Abstract
The aim of this study was to investigate the mechanisms involved in the meropenem resistance of Serratia marcescens clinical isolates. Meropenem-resistant (MIC range, 16 to 32 μg/ml) S. marcescens isolates were recovered from nine patients in a tertiary hospital in Seoul, South Korea, from June to November 2005. All the isolates shared identical or similar (>85% similarity) SpeI macrorestriction patterns, indicating clonal spread. PCR experiments did not detect any carbapenemase in those isolates. They carried the blaCTX-M-22 gene located on a 150-kbp plasmid of the incompatibility group L/M; however, the addition of clavulanic acid exhibited few effects on meropenem MICs. Although meropenem MICs were reduced 4- to 16-fold with the addition of boronic acid, no plasmid-borne AmpC β-lactamase gene was detected in PCR experiments. Real-time quantitative PCR experiments showed that expression levels of the chromosomal ampC gene in those isolates were 87.06 to 155.76 times higher than that of the reference strain ATCC 8100. SDS-PAGE showed a lack of the 42-kDa outer membrane protein (OmpF). In combination with the overproduction of the chromosomal AmpC enzyme, the loss of OmpF may have played a role in the acquisition of meropenem resistance in our isolates.
Serratia marcescens frequently exhibits resistance to extended-spectrum β-lactams due to its ability to overproduce the chromosomal AmpC enzyme and to acquire plasmid-borne extended-spectrum β-lactamases (ESBLs) (9, 10). Carbapenems have deserved special attention, as they remain the last stronghold against infections caused by Gram-negative strains resistant to oxyimino-cephalosporins. However, this has been counteracted by the emergence of carbapenem-resistant strains in diverse geographic locations (19, 20, 22, 24, 25).
Various carbapenemases of classes A and B may be involved in carbapenem resistance in S. marcescens. Variants of chromosomally encoded SME-type class A carbapenemase and the plasmid-borne carbapenemase KPC-2 have been described for S. marcescens isolates (20, 22, 25). Plasmid-borne class B metallo-β-lactamases (MBLs), including IMP-type variants and VIM-2, have been identified in S. marcescens since the first discovery of IMP-1 in an S. marcescens strain from Japan in 1991 (19, 24).
The SENTRY survey performed in 2000 to 2004 detected five S. marcescens isolates producing class A carbapenemases in nine S. marcescens isolates with decreased susceptibility to carbapenems (6). MBL-producing isolates were not detected in the survey. A nationwide survey in South Korea in 2002 reported that despite the high prevalence (17.9%; 36/201) of S. marcescens isolates with decreased carbapenem susceptibility, MBLs were detected in only two isolates, and class A carbapenemases were not detected (13). Those previous reports suggest that MBLs and class A carbapenemases may play a role in some isolates, but it leaves much to be explained regarding the mechanism of carbapenem resistance in S. marcescens.
The purpose of this study was to investigate the mechanisms involved in the meropenem resistance of an S. marcescens strain causing an outbreak of infection in a tertiary care hospital in Seoul, South Korea. Our results show that the high-level production of the chromosomal AmpC enzyme combined with the loss of an outer membrane protein (OMP) may play an important role in the acquisition of carbapenem resistance in S. marcescens.
MATERIALS AND METHODS
Bacterial strains.
From June to November 2005, clinical isolates of S. marcescens showing resistance to meropenem but susceptibility to imipenem were recovered from urinary specimens of nine patients hospitalized at a tertiary care hospital in Seoul, South Korea. Species identification was done by use of the Vitek 2 system (bioMérieux Vitek Inc., Hazelwood, MO). Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as MIC reference strains, and E. coli J53 (azide resistant) was used as a recipient strain for conjugative transfer.
Antimicrobial susceptibility testing.
Antimicrobial susceptibilities were determined by agar dilution methods according to Clinical and Laboratory Standards Institute guidelines (3). MICs of imipenem and meropenem in the presence or absence of clavulanic acid (CA) (at a fixed concentration of 4 μg/ml; Sigma, St. Louis, MO), 3-aminophenyl boronic acid (BA) (300 μg/ml; Sigma), cloxacillin (250 μg/ml; Sigma), or phenyl-arginine-β-naphthylamide (PAβN) (20 μg/ml; Sigma) were determined by Etest. CA, BA-cloxacillin, and PAβN were used as inhibitors for ESBLs, AmpC enzymes, and efflux pumps, respectively.
Phenotypic detection of β-lactamase production.
ESBL and AmpC β-lactamase production was tested by a phenotypic confirmatory test using CA and BA as inhibitors (4, 11). Carbapenemase production was screened by the modified Hodge test using an ertapenem disk on MacConkey agar plates (14). MBL production was screened by the imipenem-EDTA double-disk synergy (IEDDS) test using an imipenem disk (10 μg; Becton-Dickinson, Sparks, MD) and an EDTA (750 μg/ml) plus sodium mercaptoacetic acid (2 μg/ml; Sigma) disk spaced at a 10-mm distance from disk edge to edge on Mueller-Hinton agar plates (15).
Conjugal transfer experiments.
Conjugation experiments were carried out by the broth mating method using azide-resistant E. coli J53 as the recipient. Transconjugants were selected on MacConkey agar containing azide (100 μg/ml) and cefotaxime (2 μg/ml) or meropenem (0.5 μg/ml).
IEF of β-lactamases.
Crude extracts were prepared by the sonication of bacterial cells and were subjected to isoelectric focusing (IEF). β-Lactamase activity was visualized by staining the gel with 0.5 mM nitrocefin (Oxoid, Cambridge, United Kingdom) in 0.1 M phosphate buffer (pH 7.0). The isoelectric points (pIs) were determined after a comparison with bands of known pIs (pI 5.4, 6.0, 7.0, 7.6, and 8.0).
PCR sequencing of β-lactamase genes.
The detection of genes encoding ESBLs, AmpC β-lactamases, and class A, B, and D carbapenemases was performed by PCR amplification using pairs of previously described primers (13, 21). The major OMP genes (ompC and ompF) and the chromosomal ampC gene of S. marcescens were also amplified by using previously described primers (5). PCR products were directly sequenced with an automatic sequencer (model 3730x1; Applied Biosystems, Weiterstadt, Germany).
PFGE.
Plugs containing whole genomic DNA of the S. marcescens isolates were digested with SpeI and S1 nuclease, respectively. DNA fragments were separated by using a CHEF-DRII device (Bio-Rad, Hercules, CA). The pulsed-field gel electrophoresis (PFGE) conditions of SpeI macrorestriction analysis were 6 V/cm for 20 h with pulse times of 0.5 to 60 s at a temperature of 14°C. The pulse times for S1 nuclease restriction analysis were 9 to 90 s. The lambda ladder (Bio-Rad) was used as a DNA size marker.
Southern blotting.
The PFGE gels with plasmids linearized by S1 nuclease digestion were blotted onto a nylon membrane (Bio-Rad) and hybridized with probes specific for the genes encoding CTX-M-22 and various specific replicons, respectively. Probes were obtained by PCR experiments as described above. Probe labeling, hybridization, and detection were performed with the digoxigenin (DIG) DNA labeling and detection kit (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's protocols.
RQ-PCR.
The growth conditions used for total RNA extraction were as follows: 4 to 5 colonies of Serratia marcescens clinical isolates and strain ATCC 8100 were inoculated in 5 ml of Luria-Bertani broth (∼0.5 McFarland standard) and incubated overnight at 35°C. Real-time quantitative PCR (RQ-PCR) experiments were performed to analyze the mRNA levels of the genes encoding the chromosomal AmpC β-lactamases of S. marcescens. The primers and probes used for RQ-PCR experiments using a capillary real-time thermal cycler (LightCycler; Roche Diagnostics, Indianapolis, IN) are listed in Table 1. Amplification was carried out in a 20-μl final volume containing 4 μl of premix (LightCycler TaqMan Master kit; Roche Diagnostics), 1 μl of primers (final concentration, 0.5 μM), 0.1 μl of probe (final concentration, 0.1 μM), 9.9 μl of water, and 5 μl of cDNA. The reaction conditions were 95°C for 10 min; 45 cycles of 95°C for 10 s, 50°C for 30 s, and 72°C for 1 min; and cooling for 30 min at 40°C. Expression levels of the ampC gene were normalized against the 16S rRNA gene. The expression level of the ampC gene in S. marcescens ATCC 8100 was also measured for comparison.
TABLE 1.
Primers for chromosomal 16S rRNA and ampC genes for RQ-PCR
| Target | Primer | Sequence (5′-3′) |
|---|---|---|
| ampC gene | Sense | GCG ATC GCC GTG TCG |
| Antisense | GTC TGC TCG GTG ATC GGT TT | |
| Probe | TCG ACG GCA AAC AGC | |
| 16S rRNA gene | Sense | CCC AGA TGG GAT TAG CTA GTA GGT |
| Antisense | TGG CTG GTC ATC CTC TCA GA | |
| Probe | CTA GGC GAC GAT CCC T |
Analysis of OMPs.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out to investigate alterations in OMPs as described previously (2). Briefly, bacterial cells of S. marcescens were disrupted by ultrasonic disintegration, and the supernatants were treated with 30% Sarkosyl (Sigma-Aldrich). OMPs were collected by centrifugation at 45,000 × g for 1 h at 4°C and were analyzed by SDS-PAGE with a Mini-Protean 3 cell apparatus (Bio-Rad). The 12.5% (wt/vol) polyacrylamide gels were visualized by staining with Coomassie brilliant blue.
Nucleotide sequence accession number.
The nucleotide data reported in this paper are available in the GenBank nucleotide database under accession number HM352623.
RESULTS
Epidemiological relationships.
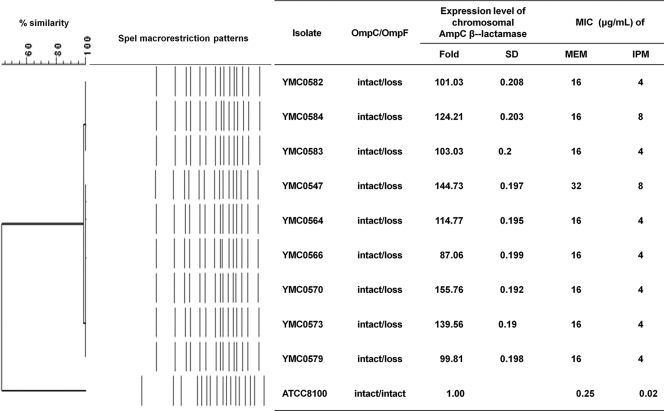
The isolates were recovered from urinary specimens of nine patients with neurological disorders such as subarachnoid hemorrhage, intracranial hemorrhage, astrocytoma, or Moyamoya disease. The patients were hospitalized at different units, such as an intensive care unit (ICU), a neurosurgical ICU, and neurological general wards, within the hospital (Table 2). However, all the isolates shared identical or similar (>85% similarity) SpeI macrorestriction patterns (Fig. 1), indicating clonal spread. The patients were treated with extended-spectrum cephalosporins or meropenem plus aminoglycosides or levofloxacin, and the condition of the patients then improved.
TABLE 2.
Description of patients and bacterial isolatesa
| Strain | Date of isolation | Specimen | Patient sex (age [yr]) | Diagnosis | Ward |
|---|---|---|---|---|---|
| YMC0547 | 12 June 2005 | Urine | F (53) | Multiple contusions | ICU |
| YMC0564 | 16 August 2005 | Urine | M (52) | Subarachnoid hemorrhage | NCU |
| YMC0566 | 18 August 2005 | Urine | M (53) | Subarachnoid hemorrhage | NCU |
| YMC0570 | 6 September 2005 | Urine | M (44) | Astrocytoma, low grade | NCU |
| YMC0573 | 17 September 2005 | Urine | F (15) | Intracerebral hemorrhage | NGW |
| YMC0579 | 12 October 2005 | Urine | F (52) | Moyamoya disease, cerebral hemorrhage | NCU |
| YMC0582 | 22 October 2005 | Urine | F (33) | Intracerebral hemorrhage | NGW |
| YMC0583 | 2 November 2005 | Urine | F (62) | Intracerebral hemorrhage | NGW |
| YMC0584 | 7 November 2005 | Urine | M (78) | Intraventricular hemorrhage | NGW |
Abbreviations: ICU, intensive care unit; NCU, neurosurgical intensive care unit; NGW, neurological general ward; F, female; M, male.
FIG. 1.
SpeI macrorestriction patterns of nine S. marcescens clinical isolates and a comparison of AmpC gene expression levels with carbapenem MICs. MEM, meropenem; IPM, imipenem.
Antimicrobial susceptibilities.
The isolates showed resistance to meropenem (MIC range, 16 to 32 μg/ml) but showed reduced susceptibility to imipenem (MIC range, 4 to 8 μg/ml) (Table 3). Despite repeated attempts, meropenem resistance was not transferred to E. coli J53 by conjugation. BA at a fixed concentration of 300 μg/ml lowered MICs of imipenem and meropenem to 0.5 to 2 μg/ml and 1 to 4 μg/ml, respectively, and cloxacillin at 250 μg/ml reduced their MICs to 0.094 to 1 μg/ml and 0.008 to 0.5 μg/ml, respectively, while the addition of CA or PAβN exhibited little effect on the MICs of those carbapenems. All nine isolates exhibited positive results in the phenotypic confirmatory test for ESBL production; however, none of them were positive in modified Hodge and IEDDS tests used to screen carbapenemase and MBL production, respectively. The results indicated that AmpC β-lactamases might be involved in acquiring carbapenem resistance in the isolates rather than ESBLs and that neither carbapenemases nor efflux pumps are involved.
TABLE 3.
MICs for S. marcescens wild strains and transconjugants
| Antimicrobial agentb | MIC (μg/ml) rangea |
||
|---|---|---|---|
| Wild strain | Transconjugant | E. coli J53 | |
| Piperacillin | >256 | 256->256 | 1 |
| Cefoxitin | >256 | 4-8 | 4 |
| Cefotaxime | >256 | 32-128 | ≤0.12 |
| Ceftazidime | 128-256 | 2-4 | 0.25 |
| Aztreonam | 64-128 | 8-16 | ≤0.12 |
| Imipenem | 4-8 | 0.12-0.25 | 0.12 |
| Imipenem + clavulanic acid | 4-8 | 0.06-0.12 | 0.12 |
| Imipenem + boronic acid | 0.5-2 | ND | ND |
| Imipenem + PAβN | 4-8 | ND | ND |
| Imipenem + cloxacillin | 0.094-1 | ND | ND |
| Meropenem | 16-32 | 0.015-0.03 | 0.03 |
| Meropenem + clavulanic acid | 16-32 | 0.015 | 0.015 |
| Meropenem + boronic acid | 1-4 | ND | ND |
| Meropenem + PAβN | 8-32 | ND | ND |
| Meropenem + cloxacillin | 0.008-0.5 | ND | ND |
| Gentamicin | >256 | 32 | 0.5 |
| Ciprofloxacin | 8-32 | 0.12 | 0.12 |
ND, not detected.
Clavulanic acid, boronic acid, cloxacillin, and PAβN were added at fixed concentrations of 4 μg/ml, 300 μg/ml, 250 μg/ml, and 20 μg/ml, respectively.
β-Lactamase characterization.
PCR and sequencing experiments detected the blaCTX-M-22 gene in all nine isolates. The probe specific for the blaCTX-M-22 gene hybridized with a 150-kbp plasmid, which was also hybridized with the probe specific for the replicon L/M. The incompatibility (Inc) group L/M plasmid was transferred to azide-resistant recipient E. coli J53 cells by conjugation. IEF of crude bacterial extracts of both wild strains and transconjugants revealed β-lactamase with a pI of 7.8. Genes encoding other types of ESBLs, plasmid-mediated AmpC β-lactamases, and class A and B carbapenemases were not detected in any of the isolates.
Expression of chromosomal AmpC β-lactamase.
Nucleotide sequences of the ampC gene PCR products from the isolates showed the closest similarity to that of S. marcescens S3 β-lactamase. The deduced sequences showed 4 amino acid differences, Q21H, E73Q, N102K, and R107H, compared to S3 β-lactamase. The deduced sequence of the ampR gene of the isolates showed 5 amino acid differences, A117T, R177H, V245A, Q249L, and R292Q, compared to that of reference S. marcescens strain ATCC 8100. RQ-PCR experiments showed that expression levels of the chromosomal ampC gene in those isolates were 87.06 to 155.76 times higher than that of reference strain ATCC 8100.
Loss of OmpF by insertion of IS1.
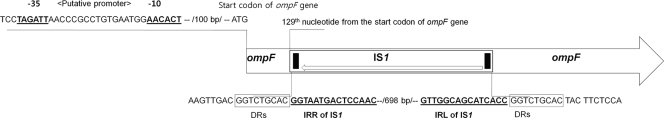
SDS-PAGE experiments showed that all nine isolates lack the 41-kDa OmpF compared to reference strain ATCC 8100 (data not shown). The 40-kDa OmpC did not show a lack of or greatly diminished expression. PCR experiments detected the ompF gene in all nine isolates. However, all the isolates yielded a ∼1.3-kb PCR product, which was larger than the expected size of 587 bp, suggesting the insertion of additional DNA. The presence of an IS1 element within the ompF gene was confirmed by direct sequencing. IS1, with 12-bp inverted repeated (IR) sequences (5′-GGTAATGACTCCAAC-3′) at each end, was inserted at the 129th base position from the start codon of the ompF gene and flanked by 9-bp direct repeated (DR) sequences (5′-GGTCTGCAC-3′) (Fig. 2).
FIG. 2.
Diagrammatic scheme of IS1 insertion into the ompF gene. The ompF gene of S. marcescens was interrupted by the insertion of the IS1 sequence at the 129th nucleotide position of the ompF gene. Putative promoter sequences of the opmF gene are indicated by boldface and underlined type. Right and left inverted repeats (IRR and IRL, respectively) of the IS1 sequence are also indicated by boldface and underlined type. Direct repeats (DRs) flanking the IS1 region are indicated by boxes.
DISCUSSION
Carbapenem resistance in the Enterobacteriaceae has been ascribed mainly to enzymatic degradation by plasmid-borne carbapenemases of classes A, B, and D (19, 20, 22, 24, 25). However, none of our meropenem-resistant S. marcescens isolates exhibited positive results in a modified Hodge test. These results indicated that they do not produce any carbapenemases and have acquired resistance by other mechanisms. Enterobacteriaceae strains have also acquired carbapenem resistance despite their lack of carbapenemase production. The loss of major OMPs has been described to play a role in the acquisition of carbapenem resistance in Enterobacteriaceae strains harboring ESBLs or plasmid-borne AmpC enzymes as well as increased efflux activity. In our study, BA and cloxacillin lowered the MICs of imipenem and meropenem for S. marcescens isolates, while CA and PAβN did not. These results suggested that AmpC enzymes are involved in the acquisition of carbapenem resistance in those isolates, whereas ESBLs and the overexpression of efflux pumps are not.
However, genes encoding plasmid-borne AmpC enzymes were not detected in all the S. marcescens isolates despite repeated experiments. Therefore, we speculated that the overproduction of the chromosomal AmpC enzyme of S. marcescens might have a role in the acquisition of carbapenem resistance in those isolates. RQ-PCR experiments showed expression levels of the chromosomal ampC gene to be 87.06 to 155.76 times higher in those isolates than in reference strain ATCC 8100. The expression of ampC is controlled by the binding of the transcriptional regulator AmpR (6). The meropenem-resistant S. marcescens clinical isolates showed 5 amino acid differences, A117T, R177H, V245A, Q249L, and R292Q, in the deduced ampR gene sequence compared to that of the susceptible reference S. marcescens strain ATCC 8100. The amino acid changes in these residues may possibly have an effect on the transcriptional regulation of chromosomal ampC expression.
S. marcescens overproducing the chromosomal AmpC enzymes exhibits a high level of resistance to oxyimino-cephalosporins, except to ceftazidime, cefepime, and cefpirome. The expansion of hydrolytic activities to ceftazidime of those enzymes can be achieved by (i) the substitution of an amino acid located in the omega loop, such as S220Y (8); (ii) the substitution of an amino acid located in the conserved residues of class C β-lactamases, such as E235K (23); and (iii) a 4-amino-acid deletion located in the H-10 helix of the β-lactamase (16, 17). The deduced sequence of the ampC gene of our isolates showed previously described amino acid differences in peripheral positions, suggesting that the AmpC enzyme in our isolates does not have expanded hydrolytic activity against ceftazidime (8, 16, 17, 18, 23).
All our isolates lack the 41-kDa OmpF protein due to the disruption of the ompF gene by the insertion of the IS1 element into the open reading frame of the gene. Various insertion sequences (ISs) have been reported to cause porin alterations (7). In combination with the overproduction of the chromosomal AmpC enzyme, the loss of OmpF may have played a role in the acquisition of meropenem resistance in our isolates. Although plasmid-borne AmpC enzymes have frequently been involved in carbapenem resistance in members of the Enterobacteriaceae (1, 12), this is the first report of the overproduction of the chromosomal AmpC enzyme contributing to carbapenem resistance. This finding is particularly disconcerting for S. marcescens, as this organism encodes an inducible, chromosomal AmpC enzyme. Together with a selection of porin-deficient mutants under antimicrobial pressure, chromosomal AmpC overproduction may present S. marcescens as a problematic nosocomial infectious agent.
Our results suggest that members of the Enterobacteriaceae harboring chromosome-borne AmpC enzymes, such as Citrobacter freundii, Enterobacter spp., and S. marcescens, could acquire carbapenem resistance without the acquisition of R plasmids carrying the genes encoding AmpC enzymes or carbapenemases. Therefore, investigation of carbapenem resistance mechanisms in these organisms may be further expanded to comprise chromosomal AmpC overproduction, and the above-mentioned organisms should be cautiously monitored for the possible development of carbapenem resistance.
Acknowledgments
This work was supported by the Korea Research Foundation (grant KRF-2008-E00194).
We thank Eun Hee Jeon at the Research Institute of Bacterial Resistance for technical assistance.
Footnotes
Published ahead of print on 27 September 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Bradford, P. A., C. Urban, N. Mariano, S. J. Projan, J. J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the foss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlone, G. M., M. L. Thomas, H. S. Rumschlag, and F. O. Sottnek. 1986. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J. Clin. Microbiol. 24:330-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A8, 8th ed. CLSI, Wayne, PA.
- 4.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing. Nineteenth informational supplement. M100-S19. CLSI, Wayne, PA.
- 5.Deshpande, L. M., R. N. Jones, T. R. Fritsche, and H. S. Sader. 2006. Occurrence and characterization of carbapenemase-producing Enterobacteriaceae: report from the SENTRY Antimicrobial Surveillance Program (2000-2004). Microb. Drug Resist. 12:223-230. [DOI] [PubMed] [Google Scholar]
- 6.Hanson, N. D., and C. C. Sanders. 1999. Regulation of inducible AmpC β-lactamase expression among Enterobacteriaceae. Curr. Pharm. Des. 5:881-894. [PubMed] [Google Scholar]
- 7.Hernandez-Alles, S., V. J. Benedi, L. Martinez-Martinez, A. Pascual, A. Aguilar, J. M. Tomas, and S. Alberti. 1999. Development of resistance during antimicrobial therapy caused by insertion sequence interruption of porin genes. Antimicrob. Agents Chemother. 43:937-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hidri, N., G. Barnaud, D. Decre, C. Cerceau, V. Lalande, J. C. Petit, R. Labia, and G. Arlet. 2005. Resistance to ceftazidime is associated with a S220Y substitution in the omega loop of the AmpC β-lactamase of a Serratia marcescens clinical isolate. J. Antimicrob. Chemother. 55:496-499. [DOI] [PubMed] [Google Scholar]
- 9.Ivanova, D., R. Markovska, N. Hadjieva, I. Schneider, I. Mitov, and A. Bauernfeind. 2008. Extended-spectrum β-lactamase-producing Serratia marcescens outbreak in a Bulgarian hospital. J. Hosp. Infect. 70:60-65. [DOI] [PubMed] [Google Scholar]
- 10.Jacoby, G. A. 2009. AmpC β-lactamases. Clin. Microbiol. Rev. 22:161-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeong, S. H., W. Song, M. J. Park, J. S. Kim, H. S. Kim, I. K. Bae, and K. M. Lee. 2008. Boronic acid disk tests for identification of extended-spectrum β-lactamase production in clinical isolates of Enterobacteriaceae producing chromosomal AmpC β-lactamases. Int. J. Antimicrob. Agents 31:467-471. [DOI] [PubMed] [Google Scholar]
- 12.Lee, C. H., C. Chu, J. W. Liu, Y. S. Chen, C. J. Chiu, and L. H. Su. 2007. Collateral damage of flomoxef therapy: in vivo development of porin deficiency and acquisition of blaDHA-1 leading to ertapenem resistance in a clinical isolate of Klebsiella pneumoniae producing CTX-M-3 and SHV-5 β-lactamases. J. Antimicrob. Chemother. 60:410-413. [DOI] [PubMed] [Google Scholar]
- 13.Lee, H. K., Y. J. Park, J. Y. Kim, E. Chang, S. G. Cho, H. S. Chae, and C. S. Kang. 2005. Prevalence of decreased susceptibility to carbapenems among Serratia marcescens, Enterobacter cloacae, and Citrobacter freundii and investigation of carbapenemases. Diagn. Microbiol. Infect. Dis. 52:331-336. [DOI] [PubMed] [Google Scholar]
- 14.Lee, K., Y. Chong, H. B. Shin, Y. A. Kim, D. Yong, and J. H. Yum. 2001. Modified Hodge and EDTA-disk synergy tests to screen metallo-β-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 7:88-91. [DOI] [PubMed] [Google Scholar]
- 15.Lee, K., Y. S. Lim, D. Yong, J. H. Yum, and Y. Chong. 2003. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-β-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 41:4623-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mammeri, H., L. Poirel, P. Bemer, H. Drugeon, and P. Nordmann. 2004. Resistance to cefepime and cefpirome due to a 4-amino-acid deletion in the chromosome-encoded AmpC β-lactamase of a Serratia marcescens clinical isolate. Antimicrob. Agents Chemother. 48:716-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumura, N., S. Minami, and S. Mitsuhashi. 1998. Sequences of homologous β-lactamases from clinical isolates of Serratia marcescens with different substrate specificities. Antimicrob. Agents Chemother. 42:176-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nomura, K., and T. Yoshida. 1990. Nucleotide sequence of the Serratia marcescens SR50 chromosomal ampC β-lactamase gene. FEMS Microbiol. Lett. 58:295-299. [DOI] [PubMed] [Google Scholar]
- 19.Osano, E., Y. Arakawa, R. Wacharotayankun, M. Ohta, T. Horii, H. Ito, F. Yoshimura, and N. Kato. 1994. Molecular characterization of an enterobacterial metallo-β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Queenan, A. M., C. Torres-Viera, H. S. Gold, Y. Carmeli, G. M. Eliopoulos, R. C. Moellering, Jr., J. P. Quinn, J. Hindler, A. A. Medeiros, and K. Bush. 2000. SME-type carbapenem-hydrolyzing class A β-lactamases from geographically diverse Serratia marcescens strains. Antimicrob. Agents Chemother. 44:3035-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song, W., H. Lee, K. Lee, S. H. Jeong, I. K. Bae, J. S. Kim, and H. S. Kwak. 2009. CTX-M-14 and CTX-M-15 enzymes are the dominant type of extended-spectrum β-lactamase in clinical isolates of Escherichia coli from Korea. J. Med. Microbiol. 58:261-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang, Y. J., P. J. Wu, and D. M. Livermore. 1990. Biochemical characterization of a β-lactamase that hydrolyzes penems and carbapenems from two Serratia marcescens isolates. Antimicrob. Agents Chemother. 34:755-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yatsuyanagi, J., S. Saito, T. Konno, S. Harata, N. Suzuki, J. Kato, and K. Amano. 2006. Nosocomial outbreak of ceftazidime-resistant Serratia marcescens strains that produce a chromosomal AmpC variant with N235K substitution. Jpn. J. Infect. Dis. 59:153-159. [PubMed] [Google Scholar]
- 24.Yum, J. H., D. Yong, K. Lee, H. S. Kim, and Y. Chong. 2002. A new integron carrying VIM-2 metallo-β-lactamase gene cassette in a Serratia marcescens isolate. Diagn. Microbiol. Infect. Dis. 42:217-219. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, R., H. W. Zhou, J. C. Cai, and G. X. Chen. 2007. Plasmid-mediated carbapenem-hydrolysing β-lactamase KPC-2 in carbapenem-resistant Serratia marcescens isolates from Hangzhou, China. J. Antimicrob. Chemother. 59:574-576. [DOI] [PubMed] [Google Scholar]