Abstract
Candida glabrata is the second leading cause of candidemia in the United States. Its high-level resistance to triazole antifungal drugs has led to the increased use of the echinocandin class of antifungal agents for primary therapy of these infections. We monitored C. glabrata bloodstream isolates from a population-based surveillance study for elevated echinocandin MIC values (MICs of ≥0.25 μg/ml). From the 490 C. glabrata isolates that were screened, we identified 16 isolates with an elevated MIC value (2.9% of isolates from Atlanta and 2.0% of isolates from Baltimore) for one or more of the echinocandin drugs caspofungin, anidulafungin, and micafungin. All of the isolates with elevated MIC values had a mutation in the previously identified hot spot 1 of either the glucan synthase FKS1 (n = 2) or FKS2 (n = 14) gene. No mutations were detected in hot spot 2 of either FKS1 or FKS2. The predominant mutation was mutation of FKS2-encoded serine 663 to proline (S663P), found in 10 of the isolates with elevated echinocandin MICs. Two of the mutations, R631G for FKS1 and R665G for FKS2, have not been reported previously for C. glabrata. Multilocus sequence typing indicated that the predominance of the S663P mutation was not due to the clonal spread of a single sequence type. With a rising number of echinocandin therapy failures reported, it is important to continue to monitor rates of elevated echinocandin MIC values and the associated mutations.
The most recent class of antifungal agents to be introduced into clinical practice for the treatment of Candida infections is the echinocandins (4). All three echinocandin antifungal drugs, caspofungin, micafungin, and anidulafungin, have been shown to be effective in treating both invasive and esophageal candidiases caused by most Candida species, including those refractory to azole therapy (6, 10, 27, 38). When the initial breakpoints for the echinocandin drugs were proposed by the Clinical and Laboratory Standards Institute (CLSI), no breakpoint for resistance was set because in the original clinical outcome trials there were too few isolates with elevated MICs for any of the echinocandins to make a judgment (31). Since then, there has been an increasing number of case reports of clinical failure of echinocandins in patients from whom Candida isolates with elevated MICs for the echinocandins have been recovered (reviewed in reference 34).
Decreased susceptibility to the echinocandins is associated with mutations in the Fks1p and Fks2p subunits of the 1,3-β-d-glucan synthase complex, which is necessary for the production of 1,3-β-d-glucan, an essential component of the Candida cell wall (11, 12, 16, 25). Specifically, the mutations occur in two regions, of nine and eight amino acids, designated hot spot 1 and hot spot 2, respectively, that appear in both Fks1p and Fks2p (25). These mutations in the FKS1 and FKS2 genes result in the inability of echinocandins to inhibit the production of 1,3-β-d-glucan (26).
Candida glabrata has recently emerged as the second most common cause of candidemia in the United States (29, 37). C. glabrata has demonstrated decreased susceptibility to azole drugs, especially fluconazole (32). This reduced susceptibility to azoles has led to the recommendation by the Infectious Diseases Society of America (IDSA) for the preferred use of an echinocandin as primary therapy for treatment of C. glabrata infections (24, 29). While there has been largely excellent coverage of C. glabrata by the echinocandins, as measured in vitro (13, 28, 31), there are cases of clinical failure of echinocandins against C. glabrata isolates (7, 14, 19, 36). To date, there has not been an epidemiological study which estimates the prevalence of C. glabrata isolates with elevated echinocandin MICs, and there is no clear picture of the relative frequency of these isolates at the population level.
The Centers for Disease Control and Prevention (CDC) and selected Emerging Infections Program (EIP) partners conducted active population-based candidemia surveillance in the metropolitan areas of Atlanta, GA, and Baltimore, MD, between 2008 and 2010. Population-based surveillance is unique in that it includes the total population of a particular geographic area and avoids the biases associated with single or select institutional studies. Candida sp. bloodstream isolates from all hospitals within each defined geographic area were collected and identified to the species level. We used C. glabrata isolates collected in the population-based surveillance study to monitor MIC values for caspofungin, micafungin, and anidulafungin and to identify changes in the Fksp proteins associated with elevated echinocandin MIC values.
MATERIALS AND METHODS
Case and isolate definitions.
Isolates were obtained from persons with an incident episode of candidemia (defined below) identified between 1 January 2008 and 1 February 2010 who were residents of Atlanta, GA, and eight surrounding counties or who were residents of Baltimore City or Baltimore County, MD (1). The capture rate for isolates from identified cases in Atlanta was 71%, and in Baltimore it was 92%. Isolates collected at participating hospitals but from patients living outside the population-based catchment area (noncases) were included in some of the analyses, but medical information was not collected in these cases. An incident episode of candidemia was defined as the 30 days following the first positive blood culture for Candida species. Positive cultures of the same species within the 30-day period were considered part of the incident case episode and were not captured. Cultures drawn more than 30 days before or after the incident case were considered a new incident episode and assigned a new case number. If a patient had more than one incident episode of C. glabrata candidemia and both isolates had elevated echinocandin MIC values, only one isolate was used in the analysis.
Isolate storage, DNA extraction, PCR amplification, and sequencing.
Prior to use, all isolates were stored in glycerol at −70°C. Isolates were identified as C. glabrata by both conventional biochemical means at the referring institutions and by Luminex assay at the CDC (9). Unlike conventional biochemical methods, the Luminex assay is able to distinguish between C. glabrata, C. bracarensis, and C. nivariensis, so two isolates of C. bracarensis and four isolates of C. nivariensis were removed from the data set to avoid the confusion of a possible mixed-species data set. After passage of each isolate twice on Sabouraud dextrose agar plates, DNA was extracted using a Mo Bio microbial DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA) according to the manufacturer's instructions. The oligonucleotide primers used for FKS hot spot analysis are described in Table 1 and were those used by Thompson and coworkers (36). PCRs were performed in a 50-μl volume containing 10 ng of genomic DNA, 0.2 μM (each) primers, Roche Taq DNA polymerase, and Taq PCR master mix as recommended by the manufacturer (Roche Diagnostics, Indianapolis, IN). PCR products were purified using Exo SAP-IT as recommended by the manufacturer (USB, Cleveland, OH). Sequencing reactions were performed using BigDye Terminator technology (ABI, Foster City, CA) with an ABI Prism 3730 DNA sequencer. Loci were sequenced in both the forward and reverse directions, using the same primers as those used for the PCRs.
TABLE 1.
FKS1 and FKS2 hot spot primers
| Locus | Primer | Primer sequence | Annealing temp (°C) |
|---|---|---|---|
| FKS1 hot spot 1 | FKS1HS1F | CCATTGGGTGGTCTGTTCACG | 52 |
| FKS1HS1R | GATTGGGCAAAGAAAGAAATACGAC | 52 | |
| FKS1 hot spot 2 | FKS1HS2F | GGTATTTCAAAGGCTCAAAAGGG | 51 |
| FKS1HS2R | ATGGAGAGAACAGCAGGGCG | 51 | |
| FKS2 hot spot 1 | FKS2HS1F | GCTTCTCAGACTTTCACCG | 49 |
| FKS2HS1R | CAGAATAGTGTGGAGTCAAGACG | 49 | |
| FKS2 hot spot 2 | FKS2HS2F | TCTTGACTTTCTACTATGCG | 46 |
| FKS2HS2R | CTTGCCAATGTGCCACTG | 46 |
Antifungal susceptibility testing.
C. glabrata antifungal susceptibility testing was performed by broth microdilution with anidulafungin, caspofungin, and micafungin as described by CLSI document M27-A3 guidelines (8), using RPMI microbroth trays custom manufactured by Trek Diagnostics (Cleveland, OH). Results were read visually after 24 h of incubation. The breakpoint values were those established by the CLSI, with a susceptible isolate having an MIC value of ≤2 μg/ml (31). There is no breakpoint for resistance, so all isolates with an MIC value of ≥4 μg/ml were considered nonsusceptible. The epidemiological cutoff values were taken from the work of Pfaller and coworkers (28).
Sequence analysis.
Nucleotide sequences were analyzed using Sequencher 4.9 software (Genecodes Inc., Ann Arbor, MI). The protein sequence was determined using the Translate tool on the ExPASy proteomics server (Swiss Institute of Bioinformatics, Switzerland), and polymorphisms were observed visually using MEGA, version 4 (35), and ClustalW2 (20) and compared to the genome database sequences (GenBank accession number XM_446406 for FKS1 and XM_448401 for FKS2).
MLST analysis.
Multilocus sequence typing (MLST) analysis was performed as previously described (22). Neighbor-joining analysis was performed using the HyPhy software program (33).
RESULTS
Antifungal susceptibility testing of C. glabrata isolates.
Antifungal susceptibility testing was performed on 490 C. glabrata bloodstream isolates identified during the study period, with inclusion of all of the incident isolates (252 from Atlanta and 238 from Baltimore). MIC values are given in Table 2. The majority of isolates (80.4%) had MIC values for all three echinocandins of ≤0.06 μg/ml. The epidemiological cutoff values for all three echinocandins were previously calculated to be 0.125 μg/ml for caspofungin, 0.25 μg/ml for anidulafungin, and 0.03 μg/ml for micafungin (28). When these numbers were applied to our data set, 96.7% of the isolates fell at or below these values for all three echinocandins (Table 3). There were 16 isolates (3.3%), all from different patients, with an MIC value for at least one echinocandin of ≥0.25 μg/ml, including 7 that were categorized as nonsusceptible to caspofungin, 5 that were nonsusceptible to micafungin, and 2 that were nonsusceptible to anidulafungin (Table 4).
TABLE 2.
Distribution of MICs for three echinocandins for 490 C. glabrata isolates
| Antifungal | No. of strains at MIC (μg/ml) (no. of isolates with FKS mutation)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.008 | 0.015 | 0.03 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | ≥16 | |
| Anidulafungin | 1 | 12 | 164 | 246 (1) | 52 | 1 (1) | 3 (3) | 2 (2) | 7 (7) | 2 (2) | ||
| Caspofungin | 4 | 176 | 240 | 55 (1) | 2 (2) | 2 (2) | 2 (2) | 2 (2) | 1 (1) | 1 (1) | 5 (5) | |
| Micafungin | 41 | 338 | 91 | 4 | 1 (1) | 3 (3) | 2 (2) | 1 (1) | 4 (4) | 5 (5) | ||
Not all isolates with MICs of <0.125 μg/ml were screened for FKS mutations; any appearing at those values relate to a higher value for one of the other echinocandins.
TABLE 3.
MIC ranges for C. glabrata surveillance isolates
| MIC (μg/ml) | No. (%) of isolates |
||
|---|---|---|---|
| Total | Atlanta | Baltimore | |
| ≤0.06 for all 3 echinocandins | 394 (80.4) | 194 (77.0) | 200 (84.0) |
| 0.125 for at least 1 echinocandin | 80 (16.3) | 47 (18.7) | 33 (13.9) |
| ≥0.25 for at least 1 echinocandina | 16 (3.3) | 11 (4.4) | 5 (2.1) |
| Total | 490 | 252 | 238 |
Indicates the MIC at which all isolates contain an FKS mutation.
TABLE 4.
Isolates with elevated MICs and associated mutations
| Isolate | MIC (μg/ml) |
Mutation |
|||
|---|---|---|---|---|---|
| Anidulafungin | Caspofungin | Micafungin | Fks1 hot spot 1 | Fks2 hot spot 1 | |
| CAS09-1648 | 0.06 | 0.125 | 0.25 | R665G | |
| CAS09-1083 | 0.5 | 0.25 | 0.125 | S663F | |
| CAS09-1437 | 0.25 | 0.25 | 0.5 | R631G | |
| CAS08-0725 | 1 | 0.5 | 0.25 | F659Y | |
| CAS08-0016 | 1 | 0.5 | 1 | S629P | |
| CAS09-1204 | 0.5 | 1 | 2 | S663P | |
| CAS09-1616 | 2 | 1 | 2 | S663P | |
| CAS08-0311 | 2 | 2 | 0.25 | P667H | |
| CAS09-0901 | 2 | 2 | 0.5 | S663P | |
| CAS08-0037 | 2 | 4 | 2 | S663P | |
| CAS08-0209 | 2 | 8 | 4 | S663P | |
| CAS08-0425 | 2 | ≥16 | 4 | S663P | |
| CAS08-0293 | 0.5 | ≥16 | 4 | S663P | |
| CAS08-0094 | 4 | ≥16 | 4 | S663P | |
| CAS09-1225 | 4 | ≥16 | 4 | S663P | |
| CAS09-1786 | 2 | ≥16 | 2 | S663P | |
FKS hot spot mutations in bloodstream isolates.
Earlier reports have shown that specific mutations in the FKS genes of Candida spp. are responsible for elevated MIC values for the echinocandins (11, 12, 16, 18, 25). Fragments encoding hot spot 1 and hot spot 2 of FKS1 and FKS2 were amplified for the 16 isolates described above with an MIC value of ≥0.25 μg/ml for at least one of the echinocandin drugs (Table 4). In addition, we amplified the same regions from 65 isolates with an MIC value of 0.125 μg/ml for one or more echinocandin and from 34 isolates with MIC values of ≤0.06 μg/ml for all of the echinocandins. All of the mutations were in the 16 isolates with an MIC value of ≥0.25 μg/ml for at least one of the echinocandins. All 16 isolates had mutations in an Fksp hot spot region, with 2 in Fks1p hot spot 1 and 14 in Fks2p hot spot 1. There were no mutations in hot spot 2 of either Fks1p or Fks2p. Likewise, there were no hot spot mutations in any of the isolates with MIC values of ≤0.125 μg/ml. MIC values for the isolates and amino acid changes in the hot spot regions are shown in Table 4.
The most common mutation was Fks2p S663P, which was present in 10 of the 16 isolates. There were four other mutations in Fks2p hot spot 1, namely, F659Y, S663F, R665G, and P667H. There were two Fks1p hot spot 1 mutations, S629P and R631G. All of the isolates that had an MIC value for one or more echinocandin in the nonsusceptible range contained the Fks2p S663P mutation.
Population-based analysis of FKS mutations.
Of the 490 isolates screened, 405 were from patients within the two EIP catchment areas, including 205 isolates from Atlanta and 200 isolates from Baltimore. The other isolates were from hospitals within the catchment area but from patients residing outside the area. Of the 405 population-based incident isolates, 10 had mutations in the Fksp hot spot regions and higher echinocandin MIC values. This corresponded to 2.9% and 2.0% of the isolates from Atlanta and Baltimore, respectively.
MLST analysis of isolates containing FKS mutations.
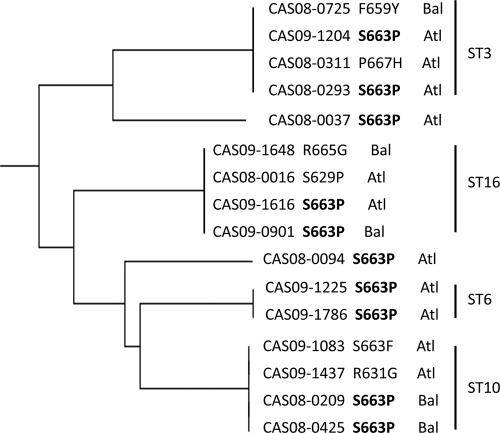
Ten of the isolates, seven from Atlanta and three from Baltimore, shared the S663P mutation. To test whether this was caused by the spread of a clonal isolate, MLST analysis was performed on all of the isolates with FKS mutations (Table 5 and Fig. 1). The seven Atlanta isolates with the Fks2p S663P mutation had five different sequence types, including two isolates with ST3 and two with ST6. The three Fks2p S663P-containing isolates from Baltimore had two different sequence types, including two isolates with ST10. Overall, 13 of the 16 isolates with high echinocandin MIC values fell into the most prominent sequence types, i.e., ST3, ST10, ST16, and ST19 (22).
TABLE 5.
MLST analysis of isolates with FKS mutations
| Isolatea | Mutation |
Site | Sequence type | |
|---|---|---|---|---|
| FKS1 hot spot 1 | FKS2 hot spot 1 | |||
| CAS09-1648* | R665G | Baltimore | ST16 | |
| CAS09-1083* | S663F | Atlanta | ST10 | |
| CAS09-1437* | R631G | Atlanta | ST10 | |
| CAS08-0725 | F659Y | Baltimore | ST3 | |
| CAS08-0016 | S629P | Atlanta | ST16 | |
| CAS09-1204 | S663P | Atlanta | ST3 | |
| CAS09-1616 | S663P | Atlanta | ST16 | |
| CAS08-0311* | P667H | Atlanta | ST3 | |
| CAS09-0901* | S663P | Baltimore | ST16 | |
| CAS08-0037* | S663P | Atlanta | ST19 | |
| CAS08-0209* | S663P | Baltimore | ST10 | |
| CAS08-0425* | S663P | Baltimore | ST10 | |
| CAS08-0293* | S663P | Atlanta | ST3 | |
| CAS08-0094 | S663P | Atlanta | ST76 | |
| CAS09-1225* | S663P | Atlanta | ST6 | |
| CAS09-1786 | S663P | Atlanta | ST6 | |
Asterisks indicate isolates included in the population-based surveillance study.
FIG. 1.
Neighbor-joining tree of C. glabrata isolates with FKS mutations.
Five hospitals, three in Atlanta and two in Baltimore, were the sources of more than one isolate of C. glabrata with an Fksp hot spot mutation, including one hospital in Atlanta with four isolates. For each of these hospitals, MLST revealed that all of the isolates with Fksp mutations were genotypically unrelated.
Epidemiological analysis of isolates with FKS mutations.
We reviewed medical chart information for the 16 case patients with isolates containing hot spot mutations. Of those 16 patients, 7 (44%) had a previous episode of candidemia more than 30 days prior to this incident case. Data were available for six of these seven patients; four patients had received caspofungin therapy, and two patients had received micafungin therapy to treat the previous candidemia infection. An additional two (13%) patients had not had a previous case of candidemia but had received echinocandin therapy prior to their incident C. glabrata candidemia. Detailed medical information was available for nine (56%) patients, and all had received an echinocandin (6 received caspofungin, while 3 received micafungin) to treat the candidemia episode caused by the C. glabrata isolate with the FKS mutation, for durations ranging from 1 to 14 days. Six (67%) of these patients survived the 30-day episodic period; three patients died. Only one of the patients who died had a C. glabrata isolate with an echinocandin MIC value in the nonsusceptible range (≥16 μg/ml).
DISCUSSION
C. glabrata has emerged as a major cause of bloodstream infection in the United States. Its decreased susceptibility to azole antifungal agents has led to the increased use of the newest antifungal agents, the echinocandins, for standard therapy of C. glabrata infections, as recommended in the IDSA guidelines for management of candidiasis (24). In our surveillance, C. glabrata remained largely susceptible to all three of the echinocandins, with susceptibility rates of 98.6%, 98.9%, and 99.6% for caspofungin, micafungin, and anidulafungin, respectively, assuming the current breakpoint of 2 μg/ml. However, if the breakpoint for susceptibility were lowered to ≤0.125 μg/ml to account for the MIC values for isolates with known FKS mutations, then 96.9% of our isolates would fall within the susceptible range, regardless of which echinocandin was considered. Among the 16 isolates with FKS mutations, the highest MIC values were for caspofungin, while the lowest MIC values were for anidulafungin, similar to the results reported by Perlin for C. albicans isolates with FKS1 mutations (26). Isolate CAS09-1648 (with R665G mutation) had MIC values of only 0.06 μg/ml for anidulafungin and 0.125 μg/ml for caspofungin, both of which are within the epidemiological cutoff range, but had an MIC of 0.25 μg/ml for micafungin. This mutation would have been missed if anidulafungin or caspofungin alone had been used to screen for mutations. It is quite possible that this mutation confers a response specific to micafungin, but at this time we do not have any information that would allow us to predict the clinical significance of such a mutation and the outcome of usage of any of the three echinocandins for this isolate. The hot spot 1 mutations R631G in Fks1p and R665G in Fks2p, which are structurally homologous mutations in the two Fks proteins, have not been reported previously for clinical C. glabrata isolates. Because they are not associated with highly elevated MIC values, perhaps patients harboring C. glabrata isolates with either of these two mutations would be less likely to fail therapy and the MIC values would not be noted because they fall under the current breakpoint for susceptible isolates.
There was no temporal difference in the number of isolates with FKS mutations collected over the 2 years of surveillance: eight isolates were collected in 2008, and eight isolates were collected in 2009. It is interesting that despite receiving isolates from 40 hospitals, 12 of the 16 isolates were clustered in five hospitals, two in Baltimore and three in Atlanta, and 25% of the isolates, although genetically unrelated to one another, came from a single hospital in Atlanta. While in at least two cases in Atlanta we cannot rule out clonality among isolates with identical Fks2p mutations and sequence types, MLST analysis does not support the hypothesis of clonal spread of a single resistance phenotype.
In looking for spontaneously derived caspofungin-resistant isolates of C. albicans on plates containing 4 μg/ml of caspofungin, Balashov and coworkers (2) found that the overwhelming majority of mutations (86%) occurred at serine 645 of Fks1p. This is structurally homologous to C. glabrata serine 663 of Fks2p. Their distribution of 62% of isolates having the S645P mutation and 8% of isolates having the S645F mutation closely parallels our findings for C. glabrata, with 63% of our high-MIC isolates having the S663P mutation and 6% having the S663F mutation. In the Balashov study, it was also found that 22% of the isolates had the S645Y mutation, one that we did not find in our study. While their study was not designed to detect mutations in Fks2p, we did find one structurally homologous mutation in Fks1p, S629P. The Fks2p S663P mutation in C. glabrata has previously been reported for at least one patient who failed anidulafungin therapy (14) and in two other surveillance reports on FKS mutations (5, 15). In a collection of random C. glabrata clinical isolates with FKS mutations, the majority of the isolates (69%) had mutations in FKS2 (15). One of the most interesting aspects of the frequency of the S663P mutation in our surveillance is that this mutation is not clonal in origin in our isolates, since it is spread among five different sequence types. Our data and previously published data (2, 25) indicate that there is strong pressure for mutation at this particular amino acid position in Fks1p of C. albicans and in Fks2p of C. glabrata, such that the emergence of this mutation in multiple C. glabrata clones is not surprising.
One of the more unfortunate aspects of the relative abundance of the S663P mutation is that it is associated with the highest echinocandin MIC values. However, not all of the isolates with the S663P mutation had the same MIC values. This may be a reflection of the expression level of the FKS2 gene compared to that of the FKS1 gene. It has been shown that in C. glabrata the FKS2 gene is expressed at a higher level than FKS1 in wild-type isolates but that the expression levels change in isolates with FKS mutations (15, 16). We have not determined the expression levels of FKS genes in our isolates, but it will be interesting to do so in the future to assess the possible role of FKS gene expression levels in MIC values for isolates with identical mutations.
All of our isolates with FKS mutations for which we had epidemiological data came from patients who had previously been treated with an echinocandin. The clinical significance of elevated MIC values for the echinocandins is unclear. Although there is a clear link between echinocandin therapy, elevated echinocandin MIC values, and treatment failure in a limited number of case reports (7, 14, 21, 23, 36), many patients with Candida isolates having elevated echinocandin MIC values respond to echinocandin therapy, and elevated MICs may not be a good predictor of outcome (3, 17, 30). If the breakpoints for the echinocandins and C. glabrata were lowered to reflect the epidemiological cutoff value for this organism, our data support the lowered values as being able to distinguish between wild-type isolates and those carrying mutations in their FKS genes that affect susceptibility.
Echinocandins are now the IDSA-recommended first-line therapy for C. glabrata candidemia (24). While only a small proportion of the C. glabrata isolates in this study had elevated echinocandin MIC values, it is likely that these isolates will continue to emerge and increase in frequency as echinocandin usage increases. It is important to continue surveillance for FKS mutations associated with elevated echinocandin MIC values and to monitor the impact on clinical outcomes.
Acknowledgments
We acknowledge Joyce Peterson, Randy Kuykendall, Lauren Smith, Mary Brandt, and Eszter Deak at the Centers for Disease Control, Betsy Siegel and Wendy Baughman in Atlanta, and Sandra Muhanuka and Rosemary Hollick in Baltimore for their contributions. In addition, we thank all of the hospitals and laboratories in Atlanta and Baltimore that contributed isolates.
A.J.Z. was supported by the Association of Public Health Laboratories (APHL).
The findings and conclusions of this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 13 September 2010.
REFERENCES
- 1.Ahlquist, A., M. M. Farley, L. H. Harrison, W. Baughman, B. Siegel, R. Hollick, S. R. Lockhart, S. S. Magill, and T. Chiller. 2009. Epidemiology of candidemia in metropolitan Atlanta and Baltimore city and county: preliminary results of population-based, active laboratory surveillance—2008-2010, abstr. M-1241. Abstr. 49th Int. Conf. Antimicrob. Agents Chemother., San Francisco, CA. American Society for Microbiology, Washington, DC.
- 2.Balashov, S. V., S. Park, and D. S. Perlin. 2006. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob. Agents Chemother. 50:2058-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, J. E. 2006. Echinocandins for candidemia in adults without neutropenia. N. Engl. J. Med. 355:1154-1159. [DOI] [PubMed] [Google Scholar]
- 4.Cappelletty, D., and K. Eiselstein-McKitrick. 2007. The echinocandins. Pharmacotherapy 27:369-388. [DOI] [PubMed] [Google Scholar]
- 5.Castanheira, M., L. N. Woosley, D. J. Diekema, S. A. Messer, R. N. Jones, and M. A. Pfaller. 2010. Low prevalence of fks1 hotspot 1 mutations in a worldwide collection of Candida strains. Antimicrob. Agents Chemother. 54:2655-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandrasekar, P. H., and J. D. Sobel. 2006. Micafungin: a new echinocandin. Clin. Infect. Dis. 42:1171-1178. [DOI] [PubMed] [Google Scholar]
- 7.Cleary, J. D., G. Garcia-Effron, S. W. Chapman, and D. S. Perlin. 2008. Reduced Candida glabrata susceptibility secondary to an FKS1 mutation developed during candidemia treatment. Antimicrob. Agents Chemother. 52:2263-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical Laboratory Standards Institute. 2008. M27-A3. Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard, 3rd ed. Clinical Laboratory Standards Institute, Wayne, PA.
- 9.Deak, E., K. A. Etienne, S. R. Lockhart, L. Gade, T. Chiller, and S. A. Balajee. 2010. Utility of a Luminex-based assay for multiplexed, rapid species identification of Candida isolates from an ongoing candidemia surveillance. Can. J. Microbiol. 56:348-351. [DOI] [PubMed] [Google Scholar]
- 10.Deresinski, S. C., and D. A. Stevens. 2003. Caspofungin. Clin. Infect. Dis. 36:1445-1457. [DOI] [PubMed] [Google Scholar]
- 11.Desnos-Ollivier, M., S. Bretagne, D. Raoux, D. Hoinard, F. Dromer, E. Dannaoui, and the European Committee on Antibiotic Susceptibility Testing. 2008. Mutations in the fks1 gene in Candida albicans, C. tropicalis, and C. krusei correlate with elevated caspofungin MICs uncovered in AM3 medium using the method of the European Committee on Antibiotic Susceptibility Testing. Antimicrob. Agents Chemother. 52:3092-3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douglas, C. M., J. A. D'Ippolito, G. J. Shei, M. Meinz, J. Onishi, J. A. Marrinan, W. Li, G. K. Abruzzo, A. Flattery, K. Bartizal, A. Mitchell, and M. B. Kurtz. 1997. Identification of the FKS1 gene of Candida albicans as the essential target of 1,3-beta-d-glucan synthase inhibitors. Antimicrob. Agents Chemother. 41:2471-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espinel-Ingroff, A. 2003. In vitro antifungal activities of anidulafungin and micafungin, licensed agents and the investigational triazole posaconazole as determined by NCCLS methods for 12,052 fungal isolates: review of the literature. Rev. Iberoam. Micol. 20:121-136. [PubMed] [Google Scholar]
- 14.Garcia-Effron, G., D. J. Chua, J. R. Tomada, J. Dipersio, D. S. Perlin, M. Ghannoum, and H. Bonilla. 2010. Novel FKS mutations associated with echinocandin resistance in Candida species. Antimicrob. Agents Chemother. 54:2225-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Effron, G., S. Lee, S. Park, J. Cleary, and D. S. Perlin. 2009. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-β-d-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob. Agents Chemother. 53:3690-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Effron, G., S. K. Katiyar, S. Park, T. D. Edlind, and D. S. Perlin. 2008. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 52:2305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kartsonis, N., J. Killar, L. Mixson, C. M. Hoe, C. Sable, K. Bartizal, and M. Motyl. 2005. Caspofungin susceptibility testing of isolates from patients with esophageal candidiasis or invasive candidiasis: relationship of MIC to treatment outcome. Antimicrob. Agents Chemother. 49:3616-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katiyar, S., M. A. Pfaller, and T. D. Edlind. 2006. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 50:2892-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogh-Madsen, M., M. C. Arendrup, L. Heslet, and J. D. Knudsen. 2006. Amphotericin B and caspofungin resistance in Candida glabrata isolates recovered from a critically ill patient. Clin. Infect. Dis. 42:938-944. [DOI] [PubMed] [Google Scholar]
- 20.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. ClustalW and ClustalX version 2. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 21.Laverdière, M., R. G. Lalonde, J. G. Baril, D. C. Sheppard, S. Park, and D. S. Perlin. 2006. Progressive loss of echinocandin activity following prolonged use for treatment of Candida albicans oesophagitis. J. Antimicrob. Chemother. 57:705-708. [DOI] [PubMed] [Google Scholar]
- 22.Lott, T. J., J. P. Frade, and S. R. Lockhart. 2010. Multilocus sequence type analysis reveals both clonality and recombination in populations of Candida glabrata bloodstream isolates from U.S. surveillance studies. Eukaryot. Cell 9:619-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, C. D., B. W. Lomaestro, S. Park, and D. S. Perlin. 2006. Progressive esophagitis caused by Candida albicans with reduced susceptibility to caspofungin. Pharmacotherapy 26:877-880. [DOI] [PubMed] [Google Scholar]
- 24.Pappas, P. G., C. A. Kauffman, D. Andes, D. K. Benjamin, Jr., T. F. Calandra, J. E. Edwards, Jr., S. G. Filler, J. F. Fisher, B. J. Kullberg, L. Ostrosky-Zeichner, A. C. Reboli, J. H. Rex, T. J. Walsh, and J. D. Sobel. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 48:503-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park, S., R. Kelly, N. Kahn, J. Robles, M. J. Hsu, E. Register, W. Li, V. Vyas, H. Fan, G. Abruzzo, A. Flattery, C. Gill, G. Chrebet, S. A. Parent, M. Kurtz, H. Teppler, C. M. Douglas, and D. S. Perlin. 2005. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 49:3264-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perlin, D. S. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10:121-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfaller, M. A. 2004. Anidulafungin: an echinocandin antifungal. Expert Opin. Invest. Drugs 13:1183-1197. [DOI] [PubMed] [Google Scholar]
- 28.Pfaller, M. A., L. Boyken, R. J. Hollis, J. Kroeger, S. A. Messer, S. Tendolkar, R. N. Jones, J. Turnidge, and D. J. Diekema. 2010. Wild-type MIC distributions and epidemiological cutoff values for the echinocandins and Candida spp. J. Clin. Microbiol. 48:52-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfaller, M. A., D. J. Diekema, L. Boyken, S. A. Messer, S. Tendolkar, R. J. Hollis, and B. P. Goldstein. 2005. Effectiveness of anidulafungin in eradicating Candida species in invasive candidiasis. Antimicrob. Agents Chemother. 49:4795-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfaller, M. A., D. J. Diekema, L. Ostrosky-Zeichner, J. H. Rex, B. D. Alexander, D. Andes, S. D. Brown, V. Chaturvedi, M. A. Ghannoum, C. C. Knapp, D. J. Sheehan, and T. J. Walsh. 2008. Correlation of MIC with outcome for Candida species tested against caspofungin, anidulafungin, and micafungin: analysis and proposal for interpretive MIC breakpoints. J. Clin. Microbiol. 46:2620-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfaller, M. A., S. A. Messer, R. J. Hollis, L. Boyken, S. Tendolkar, J. Kroeger, and D. J. Diekema. 2009. Variation in susceptibility of bloodstream isolates of Candida glabrata to fluconazole according to patient age and geographic location in the United States in 2001 to 2007. J. Clin. Microbiol. 47:3185-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pond, S. L., S. D. Frost, and S. V. Muse. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676-679. [DOI] [PubMed] [Google Scholar]
- 34.Sun, H. Y., and N. Singh. 2010. Characterisation of breakthrough invasive mycoses in echinocandin recipients: an evidence-based review. Int. J. Antimicrob. Agents 35:211-218. [DOI] [PubMed] [Google Scholar]
- 35.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, G. R., N. P. Wiederhold, A. C. Vallor, N. C. Villareal, J. S. Lewis, and T. F. Patterson. 2008. Development of caspofungin resistance following prolonged therapy for invasive candidiasis secondary to Candida glabrata infection. Antimicrob. Agents Chemother. 52:3783-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309-317. [DOI] [PubMed] [Google Scholar]
- 38.Zaas, A. K., and B. D. Alexander. 2005. Echinocandins: role in antifungal therapy, 2005. Expert Opin. Pharmacother. 6:1657-1668. [DOI] [PubMed] [Google Scholar]