Abstract
Staphylococcus sciuri strains were unexpectedly cultured from healthy persons and patients from Indonesia during a population-based survey on nasal Staphylococcus aureus carriage. Fifty-one S. sciuri isolates were further characterized. The S. aureus mecA gene was detected by PCR in 22 isolates (43.1%), whereas S. sciuri mecA was found in 33 isolates (64.7%). The staphylococcal cassette chromosome mec (SCCmec) regions of S. aureus mecA-positive isolates contained elements of classical S. aureus SCCmec types II and/or III.
Staphylococcus sciuri is an oxidase-positive, novobiocin-resistant Staphylococcus species that is associated mainly with animals (1, 5, 24, 26). Infection and colonization of humans with S. sciuri have been described as rare phenomena (2, 3, 9, 11, 28, 29, 32, 33, 36).
The bacterium has recently gained interest after it was discovered that S. sciuri strains ubiquitously carry a genetic element (S. sciuri mecA) that is closely related to the mecA gene found in methicillin-resistant Staphylococcus aureus (MRSA) strains (8, 35). This finding led to the proposal that S. sciuri mecA might be the evolutionary origin of the mecA element carried by MRSA. In S. sciuri, however, the mecA gene exists as a silent gene of unknown function, since it does not confer resistance to methicillin. Some S. sciuri strains also carry a second copy of the gene, identical to S. aureus mecA. Only isolates with both mecA genes are phenotypically methicillin resistant (9).
During a population-based survey on nasal S. aureus carriage among 3,995 individuals on the island of Java, Indonesia, we unexpectedly cultured S. sciuri from both healthy persons and patients (17, 18). In this work, we characterized these isolates, with a focus on their susceptibility to methicillin.
The survey was carried out by culturing nasal swabs on phenol red mannitol agar (PHMA; Becton Dickinson, Heidelberg, Germany), on which S. aureus produces yellow colonies due to its ability to ferment mannitol. Mannitol-fermenting bacteria were identified to the species level with the Slidex Staph Plus agglutination test (SSP) (bioMérieux, Marcy l'Etoile, France) and the Vitek 2 system (bioMérieux). During the first phase of the study, both SSP-negative and -positive isolates were identified to the species level using the Vitek 2 system. Later, this was performed only with SSP-positive isolates. Additional phenotypic tests for the identification of S. sciuri isolates included an oxidase test (BBL DrySlide oxidase; Becton Dickinson) and a novobiocin susceptibility test (30). For confirmation purposes, sequence analysis of the 16S rRNA gene was carried out with 13 randomly chosen isolates using primers EUB-L (5′-CTTTACGCCCA[AG]T[AG]A[AT]TCCG-3′) and EUB-R (5′-AGAGTTTGATC[AC]TGG[CT]TCAG-3′).
Methicillin susceptibility testing was performed by cefoxitin disk diffusion, according to the CLSI criteria (7). Antimicrobial susceptibility of additional antibiotics was determined using the Vitek 2 system (card AST-P549).
Molecular typing of the isolates was performed by pulsed-field gel electrophoresis (PFGE), as described previously for S. aureus (16, 27). The presence of the S. aureus mecA and S. sciuri mecA genes was determined as described previously (9, 22). The presence of areas homologous to regions of the S. aureus staphylococcal cassette chromosome mec (SCCmec) types I to VI was examined in a randomly chosen subset of S. aureus mecA-positive and S. aureus mecA-negative S. sciuri isolates. The S. aureus mecA-positive isolates were analyzed using the primer sets for detection of loci A to H, as described by Oliveira and de Lencastre (23), and using PCRs for cassette chromosome recombinase (ccr) genes (12-14, 19, 23). The S. aureus mecA-negative S. sciuri isolates were exclusively subjected to the PCR method of Oliveira and de Lencastre (23).
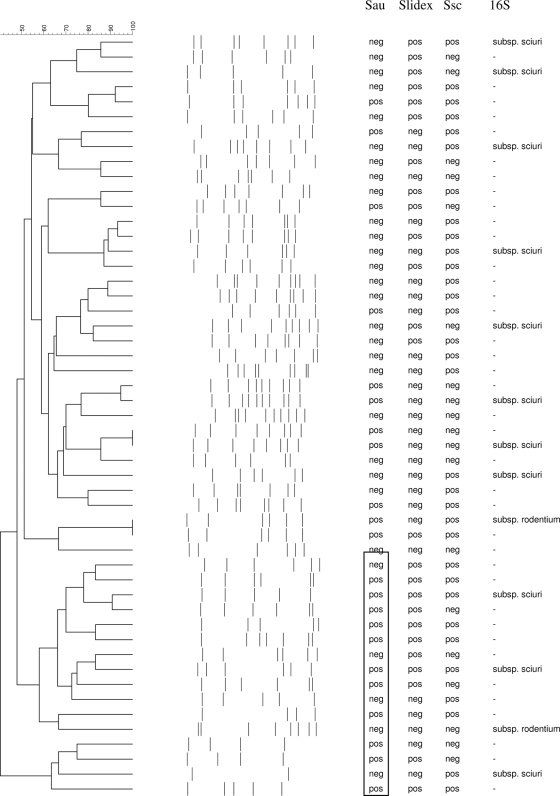
We found 55 mannitol-fermenting bacteria that were identified as S. sciuri bacteria by the Vitek 2 system. Four S. sciuri isolates were lost during storage. Thus, 51 isolates were available for further analyses. All 51 isolates were oxidase positive and resistant to novobiocin. Twenty-five strains (49.0%) gave positive SSP agglutination test results. As a check for the phenotypic identification, the 16S rRNA gene from 13 randomly chosen isolates was sequenced, and all appeared specific for S. sciuri. Eleven isolates belonged to S. sciuri subsp. sciuri, and two belonged to S. sciuri subsp. rodentium. Molecular typing revealed a high degree of genomic diversity among the S. sciuri isolates (Fig. 1).
FIG. 1.
Dendrogram based on PFGE SmaI restriction pattern analysis of 51 nares-colonizing S. sciuri isolates. Similarity analysis was performed with Dice's coefficient, and clustering was done by using the unweighted-pair group method using average linkages (UPGMA) method. The scale at the top shows percentages of similarity. Further information is shown on the right, including the presence (pos) or absence (neg) of the S. aureus mecA gene (Sau), positive (pos) or negative (neg) Slidex Staph Plus agglutination test (Slidex) results, presence (pos) or absence (neg) of the S. sciuri mecA gene (Ssc), and results of 16S rRNA gene sequencing (16S). The rectangle highlights a clustering of S. aureus mecA-positive S. sciuri strains.
Thus, using PHMA for the detection of mannitol-fermenting bacteria as S. aureus, we found a prevalence of nasal carriage of S. sciuri in Java, Indonesia, of at least 51/3,995 (1.3%). We did not identify to the species level all SSP-negative staphylococci found in the survey, and the actual prevalence may, thus, be higher. Both S. aureus and S. sciuri bacteria are mannitol-fermenting bacteria, producing yellow colonies on PHMA. S. sciuri, like S. aureus, may grow as yellow colonies on blood agar and may give positive SSP and Staphaurex test (Remel) results, which may lead to false species identification (11, 28, 36). The differences between the two species are, among others, novobiocin susceptibility and oxidase and coagulase production. Identification of S. aureus by making use of only SSP or the Staphaurex test may result in misclassification of S. sciuri as S. aureus.
The results of PCR for the S. aureus and S. sciuri mecA genes, cefoxitin disk diffusion, and antimicrobial susceptibility using Vitek 2 are shown in Table 1. The S. sciuri mecA gene was found in 33 isolates (64.7%). Although S. sciuri is described to always contain the S. sciuri mecA gene, we were unable to demonstrate the presence of the gene in 18 strains (35.3%). False-negative mecA PCRs were ruled out by including a 16S rRNA PCR as an internal control. This suggests that a genetic event such as deletion of the gene or mutation or deletion at the primer binding site may have occurred in these isolates. The latter possibility seems more likely, since sequence diversity in the mecA homologue of S. sciuri has been documented previously (26). On the other hand, the loss of the putative native mecA gene has been reported previously in a strain isolated from a rodent (34). Our findings are supported by the study of Marsou et al., in which they were also unable to detect S. sciuri mecA by PCR in 10 out of 30 isolates (20). The S. aureus mecA gene was carried by a high percentage of isolates in our collection (43.1%). Similar results were found among isolates obtained from healthy Portuguese carriers (47.8%) and strains obtained from a hospital environment in Serbia (38.1%) (10). Among clinical human isolates and animal isolates, the prevalence was lower, 28.6% and 26.5%, respectively (31). In general, S. sciuri strains with both mecA genes are oxacillin resistant, and strains without the S. aureus mecA gene are oxacillin susceptible (9). This was largely corroborated in the present study. We found three S. aureus mecA-positive strains with cefoxitin disk diffusion zones of ≥25 mm. In these strains, the S. aureus mecA gene is probably not expressed at significant levels. Such strains have been identified previously (15).
TABLE 1.
Correlation between the presence of the S. aureus mecA and S. sciuri mecA genes, cefoxitin disk diffusion susceptibility, and antimicrobial susceptibility of 51 isolates of S. sciuri
| PCR result |
No. of isolates with indicated cefoxitin zone diam (mm) |
% resistant toa: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. aureus mecA | S. sciuri mecA | ≤24 | ≥25 | ERY | FUS | GEN | NOR | RIF | SXT | TET | VAN |
| Negative | Negative | 0 | 9b | 0 | 55.6 | 0 | 0 | 0 | 0 | 11.1 | 0 |
| Negative | Positive | 0 | 20 | 0 | 60.0 | 0 | 0 | 0 | 0 | 5.0 | 0 |
| Positive | Negative | 7 | 2b | 33.3 | 44.4 | 55.5 | 66.7 | 11.1 | 22.2 | 77.8 | 0 |
| Positive | Positive | 12 | 1 | 23.1 | 69.2 | 38.5 | 15.4 | 15.4 | 7.7 | 46.2 | 0 |
| Total | 19 | 32 | 11.8 | 58.8 | 19.6 | 15.7 | 5.9 | 5.9 | 29.4 | 0 | |
Rates of resistance include resistant as well as intermediate-susceptible isolates, as tested by Vitek 2. Abbreviations: ERY, erythromycin; FUS, fusidic acid; GEN, gentamicin; NOR, norfloxacin; RIF, rifampin; SXT, trimethoprim-sulfamethoxazole; TET, tetracycline; VAN, vancomycin.
These included two S. sciuri subsp. sciuri isolates.
Antimicrobial susceptibility to other antibiotics is shown in Table 1. Overall, the resistance rates of S. aureus mecA-positive strains were higher than those of S. aureus mecA-negative strains.
Coagulase-negative staphylococci (CoNS) are believed to constitute a reservoir of resistance genes and SCCmec elements for S. aureus. Therefore, we analyzed the SCCmec regions of 18 S. aureus mecA-positive S. sciuri isolates and 10 S. aureus mecA-negative isolates. None of the SCCmec regions of the 18 S. aureus mecA-positive S. sciuri isolates were typeable by the current classification scheme, but most of the strains contained elements of classical SCCmec types II (loci C, D, and G) and/or III (ccr3) (Table 2). Although we did not search for all elements of SCCmec in our isolates, we speculate that SCCmec in S. sciuri is composed of elements of different classical SCCmec types, giving rise to mosaic-like structures. This is in agreement with previous reports and has also been demonstrated for S. epidermidis (15, 21). Furthermore, MRSA strains with variable elements in SCCmec, possibly originating from CoNS, have been described (25). Loci A to H could not be detected in our S. aureus mecA-negative strains, which is in contrast to the report by Juuti et al., in which they describe the presence of loci G and H in 4 out of 7 S. sciuri strains (15). When comparing data on SCCmec regions of MRSA strains from Indonesia to those of S. sciuri strains, differences and similarities can be noted. Sixty MRSA strains from Indonesia (University of Indonesia, Jakarta, Indonesia) that were analyzed thoroughly by Chongtrakool et al. carried SCCmec type IIIA, including the ccrC locus (6). Although parts of the type III cassette were found in our study, ccrC was absent in all S. aureus mecA-positive strains. The two MRSA strains that were found concurrently in our survey were classified as carrying types III and V by the method of Boye et al. (4). The type III strain was positive for the ccrC target, and the type V strain for was positive for both the ccrC and mecA-IS431 targets. Thus, our data do not support the hypothesis of direct genetic exchange of SCCmec elements between MRSA and S. sciuri in this specific setting.
TABLE 2.
SCCmec typing of 18 S. aureus mecA-positive S. sciuri strains
| Profile | No. of isolates | No. of isolates with fox zone diam of ≤24 mma | Presence/absence of: |
No. of isolates with positive S. sciuri mecA PCR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Loci described by Oliveira and de Lencastreb |
ccr loci |
|||||||||||||||
| A | B | C | D | E | F | G | H | ccr1 | ccr2 | ccr3 | ccr4 | ccrC | ||||
| a | 6 | 3 | − | − | + | + | − | − | + | − | − | − | + | − | − | 3 |
| b | 4 | 4 | − | − | + | − | − | − | − | − | − | − | − | − | − | 2 |
| c | 3 | 3 | − | − | − | − | − | − | − | − | − | − | − | − | − | 1 |
| d | 2 | 2 | − | − | + | − | − | − | − | − | − | − | + | − | − | 2 |
| e | 1 | 1 | − | − | − | + | − | − | − | − | − | − | − | − | − | 1 |
| f | 1 | 1 | − | − | − | + | − | − | − | − | − | − | + | − | − | 0 |
| g | 1 | 1 | − | − | − | − | − | − | − | − | − | − | + | − | − | 1 |
fox, cefoxitin.
Loci described by Oliveira and de Lencastre (31) are as follows: A, downstream of pls gene; B, kdp operon; C, mecI gene; D, dcs gene; E, region between pI258 and Tn554; F, region between Tn554 and orfX; G, left junction between IS431 and pUB110; and H, left junction between IS431 and pT181.
In summary, S. sciuri is a colonizer of the nares of people in Indonesia. This bacterial species may be misidentified as S. aureus. Further research is needed to investigate the clinical significance of this Staphylococcus species in Indonesia. S. sciuri may serve as a reservoir of the S. aureus mecA gene for S. aureus. This potential interaction with S. aureus should be investigated as well.
Acknowledgments
We thank the directors of the Dr. Soetomo Academic Hospital, Surabaya, Indonesia, and the directors of the Dr. Kariadi Academic Hospital, Semarang, Indonesia, who have facilitated our work in these hospitals. We also thank all staff members who have been involved in the isolation of bacteria. We gratefully acknowledge the contribution of medical students Diana Huis in ′t Veld, Suzanne Werter, Rianne de Jong, and Rozemarijn van der Meulen from the Radboud University Medical Center, Nijmegen, Netherlands, who helped us collect the specimens in Indonesia. Furthermore, we express our gratitude to Willemien Zandijk and Neeltje Carpaij for excellent technical assistance.
This work was facilitated by grant 99-MED-03 from the Royal Netherlands Academy of Arts and Sciences in the framework of the Scientific Program Indonesia-Netherlands (SPIN), Amsterdam, Netherlands.
Members of the Antimicrobial Resistance in Indonesia, Prevalence, and Prevention (AMRIN) Study Group are as follows. Members at the Dr. Soetomo Academic Hospital—School of Medicine, Airlangga University, Surabaya, Indonesia, include Widjoseno Gardjito, Erni P. Kolopaking, Karjadi Wirjoatmodjo, Djoko Roeshadi, Eddy Suwandojo, Eddy Rahardjo, Ismoedijanto, Paul Tahalele, Hendromartono, Hari Parathon, Usman Hadi, Nun Zairina, Mariyatul Qibtiyah, Endang Isbandiati, Kartuti Deborah, K. Kuntaman, Ni Made Mertaniasih, Marijam Purwanta, Lindawati Alimsardjono, and Maria Inge Lusida. Members at the Dr. Kariadi Academic Hospital—School of Medicine, Diponegoro University, Semarang, Indonesia, include Ariawan Soejoenoes, Budi Riyanto, Hendro Wahjono, Musrichan Adhisaputro, Winarto, Subakir, Bambang Isbandrio, Bambang Triwara, Johnny Syoeib, Endang Sri Lestari, Bambang Wibowo, Muchlis A. U. Sofro, Helmia Farida, M. M. D. E. A. H. Hapsari, and Tri Laksana Nugraha. Members at the Leiden University Medical Center, Leiden, Netherlands, include Peterhans van den Broek and D. Offra Duerink. Members at the Erasmus MC, University Medical Center, Rotterdam, Netherlands, include Henri A. Verbrugh and Inge C. Gyssens. The member at the Radboud University Medical Center, Nijmegen, Netherlands, is Monique Keuter.
Footnotes
Published ahead of print on 13 September 2010.
REFERENCES
- 1.Adegoke, G. O. 1986. Comparative characteristics of Staphylococcus sciuri, Staphylococcus lentus and Staphylococcus gallinarum isolated from healthy and sick hosts. Vet. Microbiol. 11:185-189. [DOI] [PubMed] [Google Scholar]
- 2.Aires De Sousa, M., I. Santos Sanches, M. L. Ferro, and H. de Lencastre. 2000. Epidemiological study of staphylococcal colonization and cross-infection in two West African Hospitals. Microb. Drug Resist. 6:133-141. [DOI] [PubMed] [Google Scholar]
- 3.Benz, M. S., I. U. Scott, H. W. Flynn, Jr., N. Unonius, and D. Miller. 2004. Endophthalmitis isolates and antibiotic sensitivities: a 6-year review of culture-proven cases. Am. J. Ophthalmol. 137:38-42. [DOI] [PubMed] [Google Scholar]
- 4.Boye, K., M. D. Bartels, I. S. Andersen, J. A. Moller, and H. Westh. 2007. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I-V. Clin. Microbiol. Infect. 13:725-727. [DOI] [PubMed] [Google Scholar]
- 5.Chen, S., Y. Wang, F. Chen, H. Yang, M. Gan, and S. J. Zheng. 2007. A highly pathogenic strain of Staphylococcus sciuri caused fatal exudative epidermitis in piglets. PLoS One 2:e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chongtrakool, P., T. Ito, X. X. Ma, Y. Kondo, S. Trakulsomboon, C. Tiensasitorn, M. Jamklang, T. Chavalit, J. H. Song, and K. Hiramatsu. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 50:1001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute (CLSI). 2006. Performance standards for antimicrobial susceptibility testing: 16th informational supplement. M100-S16. CLSI, Wayne, PA.
- 8.Couto, I., H. de Lencastre, E. Severina, W. Kloos, J. A. Webster, R. J. Hubner, I. S. Sanches, and A. Tomasz. 1996. Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb. Drug Resist. 2:377-391. [DOI] [PubMed] [Google Scholar]
- 9.Couto, I., I. S. Sanches, R. Sá-Leão, and H. de Lencastre. 2000. Molecular characterization of Staphylococcus sciuri strains isolated from humans. J. Clin. Microbiol. 38:1136-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dakić, I., D. Morrison, D. Vuković, B. Savić, A. Shittu, P. Ježek, T. Hauschild, and S. Stepanović. 2005. Isolation and molecular characterization of Staphylococcus sciuri in the hospital environment. J. Clin. Microbiol. 43:2782-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedin, G., and M. Widerström. 1998. Endocarditis due to Staphylococcus sciuri. Eur. J. Clin. Microbiol. Infect. Dis. 17:673-675. [DOI] [PubMed] [Google Scholar]
- 12.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jansen, M. D., A. T. Box, and A. C. Fluit. 2009. SCCmec typing in methicillin-resistant Staphylococcus aureus strains of animal origin. Emerg. Infect. Dis. 15:136-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juuti, K., S. Ibrahem, A. Virolainen-Julkunen, J. Vuopio-Varkila, and P. Kuusela. 2005. The pls gene found in methicillin-resistant Staphylococcus aureus strains is common in clinical isolates of Staphylococcus sciuri. J. Clin. Microbiol. 43:1415-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koning, S., A. van Belkum, S. Snijders, W. van Leeuwen, H. Verbrugh, J. Nouwen, M. Op 't Veld, L. W. van Suijlekom-Smit, J. C. van der Wouden, and C. Verduin. 2003. Severity of nonbullous Staphylococcus aureus impetigo in children is associated with strains harboring genetic markers for exfoliative toxin B, Panton-Valentine leukocidin, and the multidrug resistance plasmid pSK41. J. Clin. Microbiol. 41:3017-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lestari, E. S., D. O. Duerink, U. Hadi, J. A. Severin, N. J. Nagelkerke, K. Kuntaman, H. Wahjono, W. Gardjito, A. Soejoenoes, P. J. van den Broek, M. Keuter, I. C. Gyssens, and H. A. Verbrugh. 27 July 2010, posting date. Determinants of carriage of resistant Staphylococcus aureus among S. aureus carriers in the Indonesian population inside and outside hospitals. Trop. Med. Int. Health doi: 10.1111/j.1365-3156.2010.02600.x. [DOI] [PubMed]
- 18.Lestari, E. S., J. A. Severin, P. M. Filius, K. Kuntaman, D. O. Duerink, U. Hadi, H. Wahjono, and H. A. Verbrugh. 2008. Antimicrobial resistance among commensal isolates of Escherichia coli and Staphylococcus aureus in the Indonesian population inside and outside hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 27:45-51. [DOI] [PubMed] [Google Scholar]
- 19.Lim, T. T., F. N. Chong, F. G. O'Brien, and W. B. Grubb. 2003. Are all community methicillin-resistant Staphylococcus aureus related? A comparison of their mec regions. Pathology 35:336-343. [PubMed] [Google Scholar]
- 20.Marsou, R., M. Bes, M. Boudouma, Y. Brun, H. Meugnier, J. Freney, F. Vandenesch, and J. Etienne. 1999. Distribution of Staphylococcus sciuri subspecies among human clinical specimens, and profile of antibiotic resistance. Res. Microbiol. 150:531-541. [DOI] [PubMed] [Google Scholar]
- 21.Miragaia, M., J. C. Thomas, I. Couto, M. C. Enright, and H. de Lencastre. 2007. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 189:2540-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami, K., W. Minamide, K. Wada, E. Nakamura, H. Teraoka, and S. Watanabe. 1991. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 29:2240-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poutrel, B. 1984. Staphylococcus sciuri subsp lentus associated with goat mastitis. Am. J. Vet. Res. 45:2084-2085. [PubMed] [Google Scholar]
- 25.Qi, W., M. Ender, F. O'Brien, A. Imhof, C. Ruef, N. McCallum, and B. Berger-Bachi. 2005. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Zürich, Switzerland (2003): prevalence of type IV SCCmec and a new SCCmec element associated with isolates from intravenous drug users. J. Clin. Microbiol. 43:5164-5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahman, M. T., N. Kobayashi, M. M. Alam, and M. Ishino. 2005. Genetic analysis of mecA homologues in Staphylococcus sciuri strains derived from mastitis in dairy cattle. Microb. Drug Resist. 11:205-214. [DOI] [PubMed] [Google Scholar]
- 27.Severin, J. A., E. S. Lestari, K. Kuntaman, D. C. Melles, M. Pastink, J. K. Peeters, S. V. Snijders, U. Hadi, D. O. Duerink, A. van Belkum, and H. A. Verbrugh. 2008. Unusually high prevalence of Panton-Valentine leukocidin genes among methicillin-sensitive Staphylococcus aureus strains carried in the Indonesian population. J. Clin. Microbiol. 46:1989-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shittu, A., J. Lin, D. Morrison, and D. Kolawole. 2004. Isolation and molecular characterization of multiresistant Staphylococcus sciuri and Staphylococcus haemolyticus associated with skin and soft-tissue infections. J. Med. Microbiol. 53:51-55. [DOI] [PubMed] [Google Scholar]
- 29.Stepanović, S., I. Dakić, S. Djukić, B. Lozuk, and M. Svabić-Vlahović. 2002. Surgical wound infection associated with Staphylococcus sciuri. Scand. J. Infect. Dis. 34:685-686. [DOI] [PubMed] [Google Scholar]
- 30.Stepanović, S., I. Dakić, D. Morrison, T. Hauschild, P. Ježek, P. Petráš, A. Martel, D. Vuković, A. Shittu, and L. A. Devriese. 2005. Identification and characterization of clinical isolates of members of the Staphylococcus sciuri group. J. Clin. Microbiol. 43:956-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stepanović, S., T. Hauschild, I. Dakić, Z. Al-Doori, M. Švabić-Vlahović, L. Ranin, and D. Morrison. 2006. Evaluation of phenotypic and molecular methods for detection of oxacillin resistance in members of the Staphylococcus sciuri group. J. Clin. Microbiol. 44:934-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stepanović, S., P. Ježek, D. Vuković, I. Dakić, and P. Petráš. 2003. Isolation of members of the Staphylococcus sciuri group from urine and their relationship to urinary tract infections. J. Clin. Microbiol. 41:5262-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallet, F., L. Stuit, E. Boulanger, M. Roussel-Delvallez, P. Dequiedt, and R. J. Courcol. 2000. Peritonitis due to Staphylococcus sciuri in a patient on continuous ambulatory peritoneal dialysis. Scand. J. Infect. Dis. 32:697-698. [DOI] [PubMed] [Google Scholar]
- 34.Wu, S., H. de Lencastre, and A. Tomasz. 1998. Genetic organization of the mecA region in methicillin-susceptible and methicillin-resistant strains of Staphylococcus sciuri. J. Bacteriol. 180:236-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, S., C. Piscitelli, H. de Lencastre, and A. Tomasz. 1996. Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri. Microb. Drug Resist. 2:435-441. [DOI] [PubMed] [Google Scholar]
- 36.Wulf, M., A. van Nes, A. Eikelenboom-Boskamp, J. de Vries, W. Melchers, C. Klaassen, and A. Voss. 2006. Methicillin-resistant Staphylococcus aureus in veterinary doctors and students, The Netherlands. Emerg. Infect. Dis. 12:1939-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]