raise the intriguing possibility that carvacrol may affect the TOR pathway, either independently or through calcium signaling. A recent screen of >3,500 compounds identified amiodarone as an inhibitor of mTORC1, based on the induction of autophagy in nutrient-rich medium (5); a similar effect may be predicted with carvacrol, given the robust induction of autophagy genes observed (Table 2).
FIG. 4.
Carvacrol elicits transcriptional response similar to calcium stress and rapamycin. (A) Hierarchical clustering of transcriptional response to carvacrol (Carv) (15 min [15′]), CaCl2 (Ca; 5 and 30 min), rapamycin (30 min), thymol (90 min), time course series for diauxic shift, amiodarone, nitrogen depletion, and growth in YPD (d, days) as described in Materials and Methods. Data available for 5,468 genes were included in the clustering analysis. The fold change values under each condition were log2 transformed and clustered with Gene Cluster 3.0. Distances between genes and arrays were computed based on correlation (centered). Both genes and arrays were clustered with the average linkage method. The data were visualized with Java Tree View. (B and C) Venn diagrams show the extent of overlap in the transcriptional response to carvacrol (0.005%, 15 min), calcium stress (30 min), and rapamycin (30 min) for upregulated (B) and downregulated genes (C).
Recently, Bi et al. (7) described the transcriptional response of S. cerevisiae to thymol, the structural isomer of carvacrol. As shown by hierarchical clustering analysis (Fig. 4A), the transcriptional responses to carvacrol and thymol were similar. As both compounds induce a rapid and robust cytosolic calcium surge (Fig. 1A and B), the mechanisms by which these two phenolic isomers kill fungal cells are likely to be substantially similar. We also noted differences in the transcriptional response to the two isomers. Thymol repressed multiple genes implicated in thiamine (vitamin B1) biosynthesis (THI4, THI6, THI12, THI20, THI21, SNZ2, SNZ3, and PET18) and sulfur metabolism, while this response was not observed after carvacrol treatment. It is possible that these differences represent distinct cellular responses to the two isomers. However, given the different times of assessment of the transcriptional responses to thymol (90 min; 7) and carvacrol (15 min, this study), it may well be that the induction of sulfur metabolism and the repression of thiamine biosynthesis are both late-stage transcriptional responses to these compounds. Furthermore, the size of the transcriptional response appears to diminish substantially with time, with differential regulation of 922 genes by at least 2-fold after a 15-min exposure to carvacrol (0.005%) (Table 1) and 305 genes after a 90-min treatment with thymol (7) at equivalent drug concentrations. A similar transient transcriptional response has been previously observed for the membrane-active drug amiodarone, which also elicits a rapid transcriptional response peaking at between 10 and 15 min, followed by significant decay at 30 min (13). Thus, we speculate that the study of Bi et al. provides insight into a later window of cellular response that is distinct from the early observations described in this work.
Mechanism of action of terpenoid phenols.
The hydrophobic nature of terpenoid phenols ensures their preferential partition into the lipid membrane; thus, carvacrol has a log P value of 3.26 for partition into phosphatidylethanolamine membranes relative to that of buffer (28). However, hydrophobicity alone does not ensure toxicity, since p-cymene, a precursor of carvacrol, has a higher partition coefficient for lipid membranes but is nontoxic. The presence of the hydroxyl group is critical for toxicity, as seen by the lack of microbicidal effects of p-cymene and carvacrol methylesters (6, 28). It has been proposed that the delocalized electron system in carvacrol facilitates the dissociation of H+ from the −OH group. This, in turn, would allow carvacrol to shuttle H+ and monovalent cations, such as K+, across membranes, dissipating pH and K+ gradients across cell membranes (28). Consistent with this mechanism, carvacrol was also shown to depolarize bacterial cell membranes and decrease the accumulation of the fluorescent dye 5(6)-carboxyfluorescein diacetate, suggestive of an increase in membrane permeability (30). Such a mechanism, however, does not explain the transient Ca2+ bursts associated with cellular interaction with carvacrol. It may be that effects on membrane expansion and fluidity (28) cause the opening of ion channels, followed by their rapid desensitization.
The distinct phases of Ca2+ and pH transients argue against a simple mechanism involving catastrophic membrane lesion, as has been previously proposed based on the uptake of propidium iodide (21, 24). Propidium iodide staining of cells at MIC confirms cell death but does not elucidate the mechanisms leading up to cell death and loss of membrane integrity. The transient nature of the cytosolic Ca2+ surge upon exposure to carvacrol indicates that cells maintain their ability to regulate ion flux (Fig. 2A). In addition, the robust transcriptional responses that largely overlap Ca2+ stress and nutrient starvation point to the activation of specific signaling pathways downstream of membrane interaction with terpenoid phenols. These signaling cascades reveal additional fungal targets that can be used in combination with carvacrol and similar essential oil components. Recent studies have shown that azole drugs inhibit vacuolar acidification and exacerbate the Ca2+ transients elicited by amiodarone, consistent with synergic effects of these drugs (33). Future studies could examine potential interactions between essential oils and azoles. In addition, calcineurin inhibitors (cyclosporine A and FK506) that enhance Ca2+ dysregulation and rapamycin analogs that block TOR signaling could be tested for drug interactions with carvacrol against pathogenic fungi.
Acknowledgments
This work was supported by a grant from the National Institutes of Health, NIAID R01AI065983, to R.R.
Footnotes
Published ahead of print on 4 October 2010.
REFERENCES
- 1.Abdel-Massih, R. M., R. Fares, S. Bazzi, N. El-Chami, and E. Baydoun. 2010. The apoptotic and anti-proliferative activity of Origanum majorana extracts on human leukemic cell line. Leuk. Res. 34:1052-1056. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal, A. K., P. D. Rogers, S. R. Baerson, M. R. Jacob, K. S. Barker, J. D. Cleary, L. A. Walker, D. G. Nagle, and A. M. Clark. 2003. Genome wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278:34998-35015. [DOI] [PubMed] [Google Scholar]
- 3.Ali, R., C. L. Brett, S. Mukherjee, and R. Rao. 2004. Inhibition of sodium/proton exchange by a Rab-GTPase-activating protein regulates endosomal traffic in yeast. J. Biol. Chem. 279:4498-4506. [DOI] [PubMed] [Google Scholar]
- 4.Alma, M. H., A. Mavi, A. Yildirim, M. Digrak, and T. Hirata. 2003. Screening chemical composition and in vitro antioxidant and antimicrobial activities of the essential oils from Origanum syriacum L. growing in Turkey. Biol. Pharm. Bull. 26:1725-1729. [DOI] [PubMed] [Google Scholar]
- 5.Balgi, A. D., B. D. Fonseca, E. Donohue, T. C. F. Tsang, P. Lajoie, C. G. Proud, I. R. Nabi, and M. Roberge. 2009. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One 4:e7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben Arfa, A., S. Combes, L. Preziosi-Belloy, N. Gontard, and P. Chalier. 2006. Antimicrobial activity of carvacrol related to its chemical structure. Lett. Appl. Microbiol. 43:149-154. [DOI] [PubMed] [Google Scholar]
- 7.Bi, X., N. Guo, J. Jin, J. Liu, H. Feng, J. Shi, H. Xiang, X. Wu, J. Dong, H. Hu, S. Yan, C. Yu, X. Wang, X. Deng, and L. Yu. 2010. The global gene expression profile of the model fungus Saccharomyces cerevisiae induced by thymol. J. Appl. Microbiol. 108:712-722. [DOI] [PubMed] [Google Scholar]
- 8.Brett, C. L., D. N. Tukaye, S. Mukherjee, and R. Rao. 2005. The yeast endosomal Na+K+/H+ exchanger Nhx1 regulates cellular pH to control vesicle trafficking. Mol. Biol. Cell 16:1396-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalleau, S., E. Cateau, T. Bergès, J. M. Berjeaud, and C. Imbert. 2008. In vitro activity of terpenes against Candida biofilms. Int. J. Antimicrob. Agents 31:572-576. [DOI] [PubMed] [Google Scholar]
- 10.De Martino, L., V. De Feo, F. Fratianni, and F. Nazzaro. 2009. Chemistry, antioxidant, antibacterial and antifungal activities of volatile oils and their components. Nat. Prod. Commun. 4:1741-1750. [PubMed] [Google Scholar]
- 11.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 12.Forster, C., and P. M. Kane. 2000. Cytosolic Ca2+ homeostasis is a constitutive function of the V-ATPase in Saccharomyces cerevisiae. J. Biol. Chem. 275:38245-38253. [DOI] [PubMed] [Google Scholar]
- 13.Gamarra, S., E. M. Rocha, Y. Q. Zhang, S. Park, R. Rao, and D. S. Perlin. 2010. Mechanism of the synergistic effect of amiodarone and fluconazole in Candida albicans. Antimicrob. Agents Chemother. 54:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta, S. S., V. K. Ton, V. Beaudry, S. Rulli, K. Cunningham, and R. Rao. 2003. Antifungal activity of amiodarone is mediated by disruption of calcium homeostasis. J. Biol. Chem. 278:28831-28839. [DOI] [PubMed] [Google Scholar]
- 16.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin modulated transcription defines the subset of nutrient sensitive signaling pathways directly controlled by the TOR proteins. Proc. Natl. Acad. Sci. U. S. A. 96:14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez-Muñoz, G. A., and P. Kane. 2008. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J. Biol. Chem. 283:20309-20319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mastelić, J., I. Jerković, I. Blazević, M. Poljak-Blazi, S. Borović, I. Ivancić-Baće, V. Smrecki, N. Zarković, K. Brcić-Kostic, D. Vikić-Topić, and N. Müller. 2008. Comparative study on the antioxidant and biological activities of carvacrol, thymol, and eugenol derivatives. J. Agric. Food Chem. 56:3989-3996. [DOI] [PubMed] [Google Scholar]
- 19.Millard, P. I., B. L. Roth, H. P. Thi, S. T. Yue, and P. R. Haugland. 1997. Development of the FUN-1 family of fluorescent probes for vacuole labeling and viability testing of yeasts. Appl. Environ. Microbiol. 63:2897-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muend, S., and R. Rao. 2008. Fungicidal activity of amiodarone is tightly coupled to calcium influx. FEMS Yeast Res. 8:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinto, E., L. Vale-Silva, C. Cavaleiro, and L. Salgueiro. 2009. Antifungal activity of the clove essential oil from Syzygium aromaticum on Candida, Aspergillus and dermatophyte species. J. Med. Microbiol. 58:1454-1462. [DOI] [PubMed] [Google Scholar]
- 22.Pozzatti, P., L. A. Scheid, T. B. Spader, M. L. Atayde, J. M. Santurio, and S. H. Alves. 2008. In vitro activity of essential oils extracted from plants used as spices against fluconazole-resistant and fluconazole-susceptible Candida spp. Can. J. Microbiol. 54:950-956. [DOI] [PubMed] [Google Scholar]
- 23.Russo, M., G. C. Galletti, P. Bocchini, and A. Carnacini. 1998. Essential oil chemical composition of wild populations of Italian oregano spice (Origanum vulgare ssp. hirtum (Link) Ietswaart): a preliminary evaluation of their use in chemotaxonomy by cluster analysis. 1. Inflorescences. J. Agric. Food Chem. 46:3741-3746. [Google Scholar]
- 24.Salgueiro, L. R., C. Cavaleiro, E. Pinto, C. Pina-Vaz, A. G. Rodrigues, A. Palmeira, C. Tavares, S. Costa-de-Oliveira, M. J. Gonçalves, and J. Martinez-de-Oliveira. 2003. Chemical composition and antifungal activity of the essential oil of Origanum virens on Candida species. Planta Med. 69:871-874. [DOI] [PubMed] [Google Scholar]
- 25.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 26.Slamenova, D., E. Horvathova, L. Marsalkova, and L. Wsolova. 2008. Carvacrol given to rats in drinking water reduces the level of DNA lesions induced in freshly isolated hepatocytes and testicular cells by H2O2. Neoplasma 55:394-399. [PubMed] [Google Scholar]
- 27.Tampieri, M. P., R. Galuppi, F. Macchioni, M. S. Carelle, L. Falcioni, P. L. Cioni, and I. Morelli. 2005. The inhibition of Candida albicans by selected essential oils and their major components. Mycopathologia 159:339-345. [DOI] [PubMed] [Google Scholar]
- 28.Ultee, A., M. H. Bennik, and R. Moezelaar. 2002. The phenolic hydroxyl group of carvacrol is essential for action against the food-borne pathogen Bacillus cereus. Appl. Environ. Microbiol. 68:1561-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 30.Xu, J., F. Zhou, B. P. Ji, R. S. Pei, and N. Xu. 2008. The antibacterial mechanism of carvacrol and thymol against Escherichia coli. Lett. Appl. Microbiol. 47:174-179. [DOI] [PubMed] [Google Scholar]
- 31.Yoshimoto, H., K. Saltsman, A. P. Gasch, H. X. Li, N. Ogawa, D. Botstein, P. O. Brown, and M. S. Cyert. 2002. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277:31079-31088. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Y. Q., and R. Rao. 2007. Global disruption of cell cycle progression and nutrient response by the antifungal agent amiodarone. J. Biol. Chem. 282:37844-37853. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, Y. Q., S. Gamarra, G. Garcia-Effron, S. Park, D. S. Perlin, and R. Rao. 2010. Requirement for ergosterol in V-ATPase function underlies antifungal activity of azole drugs. PLoS Pathog. 6:e1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]