Abstract
HIV replication assays are important tools for HIV drug discovery efforts. Here, we present a full HIV replication system (EASY-HIT) for the identification and analysis of HIV inhibitors. This technology is based on adherently growing HIV-susceptible cells, with a stable fluorescent reporter gene activated by HIV Tat and Rev. A fluorescence-based assay was designed that measures HIV infection by two parameters relating to the early and the late phases of HIV replication, respectively. Validation of the assay with a panel of nine reference inhibitors yielded effective inhibitory concentrations consistent with published data and allowed discrimination between inhibitors of early and late phases of HIV replication. Finer resolution of the effects of reference drugs on different steps of HIV replication was achieved in secondary time-of-addition assays. The EASY-HIT assay yielded high Z′ scores (>0.9) and signal stabilities, confirming its robustness. Screening of the LOPAC1280 library identified 10 compounds (0.8%), of which eight were known to inhibit HIV, validating the suitability of this assay for screening applications. Studies evaluating anti-HIV activities of natural products with the EASY-HIT technology led to the identification of three novel inhibitory compounds that apparently act at different steps of HIV-1 replication. Furthermore, we demonstrate successful evaluation of plant extracts for HIV-inhibitory activities, suggesting application of this technology for the surveillance of biological extracts with anti-HIV activities. We conclude that the EASY-HIT technology is a versatile tool for the discovery and characterization of HIV inhibitors.
The implementation of highly active antiretroviral therapy (HAART) considerably reduces virus propagation in HIV-infected individuals and substantially extends their life expectancies (50). However, HAART still has various shortcomings, including the emergence of resistant virus strains, severe side effects of drugs, suboptimal penetration of the central nervous system, failure to completely suppress viremia, and the requirement for life-long treatment (1, 31, 35, 46). In addition, HAART is very cost intensive and thus not affordable for the majority of HIV-infected individuals in developing countries (46, 52). Achieving widespread antiretroviral treatment is a major goal for the prevention of virus-induced diseases in infected individuals and also has the potential to reduce HIV transmission (52). The latter aspect is particularly important in light of the absence of effective prevention strategies like protective HIV vaccines (5). Altogether, there is an urgent need for the continuous development of novel anti-HIV drugs and their implementation into affordable and efficient therapies (12, 21).
Different strategies have been used for the discovery and the development of anti-HIV drugs (56). One of these is the rational design of molecules that interfere with the activity of a preselected target and their testing in biochemical assays. In the past, this approach has been successful in the development of inhibitors of the HIV-1 protease (13), of which at least 10 have been approved as anti-HIV drugs (http://www.fda.gov/). However, development of new anti-HIV drugs by rational design is limited to targets for which detailed structural data exist and whose functions are amenable to biochemical testing. This is the case for only a few HIV proteins, leaving many other promising but less well-studied anti-HIV targets inaccessible to this approach (21). A further limitation of target-driven screening with biochemical assays is the requirement for highly pure, preferably single, molecules for testing, precluding the rapid exploratory appraisal of extract mixtures (47). Another strategy that has led to the successful development of anti-HIV drugs in the past is the testing of inhibitory effects in cell-based assays that evaluate the effects of compounds on replication of HIV in cultured cells (13, 43). This approach has the potential to identify inhibitors against multiple viral and cellular functions essential for HIV replication and to detect novel inhibitory mechanisms. In the past, an HIV replication assay that monitored HIV-induced cytopathic effects in human T-lymphoma cells was used for the discovery of the first nonnucleoside reverse transcriptase inhibitors (43). Since then, various other HIV replication assays have been developed, particularly assays using infection-dependent reporters (reviewed in reference 56). Reporters are either expressed as part of the virus (i.e., reporter viruses) or from reporter genes activated during infection of HIV-susceptible cells. Reporter proteins expressed by HIV indicator cell lines include various enzymes (7, 8, 17, 28) and the green fluorescent protein (GFP) (15, 19, 41, 59). Transcription of these reporter genes is generally directed by the HIV long terminal repeat (LTR) and therefore activated by the HIV-1 Tat protein. More recently, an HIV-reporter gene was developed that is dependent on both Rev and Tat for expression (59).
HIV replication assays have a great potential to expedite the development of novel anti-HIV therapies. Therefore, it is worthwhile to continue the development of these assays, in particular, to broaden their capacity to discover HIV inhibitors with novel modes of action as well as to identify new sources of HIV inhibitors. Many replication assays cover specific aspects of the replication cycle. Thus, single-round infection assays, which are performed with pseudotyped virus particles bearing env-defective viral genomes, have been optimized to detect inhibitors that interfere with various cell-associated events of replication (reviewed in reference 34). However, these assays cannot detect inhibitors of virus envelopment, budding, or virion infectivity. On the other hand, reporter cell assays based on replication-competent viruses and Tat-activated reporter genes can show greater sensitivity for early- than for late-phase inhibitors (7, 11, 41) (see Discussion). It would also be highly desirable to have sturdy HIV replication assays for rapid and reliable testing of crude biological extracts for anti-HIV activities. This would greatly advance the development of HIV therapeutics from natural products, which hold great promise for drug development (30, 42, 60).
Here we present a full-replication exploratory assay system for the discovery of HIV inhibitors (EASY-HIT). We show that the assay identifies and discriminates between inhibitory activities against different phases of HIV replication. We present data demonstrating robust and reliable performance characteristics of this assay and its suitability for screening of compound libraries. Furthermore, we show that this technology can identify anti-HIV activities of natural products, both in the form of isolated compounds and as crude biological extracts. We conclude that EASY-HIT will facilitate the mining of natural and other sources for novel HIV-inhibitors.
MATERIALS AND METHODS
Cell culture.
The HIV indicator LC5-RIC cell line (where RIC is red infected cells, indicating the LC5 parental line expressing the DsRed1 reporter), the HIV1IIIB-producer T-lymphoma cell line KE37.1-IIIB, the uninfected T-lymphoma cell line KE37.1, and HEK 293T cells were cultured under standard conditions at 37°C in 5% CO2 in Dulbecco's modified Eagle medium (DMEM containing GlutaMAX-1; Gibco, Darmstadt, Germany) or very-low-endotoxin (VLE)-RPMI 1640 medium (Biochrom AG, Berlin, Germany) supplemented with 10% fetal bovine serum (Biochrom AG) and 1% antibiotic-antimycotic solution (Gibco). LC5-RIC reporter cells were maintained under selection pressure by addition of 0.74 mg/ml Geneticin (G418 sulfate; PAA Laboratories, Pasching, Austria) and 0.125 mg/ml hygromycin B (PAA Laboratories) to the cell culture medium at every second passage to retain stable expression of the CD4 receptor and to ensure stability of the reporter construct. Selection antibiotics were removed 2 days before the cells were seeded into the assay plates and were generally not used during the assay.
LC5-RIC cells were used for a maximum of 10 passages, and every new batch of cells was initially checked for the expression of CD4 and CXCR4 receptors by flow cytometry (see below) and signal performance by determination of the signal-to-background ratio. Only LC5-RIC batches showing expression of CD4 and CXCR4 on ≥80% of the cells, no background expression of the reporter, and over 100-fold induction of relative signal intensities in HIV infection assays were used for further applications.
Generation of the LC5-RIC HIV indicator cell line.
LC5-CD4 (54) is a HeLa-derived cell clone with a stably integrated expression vector containing genes encoding human CD4 and a protein conferring neomycin resistance (33). To generate LC5-RIC, LC5-CD4 cells were transfected with the reporter construct pLRed(2×INS)R (58) modified to contain the hygromycin B resistance gene, using Fugene HD (Roche, Grenzach-Whylen, Germany) transfection reagent. The reporter gene in pLRed(2×INS)R directs Rev- and Tat-dependent expression of the red fluorescent protein DsRed1. Transfected cells were cultured in standard cell culture medium supplemented with 167 μg/ml hygromycin B (Gibco) for 48 h; single-cell colonies were isolated and expanded, and 10 cell clones were selected for further analysis. Cell clones were monitored for absence of background expression and HIV-dependent induction of reporter expression, resulting in selection of the LC5-RIC clone. Flow cytometry analysis (see below) confirmed cell surface expression of the CD4 receptor and the CXCR4 coreceptor on over 80% of LC5-RIC.
Virus stock preparation.
Infectious HIV-1 stocks were generated from the HIV-1IIIB producer cell line KE37.1-IIIB (54) or by transfection of HEK 293T cells with the infectious molecular clone pLAI.2 (44).
KE37.1-IIIB cells were adjusted to 8 × 105 per milliliter and cultured for 48 h prior to virus collection. Virus-containing culture supernatants were harvested by centrifugation at 2,000 × g and passed through a 0.45-μm-pore-size filter (Sartorius, Goettingen, Germany). Viral RNA was quantified in culture supernatants by HIV reverse transcription PCR ([RT-PCR] RealTime HIV-1; Abbott, Wiesbaden, Germany) according to the manufacturer's instructions. Supernatants harvested from the parental T-lymphoma cell line KE37.1 were used as negative controls.
For the production of HIV-1LAI, HEK 293T cells were seeded into six-well plates at a density of 105 cells per well in 2 ml of culture medium. Twenty-four hours later, cells were transfected with 1 μg of pLAI.2 DNA per well using 3.8 μl of FuGene HD transfection reagent (Roche), following the manufacturer's instructions. Forty-eight hours later 1 ml of DMEM was added to each well to boost the virus production, and after an additional 24 h, the supernatant was harvested, centrifuged for 5 min at 2,000 × g, aliquoted, and frozen at −80°C.
Virus preparations were analyzed for virus levels and absence of cell-deleterious effects using an aliquot of the frozen virus stock thawed at 37°C for 1 h. Virus levels were quantified by p24Gag enzyme-linked immunosorbent assay (ELISA; Applied Biosystems, Carlsbad, CA), according to the manufacturer's instructions, and by titration on LC5-RIC cells. For virus titration, different volumes of the virus stock were added to LC5-RIC cells seeded in a 96-well plate, with five wells assayed for each volume tested. Forty-eight hours later, fluorescent signal intensities were measured, and cells were subsequently analyzed by 3-(4,5-dimethylthiazol-2-yl)2 2,5-diphenyl tetrazolium bromide (MTT) assay. The following criteria were applied to select the volumes of the virus stocks used in assays with LC5-RIC cells: (i) over 100-fold increase of the relative fluorescent signal intensities by HIV infection, (ii) relative signal induction levels below the plateau, and (iii) a reduction of the relative MTT signal by less than 10%. Figure S1 in the supplemental material shows an example in which 5 μl of virus preparation corresponded to ∼130-fold induction of the reporter signal, 28.8 ng of p24, and 90% MTT signal relative to uninfected controls. This stock was selected for use in subsequent assays.
Isolation of primary virus.
To isolate primary virus from clinical material, 50 μl of cerebrospinal fluid (CSF) samples from HIV-infected individuals was directly added to LC5-RIC cells grown in 96-well plates at a density of 104 cells per well in 50 μl of DMEM with GlutaMAX-1 (Gibco) supplemented with 10% fetal calf serum (Biochrom AG). To promote infection, DEAE-dextran was added at a concentration of 20 μg per milliliter. Supernatants were removed after 18 h and replaced by fresh culture medium. Cultures were monitored for infection by microscopic inspection for HIV reporter expression. Cultures with infected cells were harvested after 2 weeks and seeded and expanded in six-well plates. Isolation of novel virus strains was confirmed by sequence analysis of the nef gene. In brief, RNA was extracted from cells with an RNeasy Mini Kit (Qiagen, Hilden, Germany), reverse transcribed using a SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen, Darmstadt, Germany), and PCR amplified using the previously published nef-specific primers Nef-F1 and Nef-R (24) and GoTaq polymerase (Promega, Mannheim, Germany). PCR products were purified by a NucleoSpin Extract II Kit (Macherey and Nagel, Dueren, Germany) and cloned into pCRII-TOPO (Invitrogen), and four clones per isolate were sequenced in-house (ABI Prism 3100; Applied Biosystems). ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html) was used for sequence alignments. Supernatants from infected LC5-RIC cultures were harvested, filtered sterile, and frozen at −80°C.
Standard assay setup for testing of HIV inhibitors.
LC5-RIC cells were seeded into 96-well plates (μCLEAR-Plate Black; Greiner Bio-One, Kremsmuenster, Germany), using only the 60 inner wells to avoid adverse effects caused by variations in the culture conditions in the outer wells. Cells were seeded at a density of 104 per well at 24 h prior to infection. Compounds were tested either at single concentrations or in serial dilutions, with each concentration assayed in triplicate. Compound solution (100 μl) followed by 20 μl of HIV inoculum (108 RNA copies/ml inoculum for KE37.1-derived HIV-1IIIB or 28.8 ng of p24 for HIV-1LAI derived from HEK 293T cells) was added to each well. To assay maximum infection (i.e., 100%), HIV inoculum was added to LC5-RIC cells incubated with compound-free medium. Controls for background expression consisted of LC5-RIC cells cultured in conditioned medium from KE37.1 (i.e., uninfected) cells. Plates were incubated for 48 h after virus addition, after which cultures were assayed for cellular reporter expression and for titers of infectious virus in supernatant fluids. Reporter expression was determined by measuring the total fluorescent signal intensity of each culture with a fluorescence microplate reader (Fluoroskan Ascent; ThermoFisher, Schwerte, Germany) at an excitation filter wavelength of 544 nm and an emission filter wavelength of 590 nm or with a Tecan infinite M200 (Tecan, Crailsheim, Germany) at the monochromator wavelengths 552 nm for excitation and 596 for emission. To quantify infectious virus titers, supernatant fluids (30 μl of HIV-1IIIB or 20 μl for HIV-1LAI) from cultures in the first plate were transferred to a second plate with uninfected LC5-RIC cells in exact replicate, and total fluorescent signal intensities of cultures in the second plate were measured 72 h after transfer.
MTT assay.
Influence of compounds on the viability and activity of LC5-RIC cells exposed to HIV was determined by a colorimetric assay that measures the reduction of the yellow MTT to purple formazan by mitochondrial enzymes (38). Immediately following measurement of reporter expression, cultures were incubated with 100 μl of MTT solution (0.5 mg of MTT; Sigma, Taufkirchen, Germany) in 100 μl of culture medium for 2 h under standard culture conditions. MTT solution was carefully removed, and cells were lysed by addition of 100 μl of lysis solution (10% [wt/vol] SDS and 0.6% [vol/vol] acetic acid in dimethyl sulfoxide [DMSO]). The released MTT formazan crystals were dissolved by gentle agitation, and MTT formazan concentrations were determined by an ELISA plate reader (SmartSpec Plus; Bio-Rad, Muenchen, Germany) or Tecan Infinite M200 at a test wavelength of 570 nm and a reference wavelength of 630 nm. Values for treated HIV-infected cultures were related to those of untreated, HIV-infected cultures in the same plate.
Validation of the EASY-HIT system according to the guidelines of the NIH Chemical Genomics Center.
For the estimation of quality, robustness, and reproducibility of our assay, the parameters Z′, coefficient of variation (CV), signal window (SW), and signal-to-background ratio (S/B) were determined according to established guidelines (16).These parameters were assayed in three experiments performed on three different days. For each experiment, three 96-well plates were infected with HIV-1LAI using 5 μl of virus stock solution in the standard assay setup. Each 96-well plate was loaded as described in the National Institutes of Health Chemical Genomics Center (NCGC) guidelines using a plate layout (i.e., interleaved signal format) that has a combination of wells producing different signal levels. Wells for analysis of maximum signal contained virus only, wells for the middle signal contained virus and 100 nM azidothymidine (AZT), and wells for the minimum signal contained no virus. Overall, 60 wells were analyzed per signal and experiment. A corresponding calculation for the LOPAC1280 screen was performed using the data for positive (virus, DMSO, and no compound) and negative (no virus, DMSO, and no compound) controls from each plate (n = 48/step) as maximum and minimum signals, respectively (364 wells/signal). The following formulas were used for the calculation of Z′ and SW:
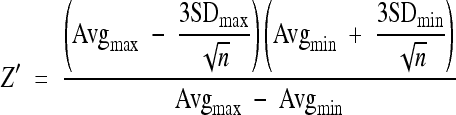 |
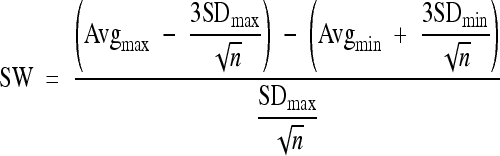 |
where SD is standard deviation, Avg is the mean signal, max and min are maximum and minimum signals, respectively, and n is the number of replicates. In addition, the following two formulas were used: CV = (SD/√n)/Avg and S/B = Avgmax/Avgmin.
Drug susceptibility assays.
To determine 50% effective concentrations (EC50) of established anti-HIV compounds for inhibition of HIV infection of LC5-RIC cells, serial dilutions of griffithsin (GRFT), enfuvirtide (T20), AZT, efavirenz (EFV), 118-D24, raltegravir (RAL), flavopiridol (FLV), saquinavir (SQV), and darunavir (DRV) in cell culture medium were tested in a standard assay setup. All test compounds were provided by the NIH AIDS Research and Reference Reagent Program (www.aidsreagent.org). Infections were carried out using HIV-1IIIB and a primary isolate of HIV.
Screening of the LOPAC1280 compound library.
The screen of the LOPAC1280 (Sigma) compound library was performed with 5 μl of HIV-1LAI stock solution. All compounds were dissolved in DMSO by the manufacturer and further diluted with standard DMEM to yield a final test concentration of 10 μM and a DMSO concentration of less than 0.2% in the assay. All compounds were tested in triplicate plates. Liquid handling was performed with a manual benchtop pipetting system (Liquidator 96; Steinbrenner Laborsysteme GmbH, Wiesenbach, Germany).
Preliminary screening of a natural product compound collection.
A preliminary screening of a collection of 400 natural compounds (Specs, Delft, Netherlands) was accomplished with HIV-1IIIB. All compounds were dissolved in DMSO and further diluted with standard DMEM to yield a final test concentration of 10 μM and a DMSO concentration of less than 0.2% in the assay. All test compounds were tested in triplicate plates.
TOA.
Time-of-addition (TOA) assays were performed in 96-well plates using a standard assay setup with some modifications. HIV-1IIIB virus preparations were added to LC5-RIC cultures at time point 0. Anti-HIV compounds were added to the cultures at different time points after addition of virus preparations. Inhibitor concentrations were ≥2× EC50 in each well. At least 15 different time points were evaluated for each compound in at least three wells per time point. Cultures were incubated with virus for 48 h and subsequently analyzed for cellular reporter gene expression and/or infectious virus titers in culture supernatants.
Flow cytometry and fluorescence microscopy.
For Gag p24 staining, 1 × 106 LC5-RIC cells were fixed, washed, and permeabilized as previously described (49). Immunostaining with a fluorescein isothiocyanate (FITC)-labeled p24-specific antibody, KC57-FITC (Becton Dickinson, Heidelberg, Germany), was carried out for 2 h in the dark, followed by three washing steps with phosphate-buffered saline (PBS) and resuspension of cells in PBS for further analysis. Immunostaining of LC5-RIC cells for CD4 and chemokine receptor CXCR4 was carried out as described by Rothenaigner et al. (49). Allophycocyanin (APC)-conjugated CD4- and CXCR4-specific antibodies (BD Pharmingen) were used to detect surface receptors CD4 and CXCR4.
Cells were analyzed by flow cytometry (FACSCalibur; Beckton Dickinson) using channels FL1 (FITC), FL3 (DsRed1), or FL4 (APC).
Fluorescence microscopic imaging was done by use of a Olympus IX81 research microscope.
Calculation of values and curve fits.
Curve fits and EC50 calculations were done with Prism, version 4 (GraphPad Software, La Jolla, CA), using the equation for sigmoidal dose response with variable slope with the following set constraint parameters: fixed top of 100 and fixed bottom of 0. As internal acceptance criteria, R2 values had to be ≥0.9.
RESULTS
Monitoring HIV infection with LC5-RIC indicator cells.
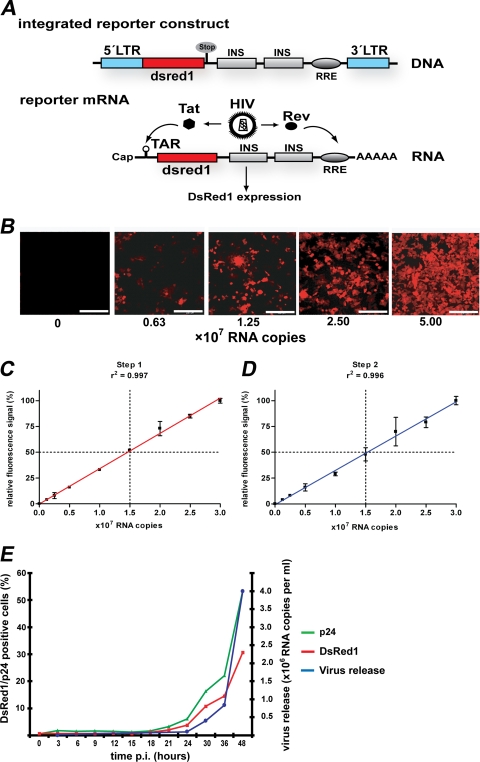
The HIV indicator LC5-RIC (where RIC is red infected cells) cell line is derived from HeLa cells transduced with a gene for stable expression of the CD4 cell surface molecule (54). In addition, LC5-RIC cells contain a reporter gene for expression of the red fluorescent protein DsRed1, which is activated by HIV-1 Tat and Rev (Fig. 1A).
FIG. 1.
Monitoring HIV infection with the HIV reporter cell line LC5-RIC. (A) Tat- and Rev-dependent HIV reporter gene in LC5-RIC cells. The integrated reporter gene contains a 5′ HIV LTR (blue) for Tat-inducible transcription, sequences encoding the red fluorescent DsRed1 reporter protein (red), and HIV-1 derived sequences for Rev-dependent expression (gray). The last consist of inhibitory sequences (INS) from the HIV-1 gag gene (p17/p24 region) and the Rev-response element (RRE) from the HIV-1 env gene (57). HIV infection of cells causes production of Tat and Rev, which is initiated in the early phase of HIV replication. Binding of Tat and Rev to reporter mRNAs at the trans-activating response region (TAR) and RRE, respectively, induces expression of the DsRed1 reporter protein. (B) HIV-dependent appearance of red-fluorescent cells in LC5-RIC cultures. LC5-RIC cultures in six-well plates were exposed to various amounts of HIV (viral RNA copy numbers indicated), and cultures were inspected by fluorescence microscopy 48 h after virus addition. Bar, 200 μm. (C) Linear correlation between the fluorescent signal intensities of infected LC5-RIC cultures and the amounts of virus (i.e., viral RNA copy numbers) used for infection (i.e., primary infection). Fluorescent signal intensities were measured at 48 h after virus addition by microplate fluorimetry. Values were normalized to the value obtained with the largest amount of virus (set at 100%). Each data point represents the mean value of six wells ± standard deviations. (D) Quantification of infectious virus titers released by LC5-RIC cultures. Aliquots of supernatants of the infected primary cultures from panel C were transferred to secondary LC5-RIC cultures, and fluorescent signal intensities of secondary cultures were measured 72 h after transfer. Values were normalized to the value obtained with the highest viral load (set at 100%). Each data point represents the mean value ± standard deviations. (E) Temporal correlation of red fluorescent signal intensities and virus production by LC5-RIC cultures in six-well plates exposed to a viral load of 5 × 107 viral RNA copies. The proportions of cells positive for DsRed1 or HIV Gag-p24 were analyzed at various time points after virus addition (p.i.) by flow cytometry. The amounts of viral RNAs in culture supernatants were determined by standardized quantitative RT-PCR. Each data point represents the mean of three wells.
To analyze the relationship between reporter expression and HIV infection, DsRed1 expression was monitored in LC5-RIC cultures exposed to different amounts of HIV-1IIIB. Microscopic analysis of LC5-RIC cultures (Fig. 1B) confirmed the absence of background expression of DsRed1 in uninfected cultures. In HIV-exposed cultures, the proportion of red fluorescent cells increased with increasing virus titers. This was confirmed by flow cytometry (data not shown).
Next, we measured the total fluorescence signal intensity of each culture by microplate fluorimetry. As demonstrated in Fig. 1C, a direct linear correlation existed between the fluorescent signal intensity of the cultured cells and the amount of virus (determined by HIV RNA copy number) used for infection. Further analysis of the effect of different amounts of input virus on the fluorescent signal intensities showed that the fluorescent signal intensity reached a plateau with increasing virus input (see Fig. S1 in the supplemental material).
We also quantified the amounts of infectious virus released by HIV-exposed cultures. To this end, aliquots of supernatant fluids from primary infected cultures were transferred to secondary uninfected LC5-RIC cultures, and fluorescence signal intensities of the secondary cultures were measured after 72 h of incubation. As shown in Fig. 1D, a linear correlation existed between the fluorescence signal intensities of the secondary cultures and the amounts of virus used for primary infection. This confirms productive infection of primary HIV-exposed LC5-RIC cells and shows that the amount of infectious virus released by the HIV-exposed LC5-RIC cells is directly proportional to the amount of virus used for primary infection.
To further validate DsRed1 reporter expression as a reliable marker for virus replication, we compared the temporal appearance of fluorescence signal intensities with quantitative HIV production parameters, i.e., percentages of Gag-positive cells in the culture and HIV RNA copy numbers in culture supernatants. As shown in Fig. 1E there was a direct and positive time-resolved correlation of reporter expression, formation of the virus antigen p24, and release of viral progeny.
To determine whether monitoring of HIV infection with LC5-RIC cells can be adapted to screening for HIV inhibitors, we assessed criteria for robustness and reproducibility of the reporter signal according to established guidelines (16). To assess reproducibility, these criteria were evaluated in experiments with multiple plates (n = 3) in multiple wells per plate and with each experiment performed on a different day (n = 3). Signal variability was tested for cultures infected with HIV (maximum signal), cultures lacking HIV (minimum signal), and cultures treated with 100 nM of the reverse transcriptase inhibitor AZT (mid signal; here, ∼37% relative fluorescent signal intensity). Table 1 summarizes the results of these validation experiments. The coefficient of variation (CV) is a statistical measure for the average deviation of signal intensities. In all validation experiments, the CV values calculated for all three signal types remained below 2%, which is 10 times lower than the maximum acceptance criterion of 20%. The Z′ factor is a statistical value that reflects the dynamic range of the assay and the variation of signal measurements (61) and should be ≥0.4 with a maximum value of 1. The Z′ factors determined for infection assays with LC5-RIC were equal to or above 0.97. The signal window (SW) values and the signal-to-background (S/B) ratios indicated a broad dynamic range of reporter signal production by LC5-RIC cells. This resulted from both very low background signals, comparable to those of the parental LC5 cells lacking the reporter gene, and high fluorescent signal intensities of infected cells (see Fig. S2 in the supplemental material). Edge and drift effects within plates were not observed.
TABLE 1.
EASY-HIT assay characteristics according to NIH NCGC guidelines
| Assay parametera | Value for the parameter in: |
NCGC acceptance criterion | |||||
|---|---|---|---|---|---|---|---|
| Validation experimentb |
LOPAC1280 library screeningc |
||||||
| Expt 1 | Expt 2 | Expt 3 | Mean | Step 1 | Step 2 | ||
| CV high (%) | 0.88 ± 0.12 | 1.09 ± 0.30 | 1.12 ± 0.15 | 1.03 ± 0.21 | 2.07 ± 0.59 | 2.06 ± 0.60 | ≤20 |
| CV middle (%) | 1.84 ± 0.30 | 1.68 ± 0.20 | 1.23 ± 0.15 | 1.59 ± 0.34 | ND | ND | |
| CV low (%) | 1.84 ± 0.25 | 1.88 ± 0.25 | 1.61 ± 0.37 | 1.78 ± 0.29 | 11.85 ± 4.01 | 6.00 ± 1.84 | |
| Z′ | 0.97 ± 0.01 | 0.97 ± 0.01 | 0.97 ± 0.00 | 0.97 ± 0.01 | 0.94 ± 0.02 | 0.94 ± 0.02 | 0.4 |
| SWc | 105 ± 7.56 | 86.13 ± 18.61 | 90.83 ± 7.08 | 93.99 ± 13.63 | 49 ± 17 | 50 ± 17 | ≥2 |
| S/Bd (fold) | 95 ± 3 | 62 ± 6 | 57 ± 4 | 71 ± 19 | 157 ± 24 | 330 ± 33 | |
CV, coefficient of variation; SW, signal window; S/B, signal-to-background ratio.
Values indicated for each experiment represent the mean ± SD of values from three plates, with 60 wells analyzed for each signal. Each experiment was performed on a different day.
Forty-eight plates were used for each step. ND, not done.
Together these results show that reporter expression in LC5-RIC cells represents a reliable quantitative parameter for HIV infection.
EASY-HIT, an exploratory assay system for the discovery of HIV inhibitors.
Our next goal was to design and validate an LC5-RIC-based full-replication assay system for the discovery of HIV inhibitors, which we called EASY-HIT.
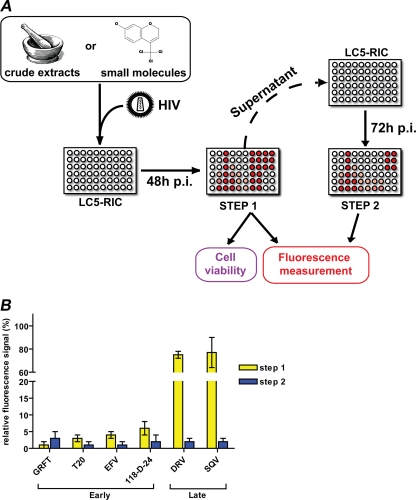
Figure 2A shows a schematic overview of the assay. In a standard assay procedure, candidate inhibitors were added to LC5-RIC cultures in microtiter plates, HIV preparations were added within 30 min of beginning treatment, and test cultures were incubated for 48 h after virus addition. Reference cultures were either exposed to the virus without inhibitor candidates (maximum signal) or not exposed to the virus (minimum signal). Effects of compounds on HIV infection of LC5-RIC cultures were determined in two steps. The first step quantified reporter expression by assaying fluorescent signal intensities of cells in test cultures. The second step quantified the infectious virus produced in test cultures by transferring aliquots of supernatants of test cultures to uninfected LC5-RIC cells, followed by measurement of fluorescent signal intensities at 72 h after transfer. Cells evaluated for reporter expression were subjected to an MTT assay to detect deleterious effects of compounds on cell viability under test conditions.
FIG. 2.
EASY-HIT standard assay for the identification of HIV inhibitors. (A) Schematic of the EASY-HIT assay. HIV indicator cells LC5-RIC are exposed to test substances (e.g., extracts or single compounds) and virus for 48 h, and inhibitory effects of compounds on HIV replication in these test cultures were evaluated in two steps. The first step measures fluorescence signal intensities of the cells in the test cultures. For the second step, aliquots of supernatants of the test cultures are transferred to a new plate with LC5-RIC cells, and fluorescent signals are measured 72 h after transfer. Effects of test substances on the viability of cells in test cultures are analyzed by MTT assay. (B) Effects of reference compounds known to interfere with the early and late phases of HIV replication on fluorescent signal intensities measured in the first and second steps of the EASY-HIT assay. Fluorescent signal intensities measured for test cultures were normalized to fluorescent signal intensities in control cultures (exposed to HIV only). Inhibitors were used at concentrations corresponding to 3× EC50. All inhibitors reduced the fluorescent signals in the second step of the assay. Early-phase inhibitors reduced the fluorescent signals in the first and second steps of the EASY-HIT assay to <10% of the signal of the infected, untreated control. Late-phase inhibitors reduced fluorescent signal intensities predominantly in the second step and showed only small effects in the first step. Mean values of three wells ± standard deviations are indicated.
For validation of the EASY-HIT system, we compiled a panel of nine HIV inhibitors known to interfere with different processes that occur in either the early or late replication phases (reviewed in reference 13; see also references in Table 2). Early-phase inhibitors were selected that interfere with virus entry (griffithsin [GRFT] and T20), reverse transcription (the nucleoside reverse transcriptase inhibitor azidothymdine [AZT] and the nonnucleoside reverse transcriptase inhibitor efavirenz [EFV]), provirus integration (raltegravir [RAL] and 118-D-24), and HIV transcription (flavopiridol [FLV]). Late-phase compounds were selected that prevent production of infectious virus particles by inhibiting protease activity (saquinavir [SQV] and darunavir [DRV]). As shown in Fig. 2B, the two-step format of the EASY-HIT assay enables detection of both early- and late-phase inhibitors and can be used to discriminate between both types of inhibitors. Whereas early-phase inhibitors (GRFT, T20, EFV, and 118-D-24) potently reduced relative fluorescent signal intensities measured in both steps of the assay, the late-phase inhibitors (DRV and SQV) reduced the signal intensity predominantly in the second step.
TABLE 2.
Antiviral activities of anti-HIV drugs that target different stages of HIV replication in the EASY-HIT system
| Phase | Classa | Compoundb | EASY-HIT EC50 (nM)c |
Reference data |
||
|---|---|---|---|---|---|---|
| HIV-1IIIB | Primary isolate | EC50 (nM) | Cell line (reference) | |||
| Early | Entry inhibitor | GRFT | 0.15 ± 0.01 | 0.19 ± 0.03 | 0.043-0.63 | CEM-SS human T4-lymphoblastoid cell line, PBMC (37) |
| Entry inhibitor | T20 | 4.58 ± 0.46 | 14.70 ± 2.34 | 0.4-190 | PBMC (27) | |
| NRTI | AZT | 146.00 ± 22.00 | ND | 15-390 | PBMC (40) | |
| NNRTI | EFV | 0.08 ± 0.01 | 0.03 ± 0.00 | 0.8-1.4 | MT-4 human leukemia T-cell line (22) | |
| Integration | RAL | 134.00 ± 72.80 | ND | 1.2-24 | PBMC (4) | |
| Integration | 118-D-24 | 4.37 ± 1.15 | 5.04 ± 0.5 | 7 | CEM-SS human T4-lymphoblastoid cell (51) | |
| Transcription | FLV | 17.17 ± 1.63 | ND | <12 | PM-1 T-cell lymphoma line (23) | |
| Late | PI | SQV | 2.96 ± 0.10 | 2.79 ± 0.27 | 8.0-220 | PBMC (40) |
| PI | DRV | 3.54 ± 0.10 | ND | 3 | MT-2 cells (29) | |
NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, nonnucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
GRFT, griffithsin; AZT, azidothymidine; EFV, efavirenz; RAL, raltegravir; FLV, flavopiridol; SQV, saquinavir; DRV, darunavir.
Mean values ± SD of three independent experiments are shown. Values for compound 118-D-24 are micromolar (μM). ND, not done.
The potencies of these inhibitors to prevent HIV-1 replication in LC5-RIC cells was determined by testing serial dilutions of the inhibitors with the standard EASY-HIT assay procedure. Tests were performed with two different HIV-1 strains: a laboratory-adapted strain (HIV-1IIIB) and a primary HIV-1 strain isolated from clinical material (for details, see Materials and Methods). All inhibitors significantly reduced replication of both HIV-1 strains. As shown in Table 2, the EC50 (half maximal effective concentrations) values determined for each inhibitor were similar for both HIV-1 strains and agreed well with the published EC50 of these inhibitors in T cells (peripheral blood mononuclear cells [PBMC] and human T-cell lines).
These results confirm that the assay procedure allows evaluation of the anti-HIV activities of inhibitors that interfere with different stages of HIV-1 replication.
Screening of a compound library with EASY-HIT.
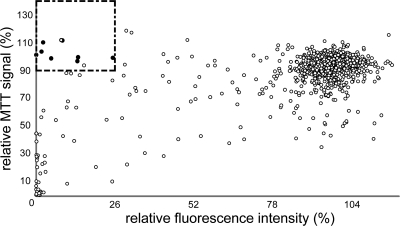
To evaluate whether this assay system can be applied to screening of small-molecule libraries for HIV inhibitors, we performed a pilot screen of a library of 1280 known pharmaceutically active compounds (LOPAC1280) for compounds with HIV-inhibitory activities. Each test compound was assayed at 10 μM in triplicate plates and analyzed for its capacity to reduce the relative fluorescent signal of HIV-exposed LC5-RIC cells to ≤26% in the second assay step (i.e., reduction of virus production). This cutoff was selected according to Jones et al. (26) and represents a decrease of the fluorescent signal by ≥3 standard deviations of the average fluorescent signal detected for all compound-treated HIV-exposed LC5-RIC samples. To avoid false-positive results, we excluded all compounds that decreased the relative MTT signal of the cells in test cultures by more than 10% from the set of potentially active compounds. Application of these criteria resulted in the selection of 10 inhibitor candidates (Fig. 3). Three of these substances turned out to be approved inhibitors of the HIV-1 reverse transcriptase (AZT, stavudine [d4T], and zalcitabine [ddC]), five additional compounds were listed in the NIAID Division of AIDS HIV/OI/Therapeutics database (http://chemdb.niaid.nih.gov/) as known inhibitors of HIV replication, and two substances had not been reported to inhibit HIV in this database to date.
FIG. 3.
Screening of a compound library for HIV-inhibitory molecules. LC5-RIC cultures were incubated in triplicates with HIV-1LAI virus and compounds of the LOPAC1280 library at 10 μM for 48 h (i.e., test cultures). Subsequently, aliquots of potentially infectious supernatants of test culture were transferred to uninfected LC5-RIC indicator cells, and fluorescent signal intensities were measured 72 h after transfer. Fluorescent signal intensities determined for samples from test cultures were normalized to fluorescent signal intensities determined for control samples (i.e., untreated HIV-infected cultures). Compound effects on the viability of cells in the test cultures were measured by MTT assay and normalized to values of control cultures. Each data point in the scatter plot indicates the value for the relative MTT signal (y axis) and the relative fluorescent signal intensity (x axis) determined for each compound. Potentially inhibitory compounds (indicated by the rectangle) were defined as compounds that decreased the relative fluorescent signal by greater than three times the standard deviation of the average relative fluorescent signal of all tested compounds (≤26%), without reducing the relative MTT signal below 90%. Filled circles represent selected compounds with known anti-HIV activities listed in the NIAID Division of AIDS HIV/OI/Therapeutics database.
Validation parameters characterizing the EASY-HIT system under these screening conditions are listed in Table 1 and confirm the excellent performance of the screening assay.
Time-of-addition assays for classification of inhibitory activities.
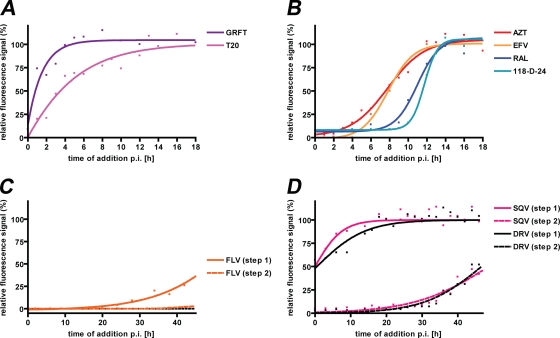
While the basic EASY-HIT discovery assay distinguishes between inhibitors of early and late phases of HIV-1 replication, characterization of inhibitory activities would benefit from the means to discriminate different steps of HIV replication at higher resolution. Time-of-addition assays monitor the inhibitory efficacies of compounds added to HIV target cells at different time points after exposure of the cells to the virus. As demonstrated in Fig. 4, time-of-addition assays with reference inhibitors (see above) yielded distinct profiles. Compounds known to interfere with very early steps of replication like virus attachment (e.g., GRFT) lost efficacy against HIV at much earlier time points than compounds that interfere with late steps of replication, like virion maturation (i.e., SQV). The profiles of these reference compounds (Fig. 4) demonstrate the temporal discrimination of HIV-1 entry (Fig. 4A), reverse transcription and integration (Fig. 4B), HIV transcription (Fig. 4C), and virus maturation processes (Fig. 4D).
FIG. 4.
Discrimination of sequential stages of HIV-1 replication by time-of-addition assays with HIV-inhibitors against known targets. HIV inhibitors that prevent HIV entry into host cells (A), reverse transcription (AZT and EFV) and integration of viral DNA into the host genome (RAL and 118-D-24) (B), HIV transcription (C), and virion maturation (D) were assayed, as indicated on the figure. HIV-1IIIB virus preparations were added to LC5-RIC cultures at time point 0. Anti-HIV compounds were added to the cultures at different time points after virus addition (i.e., p.i.). Final concentrations of inhibitors in wells were ≥2× EC50. Plates were incubated for a total period of 48 h after virus addition, and cultures were assayed for HIV reporter expression (solid lines; step 1) or for amounts of infectious virus in culture supernatants (dashed lines; step 2). Fluorescent signal intensities of treated cultures were related to those of cultures infected without inhibitors (set at 100% infection). Each data point represents the mean of values measured in at least three wells for a single time point. R2 values for each curve were ≥0.95.
Evaluation and characterization of antiviral activities of natural products.
We then addressed the usefulness of EASY-HIT to evaluate and characterize anti-HIV activities from different sources of natural products. Preliminary screening of a 400-compound natural product library (Specs, Netherlands) resulted in identification of three molecules that were found to inhibit HIV replication in our assay (i.e., abyssinone IV, 3,12-dioxocholanic acid, and 7-alpha-acetoxy-6-beta-hydroxyroyleanone). All three molecules showed no or only a minor (≤5%) decrease in the relative MTT signal of the cells in test cultures at 10 μM, indicating that they lacked cytotoxicity at this concentration. Repeated testing of these three compounds in dose-response assays yielded EC50 of ≤5 μM for all three compounds. Anti-HIV activities of these compounds were not listed in the NIAID Division of AIDS HIV/OI/TB Therapeutics database (http://chemdb.niaid.nih.gov/), suggesting that they are new HIV inhibitor candidates.
The potential of these three compounds to inhibit the early phase of HIV-1 replication was evaluated by measuring their effects on reporter expression at concentrations that completely blocked the release of infectious HIV (i.e., >95% reduction of fluorescent signal in the second step). Under these conditions, abyssinone IV and 3,12-dioxocholanic acid reduced the fluorescent signal during primary infection more strongly than 7-alpha-acetoxy-6-beta-hydroxyroyleanone, suggesting that the former two compounds interfere more strongly with the early phase of HIV-1 replication than the latter compound.
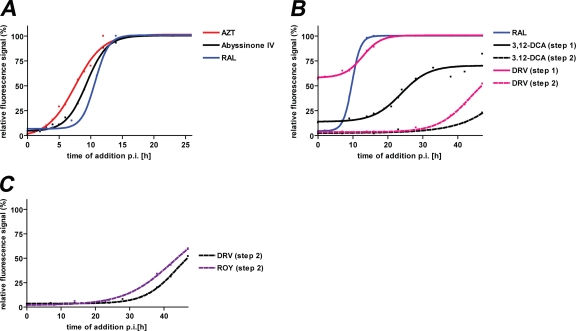
We then compared the inhibitory profiles of these compounds with those of known anti-HIV drugs in time-of-addition assays. The profile of abyssinone IV was closest to the profiles of AZT and of raltegravir (Fig. 5A). This confirms that abyssinone IV interferes with HIV-1 replication during the early phase of replication and suggests that it may affect processes temporally associated with the stages of reverse transcription or integration. The profile of 3,12-dioxocholanic acid (Fig. 5B) shows that this compound can inhibit reporter production at later time points (50% loss of inhibition at ∼26 h postinfection [p.i.]) than raltegravir (50% loss of inhibition at ∼10 h p.i.). This indicates that 3,12-dioxocholanic acid may act after integration in the late phase of HIV replication. However, compared to the late-phase inhibitor darunavir, 3,12-dioxocholanic acid showed a much more pronounced inhibitory effect on reporter production in test cultures. Furthermore, 3,12-dioxocholanic acid more strongly suppressed the production of infectious virus at later time points (>35 h p.i.) than darunavir. Together, these results indicate that the inhibitory activity of 3,12-dioxocholanic acid differs from the activity of current anti-HIV drugs, suggesting that it may represent a new class of inhibitors. The profile of 7-alpha-acetoxy-6-beta-hydroxyroyleanone resembled that of darunavir (Fig. 5C), indicating that this compound interferes with the late phase of HIV-1 replication. We speculate that it may either act at a similar stage of replication as darunavir or by binding to viral envelope proteins or viral membranes, similar to the mechanism of action of deoxycholate or related bile acid derivatives (2).
FIG. 5.
Comparisons of time-of-addition profiles of three novel anti-HIV molecules from a natural product collection with those of selected reference anti-HIV drugs. (A) Comparison of the profiles of abyssinone IV and the reference drugs AZT and RAL. (B) Comparison of the profiles of 3,12-dioxocholanic acid (3,12-DCA) and the reference drugs RAL and DRV. (C) Comparison of the profiles of 7-alpha-acetoxy-6-beta-hydroxyroyleanone (ROY) and the reference drug DRV. Data for profiles of reference anti-HIV drugs were taken from Fig. 4. Solid line, analysis of HIV reporter expression in test cultures; dotted line, analysis of amounts of infectious virus in supernatants of test cultures. Each data point represents the mean of values measured in at least three wells for a single time point.
To check whether EASY-HIT is suitable for directly measuring antiviral activities in natural materials, crude extracts from plants with established anti-HIV activities and a control plant (Nicotiana tabacum) were tested in the indicator cells. Prunella vulgaris and Arctium lappa are well-known medicinal plants of the Lamiaceae and Asteraceae plant families, respectively. Extracts from members of both plant families have been shown to inhibit HIV entry, and the hypothetical compounds responsible for the anti-HIV effects were named prunellin (P. vulgaris) and arctigenin (A. lappa) (20, 32, 45). In addition, extracts of the medicinal plant Echinacea sp. (Asteraceae) show inhibition of the HIV integrase enzyme (10, 42).
The crude extracts of P. vulgaris and A. lappa (see Fig. S3A in the supplemental material) and E. purpurea (data not shown) showed significant dose-dependent reduction of HIV infection whereas the aqueous extract of the control plant N. tabacum, had no effect on HIV infection or replication. All crude plant extracts showed no or only a very slight (≤10%) decrease in the relative MTT signal of the cells in test cultures even at the highest extract concentrations tested. Time-of-addition assays with crude extracts of P. vulgaris, A. lappa, and E. purpurea confirmed that extracts of P. vulgaris and A. lappa inhibited initial steps of virus entry, whereas the extract of Echinacea purpurea interfered with HIV replication at later stages, possibly affecting reverse transcription or provirus integration or both (see Fig. S3B in the supplemental material).
DISCUSSION
Successful anti-HIV therapy and prevention require inhibitors targeting many different stages of HIV-1 replication (50). A major objective in developing the EASY-HIT technology was to enable identification of inhibitors against all stages of the HIV-1 replication cycle. The full-replication capability of this technology was validated with a panel of nine known HIV inhibitors that target different processes of HIV replication, beginning from the very early processes of viral attachment and entry and extending through reverse transcription, integration, transcription, and production of infectious virions (Table 1). The 50% effective concentrations (EC50) of all tested drugs were consistent with published data for various T-cell lines and HIV isolates (22, 27, 29, 37, 40, 51). Testing of the reference compounds in time-of-addition assays showed that this technology can also be used in secondary assays for higher-resolution discrimination of different steps of the replication cycle, facilitating the classification of inhibitor activities.
In the following we will compare various features of the EASY-HIT technology with other HIV infection reporter systems, discuss the quality of its performance characteristics and its performance in a pilot screen of the LOPAC1280 compound library, and outline the use of this technology for identification of novel HIV inhibitors from biological sources.
Like other fluorescent HIV reporter assays, the EASY-HIT assay quantifies levels of a fluorescent reporter protein induced during the early phase of HIV replication (19, 41, 59). However, in contrast to conventional assays, the EASY-HIT assay is designed to evaluate an additional HIV infection parameter which relates to the late phase of virus replication, i.e., production of infectious virus. For this purpose, a two-step assay is employed that measures the fluorescent signals first directly in the cells of the test culture and subsequently in LC5-RIC indicator cells incubated with aliquots of supernatants of test cultures (Fig. 2). All inhibitors of HIV replication, regardless of the phase of replication at which they act, should reduce production of infectious virus and, thus, decrease the fluorescent signal in the second assay step. Compounds that also efficiently reduce the fluorescent signal in the first step are potential early-phase inhibitors while compounds that reduce the fluorescent signal more strongly in the second step than in the first are potential late-phase inhibitors. The advantages of this two-step assay design are as follows: (i) improved detection of late-phase inhibitors, (ii) availability of an additional control for unspecific effects of compound treatment on reporter production, and (iii) distinction between potential early- and late-phase inhibitors of HIV replication early in the selection process. The last enables the selection of candidate inhibitors with different modes of activity already during screening and helps to focus screening efforts, if desired. This assay design overcomes a previously described limitation of various conventional HIV reporter assays that detect late-phase inhibitors less efficiently than early-phase inhibitor (7, 11, 41). Late-phase HIV inhibitors continue to be of interest in the development of new anti-HIV drugs and may be more effective in reducing the viral burden in vivo than early-phase inhibitors (14).
Many HIV reporter systems are based on a reporter gene under the transcriptional control of the LTR, which is activated by the viral Tat protein during infection (for examples, see references 7, 8, 19, 28, and 41). However, background expression of these reporter genes has been reported to occur in HIV indicator cells (48, 53, 59). This is due to the inherent basal activity of the HIV LTR, which is essential for initiating HIV expression during replication, and the capacity of the LTR to modulate its activity in response to extracellular stimuli (reviewed in references 3 and 9). Wu et al. (59) demonstrated that background reporter expression in CEM cells can be reduced by incorporating additional regulatory sequences for Rev responsiveness into the LTR-driven reporter gene. Therefore, we based our system on a reporter gene activated by both Rev and Tat rather than by just Tat (57, 58). Indeed, the fluorescent signal measured for LC5-RIC cells in the absence of HIV was indistinguishable from that of the parental cells lacking the reporter, confirming the absence of background expression of the reporter in LC5-RIC cells.
In contrast to the other fluorescence-based reporter assays, the reporter gene in the LC5-RIC cells encodes a red fluorescent protein (DsRed1) instead of the more common GFP. This allows simultaneous visualization of GFP in infected LC5-RIC cells in future studies, e.g., as virion label (25, 39) or as cellular reporter.
HIV indicator cells used in reporter assays were developed from both T-cell lines derived from human acute T-cell leukemias (e.g., CEM and Jurkat) (19, 41, 59) and from HeLa cells (28, 36, 55). We chose HeLa cells for the generation of the LC5-RIC indicator cell line because of their adherent cell growth. Adherent cell growth facilitates accurate quantification of the fluorescent signal intensities of cultured cells by microplate fluorimetry and allows easy removal of cell culture supernatants for further analysis. In addition, adherent cell growth facilitates the use of LC5-RIC cells in future imaging applications, e.g., to analyze compound- or infection-related changes of morphological cell parameters. LC5-RIC cells stably express CD4 and CXCR4 on their cell surfaces (>80% positive cells). They are permissive for infection and replication of laboratory-adapted HIV-1 strains as well primary HIV-1 isolates. HIV infection of LC5-RIC cells with laboratory HIV-1 isolates does not require infection-enhancing agents like DEAE-dextran, which is generally used in infection protocols with HeLa-derived cell lines such as HeLa-MAGI or TZM-BL (28, 55). We recently engineered a CCR5-expressing derivative of the LC5-RIC line, enabling future studies with macrophage-tropic (R5) viruses.
Analysis of various parameters for robustness and reliability revealed that the EASY-HIT assay surpassed the minimum acceptance criteria for all parameters by a wide margin (Table 1), confirming the high quality of its performance. Furthermore, the assay retained high robustness and reliability during screening of the LOPAC1280 library, yielding a Z′ value of 0.94 (Table 1). Z′ values reported for screens performed with other cell-based HIV inhibitor assays ranged from 0.72 to 0.84 (6, 7, 18, 41). Candidate inhibitor compounds were selected from the LOPAC1280 screen by two criteria: (i) reduction of the relative fluorescent reporter signal below a threshold of three times the standard deviation of the average signal of all tested compounds and (ii) maintenance of ≥90% of the relative MTT signal in test cultures. The latter criterion was chosen to reduce false-positive results due to adverse effects of compounds on cell viabilities and activities and to thus improve identification of candidate inhibitors that specifically affect HIV replication. Application of these criteria led to the identification of a set of 10 compounds that included 8 compounds with known HIV-inhibitory activities, corresponding to an overall selection rate of 0.8% of compounds from the LOPAC1280 library. The results from this pilot screen of the LOPAC1280 library indicates that the EASY-HIT assay is suitable for screening applications. Several industrial-scale screens have been described previously (6, 7). Here, the screen with the EASY-HIT technology was performed in an academic setting with the benefit of only a manual benchtop liquid pipetting system. Under these conditions, testing of all 1280 compounds of the LOPAC1280 library in triplicate plates was completed within 6 days, indicating the potential usefulness of our technology for the screening of larger libraries.
The exploration of novel sources of bioactive structures including plants, microbes, or sea organisms for antiviral activities is becoming increasingly important (60). The results of this study indicate that the EASY-HIT technology is a useful tool to identify and develop natural products with HIV-inhibitory activities. We show that three novel candidate inhibitors in a collection of natural products interfered with different steps of HIV replication, with one candidate inhibitor acting at the early phase of HIV replication, another at the late phase, and a third targeting a postintegration mechanism not covered by conventional antiviral therapies so far. We also demonstrate that our system can be used to identify HIV-inhibitory activities in crude plant extracts, yielding defined and reproducible dose-dependent inhibition curves and distinct time-of-addition profiles. The stages of HIV replication identified by the latter as targets for interference agreed with published data (20, 32, 45). This suggests application of the EASY-HIT technology for identification of HIV inhibitors from biological sources by bioassay guided fractionation procedures in future studies.
In summary, we established a robust and reliable full HIV-1 replication system for integrated identification and characterization of novel HIV inhibitors. We demonstrate that EASY-HIT can be applied to screening of small-molecule libraries in an academic environment with limited automation. In addition, we show that EASY-HIT can be used to evaluate anti-HIV activities of natural products, including crude plant extracts. We expect our technology to be of interest for industrial settings as well as nonprofit research aimed at developing affordable therapies for HIV-infected people in resource-poor settings and may also help individuals in these areas develop novel therapeutic strategies from local biological sources.
Supplementary Material
Acknowledgments
This work was supported by the Life Science Foundation for the Promotion of Science and Research (grant to R.B.W. and J.D.).
We thank Ben Berkhout for providing pLAI.2, Manja Meggendorfer for critical reading of the manuscript, and Ingrid Huelsmeier, Susanne Stich, and Ute Finkel for expert technical assistance.
Footnotes
Published ahead of print on 27 September 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Adamson, C. S., and E. O. Freed. 2008. Recent progress in antiretrovirals-lessons from resistance. Drug Discov. Today 13:424-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Jabri, A. A., M. D. Wigg, E. Elias, R. Lambkin, C. O. Mills, and J. S. Oxford. 2000. In vitro anti-HIV-1 virucidal activity of tyrosine-conjugated tri- and dihydroxy bile salt derivatives. J. Antimicrob. Chemother. 45:617-621. [DOI] [PubMed] [Google Scholar]
- 3.Baba, M. 2004. Inhibitors of HIV-1 gene expression and transcription. Curr. Top. Med. Chem. 4:871-882. [DOI] [PubMed] [Google Scholar]
- 4.Bar-Magen, T., R. D. Sloan, V. H. Faltenbacher, D. A. Donahue, B. D. Kuhl, M. Oliveira, H. Xu, and M. A. Wainberg. 2009. Comparative biochemical analysis of HIV-1 subtype B and C integrase enzymes. Retrovirology 6:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barouch, D. H. 2008. Challenges in the development of an HIV-1 vaccine. Nature 455:613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blair, W. S., J. Isaacson, X. Li, J. Cao, Q. Peng, G. F. Kong, and A. K. Patick. 2005. A novel HIV-1 antiviral high throughput screening approach for the discovery of HIV-1 inhibitors. Antiviral Res. 65:107-116. [DOI] [PubMed] [Google Scholar]
- 7.Cao, J., J. Isaacson, A. K. Patick, and W. S. Blair. 2005. High-throughput human immunodeficiency virus type 1 (HIV-1) full replication assay that includes HIV-1 Vif as an antiviral target. Antimicrob. Agents Chemother. 49:3833-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiba-Mizutani, T., H. Miura, M. Matsuda, Z. Matsuda, Y. Yokomaku, K. Miyauchi, M. Nishizawa, N. Yamamoto, and W. Sugiura. 2007. Use of new T-cell-based cell lines expressing two luciferase reporters for accurately evaluating susceptibility to anti-human immunodeficiency virus type 1 drugs. J. Clin. Microbiol. 45:477-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copeland, K. F. 2005. Modulation of HIV-1 transcription by cytokines and chemokines. Mini Rev. Med. Chem. 5:1093-1101. [DOI] [PubMed] [Google Scholar]
- 10.Cos, P., L. Maes, D. Vanden Berghe, N. Hermans, L. Pieters, and A. Vlietinck. 2004. Plant substances as anti-HIV agents selected according to their putative mechanism of action. J. Nat. Prod. 67:284-293. [DOI] [PubMed] [Google Scholar]
- 11.Daelemans, D., C. Pannecouque, G. N. Pavlakis, O. Tabarrini, and E. De Clercq. 2005. A novel and efficient approach to discriminate between pre- and post-transcription HIV inhibitors. Mol. Pharmacol. 67:1574-1580. [DOI] [PubMed] [Google Scholar]
- 12.De Clercq, E. 2007. The design of drugs for HIV and HCV. Nat. Rev. Drug Discov. 6:1001-1018. [DOI] [PubMed] [Google Scholar]
- 13.De Clercq, E. 2009. Looking back in 2009 at the dawning of antiviral therapy now 50 years ago an historical perspective. Adv. Virus Res. 73:1-53. [DOI] [PubMed] [Google Scholar]
- 14.Donahue, D. A., R. D. Sloan, B. D. Kuhl, T. Bar-Magen, S. M. Schader, and M. A. Wainberg. 2010. Stage-dependent inhibition of HIV-1 replication by antiretroviral drugs in cell culture. Antimicrob. Agents Chemother. 54:1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorsky, D. I., M. Wells, and R. D. Harrington. 1996. Detection of HIV-1 infection with a green fluorescent protein reporter system. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13:308-313. [DOI] [PubMed] [Google Scholar]
- 16.Eli Lilly and Company and the National Institutes of Health Chemical Genomics Center. 2008. Assay guidance manual, version 5.0. National Institutes of Health Chemical Genomics Center, Bethesda, MD. http://www.ncgc.nih.gov/guidance/manual_toc.html.
- 17.Felber, B. K., and G. N. Pavlakis. 1988. A quantitative bioassay for HIV-1 based on trans-activation. Science 239:184-187. [DOI] [PubMed] [Google Scholar]
- 18.Garcia, J. M., A. Gao, P. L. He, J. Choi, W. Tang, R. Bruzzone, O. Schwartz, H. Naya, F. J. Nan, J. Li, R. Altmeyer, and J. P. Zuo. 2009. High-throughput screening using pseudotyped lentiviral particles: a strategy for the identification of HIV-1 inhibitors in a cell-based assay. Antiviral Res. 81:239-247. [DOI] [PubMed] [Google Scholar]
- 19.Gervaix, A., D. West, L. M. Leoni, D. D. Richman, F. Wong-Staal, and J. Corbeil. 1997. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc. Natl. Acad. Sci. U. S. A. 94:4653-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geuenich, S., C. Goffinet, S. Venzke, S. Nolkemper, I. Baumann, P. Plinkert, J. Reichling, and O. T. Keppler. 2008. Aqueous extracts from peppermint, sage and lemon balm leaves display potent anti-HIV-1 activity by increasing the virion density. Retrovirology 5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene, W. C., Z. Debyser, Y. Ikeda, E. O. Freed, E. Stephens, W. Yonemoto, R. W. Buckheit, J. A. Este, and T. Cihlar. 2008. Novel targets for HIV therapy. Antiviral Res. 80:251-265. [DOI] [PubMed] [Google Scholar]
- 22.Hazen, R. J., R. J. Harvey, M. H. St Clair, R. G. Ferris, G. A. Freeman, J. H. Tidwell, L. T. Schaller, J. R. Cowan, S. A. Short, K. R. Romines, J. H. Chan, and L. R. Boone. 2005. Anti-human immunodeficiency virus type 1 activity of the nonnucleoside reverse transcriptase inhibitor GW678248 in combination with other antiretrovirals against clinical isolate viruses and in vitro selection for resistance. Antimicrob. Agents Chemother. 49:4465-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heredia, A., C. Davis, D. Bamba, N. Le, M. Y. Gwarzo, M. Sadowska, R. C. Gallo, and R. R. Redfield. 2005. Indirubin-3′-monoxime, a derivative of a Chinese antileukemia medicine, inhibits P-TEFb function and HIV-1 replication. AIDS 19:2087-2095. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann, D., J. Seebach, A. Cosma, F. D. Goebel, K. Strimmer, H. M. Schatzl, and V. Erfle. 2008. Therapeutic vaccination reduces HIV sequence variability. FASEB J. 22:437-444. [DOI] [PubMed] [Google Scholar]
- 25.Hubner, W., P. Chen, A. Del Portillo, Y. Liu, R. E. Gordon, and B. K. Chen. 2007. Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J. Virol. 81:12596-12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, J., J. Rodgers, M. Heil, J. May, L. White, J. A. Maddry, T. M. Fletcher III, G. M. Shaw, J. L. Hartman IV, and O. Kutsch. 2007. High throughput drug screening for human immunodeficiency virus type 1 reactivating compounds. Assay Drug Dev. Technol. 5:181-189. [DOI] [PubMed] [Google Scholar]
- 27.Ketas, T. J., P. J. Klasse, C. Spenlehauer, M. Nesin, I. Frank, M. Pope, J. M. Strizki, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2003. Entry inhibitors SCH-C, RANTES, and T-20 block HIV type 1 replication in multiple cell types. AIDS Res. Hum. Retroviruses 19:177-186. [DOI] [PubMed] [Google Scholar]
- 28.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koh, Y., H. Nakata, K. Maeda, H. Ogata, G. Bilcer, T. Devasamudram, J. F. Kincaid, P. Boross, Y. F. Wang, Y. Tie, P. Volarath, L. Gaddis, R. W. Harrison, I. T. Weber, A. K. Ghosh, and H. Mitsuya. 2003. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 47:3123-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam, K. S. 2007. New aspects of natural products in drug discovery. Trends Microbiol. 15:279-289. [DOI] [PubMed] [Google Scholar]
- 31.Little, S. J., S. Holte, J. P. Routy, E. S. Daar, M. Markowitz, A. C. Collier, R. A. Koup, J. W. Mellors, E. Connick, B. Conway, M. Kilby, L. Wang, J. M. Whitcomb, N. S. Hellmann, and D. D. Richman. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347:385-394. [DOI] [PubMed] [Google Scholar]
- 32.Liu, S., S. Jiang, Z. Wu, L. Lv, J. Zhang, Z. Zhu, and S. Wu. 2002. Identification of inhibitors of the HIV-1 gp41 six-helix bundle formation from extracts of Chinese medicinal herbs Prunella vulgaris and Rhizoma cibotte. Life Sci. 71:1779-1791. [DOI] [PubMed] [Google Scholar]
- 33.Maddon, P. J., D. R. Littman, M. Godfrey, D. E. Maddon, L. Chess, and R. Axel. 1985. The isolation and nucleotide sequence of a cDNA encoding the T cell surface protein T4: a new member of the immunoglobulin gene family. Cell 42:93-104. [DOI] [PubMed] [Google Scholar]
- 34.McMahon, M. A., L. Shen, and R. F. Siliciano. 2009. New approaches for quantitating the inhibition of HIV-1 replication by antiviral drugs in vitro and in vivo. Curr. Opin. Infect. Dis. 22:574-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menendez-Arias, L. 2002. Targeting HIV: antiretroviral therapy and development of drug resistance. Trends Pharmacol. Sci. 23:381-388. [DOI] [PubMed] [Google Scholar]
- 36.Montefiori, D. C. 2009. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 485:395-405. [DOI] [PubMed] [Google Scholar]
- 37.Mori, T., B. R. O'Keefe, R. C. Sowder II, S. Bringans, R. Gardella, S. Berg, P. Cochran, J. A. Turpin, R. W. Buckheit, Jr., J. B. McMahon, and M. R. Boyd. 2005. Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J. Biol. Chem. 280:9345-9353. [DOI] [PubMed] [Google Scholar]
- 38.Mosmann, T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65:55-63. [DOI] [PubMed] [Google Scholar]
- 39.Muller, B., J. Daecke, O. T. Fackler, M. T. Dittmar, H. Zentgraf, and H. G. Krausslich. 2004. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J. Virol. 78:10803-10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakata, H., M. Amano, Y. Koh, E. Kodama, G. Yang, C. M. Bailey, S. Kohgo, H. Hayakawa, M. Matsuoka, K. S. Anderson, Y. C. Cheng, and H. Mitsuya. 2007. Activity against human immunodeficiency virus type 1, intracellular metabolism, and effects on human DNA polymerases of 4′-ethynyl-2-fluoro-2′-deoxyadenosine. Antimicrob. Agents Chemother. 51:2701-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ochsenbauer-Jambor, C., J. Jones, M. Heil, K. P. Zammit, and O. Kutsch. 2006. T-cell line for HIV drug screening using EGFP as a quantitative marker of HIV-1 replication. Biotechniques 40:91-100. [DOI] [PubMed] [Google Scholar]
- 42.Pal Singh, I. P., S. B. Bharate, and K. K. Bhutani. 2005. Anti-HIV natural products. Curr. Sci. 89:269-290. [Google Scholar]
- 43.Pauwels, R. 2006. Aspects of successful drug discovery and development. Antiviral Res. 71:77-89. [DOI] [PubMed] [Google Scholar]
- 44.Peden, K., M. Emerman, and L. Montagnier. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661-672. [DOI] [PubMed] [Google Scholar]
- 45.Reinke, R. A., D. J. Lee, B. R. McDougall, P. J. King, J. Victoria, Y. Mao, X. Lei, M. G. Reinecke, and W. E. Robinson, Jr. 2004. L-chicoric acid inhibits human immunodeficiency virus type 1 integration in vivo and is a noncompetitive but reversible inhibitor of HIV-1 integrase in vitro. Virology 326:203-219. [DOI] [PubMed] [Google Scholar]
- 46.Richman, D. D., D. M. Margolis, M. Delaney, W. C. Greene, D. Hazuda, and R. J. Pomerantz. 2009. The challenge of finding a cure for HIV infection. Science 323:1304-1307. [DOI] [PubMed] [Google Scholar]
- 47.Rishton, G. M. 2008. Natural products as a robust source of new drugs and drug leads: past successes and present day issues. Am. J. Cardiol. 101:43D-49D. [DOI] [PubMed] [Google Scholar]
- 48.Rocancourt, D., C. Bonnerot, H. Jouin, M. Emerman, and J. F. Nicolas. 1990. Activation of a beta-galactosidase recombinant provirus: application to titration of human immunodeficiency virus (HIV) and HIV-infected cells. J. Virol. 64:2660-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rothenaigner, I., S. Kramer, M. Ziegler, H. Wolff, A. Kleinschmidt, and R. Brack-Werner. 2007. Long-term HIV-1 infection of neural progenitor populations. AIDS 21:2271-2281. [DOI] [PubMed] [Google Scholar]
- 50.Simon, V., D. D. Ho, and Q. Abdool Karim. 2006. HIV/AIDS epidemiology, pathogenesis, prevention, and treatment. Lancet 368:489-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svarovskaia, E. S., R. Barr, X. Zhang, G. C. Pais, C. Marchand, Y. Pommier, T. R. Burke, Jr., and V. K. Pathak. 2004. Azido-containing diketo acid derivatives inhibit human immunodeficiency virus type 1 integrase in vivo and influence the frequency of deletions at two-long-terminal-repeat-circle junctions. J. Virol. 78:3210-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.UNAIDS. 2008. 2008 Report on the global HIV/AIDS epidemic. UNAIDS/08.27E /JC1511E. Joint United Nations Programme on HIV/AIDS, Geneva, Switzerland.
- 53.Vodicka, M. A., W. C. Goh, L. I. Wu, M. E. Rogel, S. R. Bartz, V. L. Schweickart, C. J. Raport, and M. Emerman. 1997. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology 233:193-198. [DOI] [PubMed] [Google Scholar]
- 54.Wachinger, M., A. Kleinschmidt, D. Winder, N. von Pechmann, A. Ludvigsen, M. Neumann, R. Holle, B. Salmons, V. Erfle, and R. Brack-Werner. 1998. Antimicrobial peptides melittin and cecropin inhibit replication of human immunodeficiency virus 1 by suppressing viral gene expression. J. Gen. Virol. 79:731-740. [DOI] [PubMed] [Google Scholar]
- 55.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Westby, M., G. R. Nakayama, S. L. Butler, and W. S. Blair. 2005. Cell-based and biochemical screening approaches for the discovery of novel HIV-1 inhibitors. Antiviral Res. 67:121-140. [DOI] [PubMed] [Google Scholar]
- 57.Wolff, H., R. Brack-Werner, M. Neumann, T. Werner, and R. Schneider. 2003. Integrated functional and bioinformatics approach for the identification and experimental verification of RNA signals: application to HIV-1 INS. Nucleic Acids Res. 31:2839-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolff, H., K. Hadian, M. Ziegler, C. Weierich, S. Kramer-Hammerle, A. Kleinschmidt, V. Erfle, and R. Brack-Werner. 2006. Analysis of the influence of subcellular localization of the HIV Rev protein on Rev-dependent gene expression by multi-fluorescence live-cell imaging. Exp. Cell Res. 312:443-456. [DOI] [PubMed] [Google Scholar]
- 59.Wu, Y., M. H. Beddall, and J. W. Marsh. 2007. Rev-dependent indicator T cell line. Curr. HIV Res. 5:394-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yasuhara-Bell, J., and Y. Lu. 2010. Marine compounds and their antiviral activities. Antiviral Res. 86:231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, J. H., T. D. Chung, and K. R. Oldenburg. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67-73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.