Abstract
We have previously reported the establishment of a Staphylococcus aureus laboratory strain, 10*3d1, having reduced susceptibility to daptomycin and heterogeneous vancomycin-intermediate S. aureus (VISA) phenotype. The strain was generated in vitro by serial daptomycin selection (Camargo, I. L., H. M. Neoh, L. Cui, and K. Hiramatsu, Antimicrob. Agents Chemother. 52:4289-4299, 2008). Here we explored the genetic mechanism of resistance in the strain by whole-genome sequencing and by producing gene-replaced strains. By genome comparison between 10*3d1 and its parent methicillin-resistant Staphylococcus aureus (MRSA) strain N315ΔIP, we identified five nonsynonymous single nucleotide polymorphisms (SNPs). One of the five mutations was found in the rpoB gene encoding the RNA polymerase β subunit. The mutation at nucleotide position 1862 substituted the 621st alanine by glutamic acid. The replacement of the intact rpoB with the mutated rpoB, designated rpoB(A621E), conferred N315ΔIP with the phenotypes of reduced susceptibility to daptomycin and hetero-VISA. The rpoB(A621E)-mediated resistance conversion was accompanied by a thickened cell wall and reduction of the cell surface negative charge. Being consistent with these phenotypic changes, microarray data showed that the expression of the dlt operon, which increases the cell surface positive charge, was enhanced in the rpoB(A621E) mutant. Other remarkable findings of microarray analysis of the rpoB(A621E) mutant included repression of metabolic pathways of purine, pyrimidine, arginine, the urea cycle, and the lac operon, enhancement of the biosynthetic pathway of vitamin B2, K1, and K2, and cell wall metabolism. Finally, mutations identified in rplV and rplC, encoding 50S ribosomal proteins L22 and L3, respectively, were found to be associated with the slow growth, but not with the phenotype of decreased susceptibility to vancomycin and daptomycin, of 10*3d1.
Daptomycin, a semisynthetic chemical derived from Streptomyces roseosporus, is a cyclic lipopeptide antibiotic having bactericidal activity against a broad range of aerobic and anaerobic Gram-positive bacteria (1, 27, 29, 48). The exact mechanism of action for daptomycin has not been fully elucidated yet. However, it is generally accepted that daptomycin binds to the cytoplasmic membrane of Gram-positive bacteria via calcium-dependent binding (27). Once bound, the lipopeptide tail of the molecule is inserted into the bacterial cell membrane. This tail serves as an ion channel through which an efflux of potassium, and potentially other ions, can pass through, thereby causing the bacterial cell to rapidly depolarize. Depolarization results in multiple failures in the DNA, RNA, and protein synthesis of the bacteria, ultimately resulting in a rapid bacterial cell death (27, 47). As daptomycin has a rapid bactericidal activity, it is approved in the United States and the European Union as an alternative option for the treatment of infections caused by S. aureus (44, 49).
Although daptomycin has a potent bactericidal activity against methicillin-resistant S. aureus (MRSA), it tends to have less antimicrobial activity against vancomycin-intermediate S. aureus (VISA) clinical strains (17, 43). Various groups reported that MRSA with reduced susceptibility to vancomycin and/or daptomycin were generated during the treatment with either vancomycin or daptomycin (21, 26, 32, 42). Recently, we reported the establishment of a laboratory MRSA strain 10*3d1 having dual heteroresistance to daptomycin and vancomycin by serial daptomycin selection (6). Despite its selection by daptomycin alone, this strain expressed raised resistance to both daptomycin and vancomycin, a thickened cell-wall, and a partially overlapped transcription profile with that of hetero-VISA (6). Here we explored the genetic mechanism for the dual heteroresistance of the strain 10*3d1. Using whole-genome sequencing and gene replacement experiments, we proved that a nonsynonymous mutation in rpoB, the gene that codes for the ß subunit of the bacterial DNA-dependent RNA polymerase, was responsible for the phenotypic conversion.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Staphylococcus aureus strain N315ΔIP is a derivative of strain N315, in which the repressor mecI was inactivated and the beta-lactamase plasmid was cured (31). The genotype of inactivated mecI and absence of the beta-lactamase plasmid is characteristic of recent Japanese methicillin-resistant S. aureus (MRSA) (23). 10*3d1 is an N315ΔIP derivative with a daptomycin-nonsusceptible and vancomycin-heteroresistant phenotype, obtained by gradual daptomycin exposure as described previously (6). Clinical VISA isolates Mu50 and MI, their vancomycin-susceptible in vitro derivatives Mu50-P35 and MI-P84, and respective VISA revertants Mu50-PR and MI-PR described elsewhere previously (13, 14) were used as control strains in some experiments. Among these two isogenic triple sets of VISA phenotype-associated S. aureus strains, all VISA strains are daptomycin nonsusceptible (17). All the strains were grown in brain heart infusion (BHI) broth (Difco, Detroit, MI) at 37°C with aeration if not otherwise indicated. For each experiment, an overnight culture was diluted 100-fold in prewarmed fresh BHI broth and further incubated with aeration to ensure exponential growth condition before sampling. The cell growth was monitored by measuring the optical density of the culture at 578 nm (OD578) with a spectrophotometer (Phamacia LKB Biotechnology, Inc., Uppsala, Sweden).
Genome sequencing and SNP detection of daptomycin-nonsusceptible S. aureus strain 10*3d1 with short reads.
10*3d1 genome sequencing was performed using the Solexa/Illumina genome analyzer (Illumina, Inc., San Diego, CA), a recently introduced highly parallel genome sequencer. The chromosomal genome sequence of N315 (accession number BA000018) was used as a scaffold to assemble and orient the reads. In a duplicated run of this instrument, a total of 5,833,472 36-bp-long reads were collected, giving a total coverage of an about 74.5 genome equivalent. Short reads were then aligned to the N315 genome using a short-read mapping program of MetaGenomeGambler PRO (MGG), version 2.1.2 (In Silico Biology, Inc., Yokohama, Japan) with parameters of a seed size of 12 bp and a maximum gap size allowed on a read of 3 bp to generate primary single nucleotide polymorphism (SNP) calls. Only uniquely mapped reads were retained by setting values of −1 for cost to both open and extend a gap and of 15 for cutoff overlap length. We filtered SNP calls and combined them into a single list using the Assemble program of MGG, and the resulting SNPs were verified by manually inspecting multiple alignments of all short reads mapping to each SNP locus. The identified SNPs were then verified by resequencing of the corresponding loci of N315 and 10*3d1 genomic DNAs, together with resequencing the corresponding loci of N315ΔIP (its whole-genome sequence was determined recently [28]), a parent strain of 10*3d1. The resequencing was performed using an Applied Biosystems 3730 capillary sequencer (Applied Biosystems Japan Ltd.) with forward and reverse primers for each locus.
Complete-genome comparisons of N315ΔIP and 10*3d1 using NimbleGen microarray.
Complete genome comparison of 10*3d1 and its parent strain N315ΔIP was performed using an array-based service (CGS) provided by Roche NimbleGen, Inc. (Madison, WI). Briefly, the test (10*3d1) and reference (N315ΔIP) genome DNA samples were separately cleaved to pools of low-molecular-weight fragments and labeled with fluorescent dyes, Cy3 and Cy5, respectively. The labeled samples were then competitively hybridized to a NimbleGen CGS whole-genome tiling array, which was generated with the S. aureus N315 genome sequence as a reference. Resulting hybridization signals were analyzed using NimbleScan, version 2.5, software, and the signal ratio of the number of reference samples to the number of test samples for all probes were plotted versus N315 genomic position. The locations of probes along the genome, which have a significant ratio shift between reference and test probes for both strands, represent the possible sequence differences, including SNPs, deletions, sites of insertion, endpoints of inversions, or translocations. Those locations indicated were checked by PCR and resequencing of both test and reference genomic DNA.
Electron microscopic evaluation of cell wall thickness.
Preparation and examination of S. aureus cells by transmission electron microscopy were performed as described previously (15). Morphometric evaluation of cell wall thickness was performed using photographic images at a ×30,000 final magnification. Thirty cells of each strain with nearly equatorial cut surfaces were measured for the evaluation of cell wall thickness, and results were expressed as mean value ± standard deviation (SD).
Antimicrobial susceptibility testing.
The MICs of several antimicrobials were measured for the constructed mutants and parent strains using Etest strips (AB Biodisk, Sweden). For the daptomycin MIC, the medium was supplemented with 50 mg/liter Ca2+ calcium according to Clinical and Laboratory Standards Institute (CLSI) criteria (7). A sterile cotton swab was immersed in a 0.5 McFarland standard of tested bacterial culture before streaking on sterile Mueller-Hinton (MH) agar plates. The antimicrobial strips were applied after 10 min. Plates were then incubated at 37°C and read after 24 h.
Analysis of daptomycin- and vancomycin-resistant subpopulations (population analysis).
The population analysis shows how many cells in a fixed number of cells (usually about 107 CFU) of each strain are resistant to various concentrations of antibiotics. Overnight cultures of tested strains in BHI broth were adjusted to an OD578 of 0.3. Tenfold dilutions of these cell suspensions were then prepared, and 0.1 ml of each suspension was spread on BHI agar plates containing various concentrations of daptomycin or vancomycin. Plates were incubated for 48 h at 37°C, and the log number of CFU was plotted against daptomycin or vancomycin concentration.
Growth curve and doubling time.
A portion of fresh bacterial culture was diluted to about 1 × 105 CFU/ml bacterium cells in 10 ml fresh BHI broth and grown at 37°C with 25-rpm shaking in a photo-recording incubator (TN-2612; Advantec, Tokyo, Japan). The OD was monitored automatically every 2 min, and cells were grown to an OD600 of 1.0. For growth curve and doubling time determination, OD versus time in the exponential growth phase of each strain was plotted. Doubling times were then calculated as follows: doubling time = [(t2 − t1) × log 2]/[log OD600 at t2 − log OD600 at t1].
Time-kill assay.
Time-kill methodology was used to test the strain's susceptibility to daptomycin. A final inoculum of about 1 × 106 CFU/ml bacterium cells was used. Prior to inoculation, each tube of fresh cation-adjusted Mueller-Hinton broth with 50 μg/ml CaCl2 was supplemented with daptomycin at concentrations of 0.5, 1, 2, 4, and 8 μg/ml. A tube without antibiotic was used as a growth control. The culture tubes were incubated in the TN-2612 incubator at 37°C for growth curve recording. Viability counts were performed at 0, 2, 4, 8, and 24 h by plating serially diluted cultures on MH agar plate. The growth inhibition effect of daptomycin was also evaluated by plotting the bacterial growth curve.
Introduction of chromosomal point mutations by a targeted sequence substitution method.
Plasmid pKOR1 constructed by Bae and Schneewind is an Escherichia coli/S. aureus shuttle vector which has been described as facilitating allelic replacement in S. aureus without the use of antibiotic markers (3). It permits rapid cloning via lambda recombination and ccdB selection in E. coli, while selection of knockout clones without plasmids in S. aureus is accomplished by anhydrotetracycline-mediated induction of pKOR1-encoded secY antisense transcripts, as secY is essential for S. aureus growth (3). To introduce the chromosomal point mutations identified in the drug-resistant mutant into its drug-sensitive parent strain, this method was utilized with some innovation as described previously (16, 38). Briefly, instead of cloning the up- and downstream fragments of a target gene into pKOR1 (as done in knockout experiments [3]), the gene in-frame containing the intended nucleotide sequence was amplified from sequence-donor strain 10*3d1 using primers that contain attB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-) and attB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGG-) sites on the respective up- and downstream sequences. This fragment was then cloned into pKOR1. Following cloning and ccdB selection in E. coli, the constructed plasmid was then introduced into strain N315ΔIP by electroporation using Bio-Rad GenePulser Xcell (Bio-Rad Laboratories, Inc., Hercules, CA) with the setting parameters described previously (31). Overnight culture of plasmid-carrying clones at 43°C selects for single crossover mutants that carry both mutated and corrected nucleotides. Single crossover mutants were then cultured in drug-free broth to facilitate plasmid excision and subjected to anhydotetracycline induction, whereby only nonplasmids carrying mutants could survive. To check for successful introduction of the mutations, resulting mutants were checked for the target sequences via sequencing the corresponding region with forward and reverse primers which were located 50 to 100 bp beyond the primers used in cloning. By using this strategy, the chromosomal point mutations identified in drug-resistant strain 10*3d1 were introduced into drug-sensitive parent strain N315ΔIP.
Determination of whole-cell zeta potential.
Zeta potential is a useful indicator of electronic charge of colloidal suspensions or emulsions (5, 51). The zeta potential was determined using zeta potential analyzer DelsaNano HC (Beckman Coulter, Inc., CA). Suspensions of 105 cells/ml in 10 mM phosphate-buffered saline (PBS), pH 7.4, were applied for triplicate analysis in each experiment. Briefly, the microelectrophoresis chamber was filled with a bacterial suspension, and a voltage difference of 100 V was applied over the chamber. Data represent the means and standard deviations from three independent experiments and are presented by the ratio of change in the zeta potential of resistant strain versus control strain. The zeta potential change ratio (percentage) was calculated as follows: {1 − [(Zt − Zc)/Zc]} × 100, where Zt and Zc represent zeta potential of test and control, respectively.
RNA preparation and microarray analysis.
Bacteria to be tested were grown in 10 ml BHI broth to exponential phase (OD600 = 0.6) before harvest. Pellets were then suspended with precooled T10E10 buffer (10 mM Tris-HCl [pH 8.0]), and lysostaphin solution (WAKO, Japan) at 3.0 μg/ml final concentration was added. The suspensions were then incubated at 37°C for 3 min until complete cell lysis occurred. Immediately, 7 ml of acidic phenol (pH 5.2, equilibrated with 20 mM sodium acetate) was added, and 600 μl 3 M sodium acetate was added. The samples were then frozen and thawed 3 times at −80°C and 65°C, respectively. Phenol-chloroform extraction and ethanol precipitation were then carried out. After that, the resulting RNA pellet was subjected to digestion with RNase-free DNase I (Roche, Mannheim, Germany) at 37°C for 30 min. The RNA samples were then purified again with phenol-chloroform extraction and ethanol precipitation. Pellets were then resuspended in 25 μl diethyl pyrocarbonate (DEPC)-treated water. The construction and analysis of the DNA microarray have been described previously (13).
Statistical analysis of data.
The statistical significance of the data was evaluated with Student's t test.
Microarray data accession number.
Transcriptional profiles of RpoB621E mutation-related strains may be found under CIBEX accession no. CBX135.
RESULTS AND DISCUSSION
Comparison of the chromosomes of 10*3d1 and N315ΔIP.
To study the mechanism of daptomycin resistance in S. aureus, we have established MRSA strain 10*3d1 with reduced susceptibility to daptomycin (6). The strain was derived from N315ΔIP, a daptomycin- and vancomycin-susceptible laboratory MRSA strain (6, 28, 31) by serial daptomycin selection. The strain unexpectedly showed decreased susceptibility to both daptomycin and vancomycin and expressed some of the VISA characteristics of thickened cell wall and decreased growth rate, as well as the transcription profile partially overlapping that of hetero-VISA (6). Since we noticed cross-resistance of VISA clinical strains to vancomycin and daptomycin in early 2006 (17), the unexpected acquisition of the dual resistance by 10*3d1 prompted us to carry out a whole-genome comparison of 10*3d1 and N315ΔIP and a gene replacement experiment to clarify the genetic basis for the phenomenon. Our strategy was (i) whole-genome sequencing of the 10*3d1 chromosome and identifying SNPs in comparison with its isogenic parent strain N315ΔIP and (ii) introduction of the SNPs into the N315ΔIP chromosome by single-gene replacement to test the effect of the mutations on vancomycin and daptomycin susceptibilities.
We employed two methods for SNP detection: whole-genome sequencing by using a Solexa/Illumina genome analyzer and competitive DNA hybridization using the NimbleGen microarray provided by Roche NimbleGen, Inc. These two methods were complementary to each other in terms of the SNP detection sensitivity and accuracy and genome scale coverage. Table 1 lists the SNPs detected by the above two methods and confirmed by manual validation resequencing. A total of 5 SNPs located in 4 distinct genes were identified, all of which caused amino acid substitution.
TABLE 1.
List of nucleotide differences between N315ΔIP and 10*3d1 chromosomes
| SNP no.a | Position on N315 genome | Nucleotide |
N315 genome information |
Amino acid change (N315→10*3d1)b | ||||
|---|---|---|---|---|---|---|---|---|
| N315 | N315ΔIP | 10*3d1 | ID | Gene | Product | |||
| S-1 | 581481 | C | C | A | SA0500 | rpoB | RNA polymerase beta chain | A621E |
| S-2 | 2065641 | G | G | T | SA1826 | Hypothetical protein | A14E | |
| S-3 | 2305476 | C | C | T | SA2042 | rplV | 50S ribosomal protein L22 | c |
| S-4 | 2307902 | C | C | G | SA2047 | rplC | 50S ribosomal protein L3 (BL3) | G120D |
| S-5 | 2307947 | C | C | T | SA2047 | rplC | 50S ribosomal protein L3 (BL3) | G135A |
“S” stands for single nucleotide polymorphism (SNP).
The number denotes the position of amino acid substitution. The left amino acid is that of N315, and the right that of 10*3d1. A, alanine; E, glutamic acid; G, glycine; D, aspartate.
The initiation codon is changed from ATG to ATA.
Exploring the effect of SNPs of 10*3d1.
To test which SNP is responsible for the raised vancomycin and daptomycin resistance, we introduced the SNPs one by one into the N315ΔIP chromosome. We used a targeted sequence substitution method for the SNP introduction (16, 38), whereby the SNPs were introduced directly on the chromosome without insertion or deletion of any additional sequence or antibiotic selection markers.
(i) The effect of the SA1826 mutation.
Open reading frame (ORF) SA1826 encodes a conserved hypothetical protein with its two domains similar to penicillinase repressor and type III secretion needle MxiH. We first introduced the SA1826 mutation into N315ΔIP, constructing N315ΔIP-SA182614E and N315ΔIP-SA182614A (Table 2). The latter was a revertant from the recombination intermediate strain regaining the original intact SA1826 gene. Such revertants were obtained in every allelic replacement procedure and used as isogenic control strains throughout this study (Table 2). The constructed mutant strains were then determined for their susceptibilities to daptomycin and vancomycin. Etest determinations found no difference of daptomycin and vancomycin susceptibilities between N315ΔIP-SA182614E and N315ΔIP-SA182614A (Table 2), indicating that the SA1826 mutation was not responsible for the altered drug resistance phenotype in 10*3d1.
TABLE 2.
SNP type, antibiotic susceptibility profile, and doubling time of S. aureus strains used in this study
| Strain | Sequence typea |
MIC (mg/liter) by Etestb |
Doubling time (min) | Reference for strains | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RpoB621 | SA182614 | rplV1∼3 | RplC120 | RplC135 | Daptomycin | Vancomycin | Oxacillin | Rifampin | |||
| N315ΔIP | A | A | ATG | G | G | 0.38 | 0.5 | 48 (256)c | 0.006 | 35.73 | 6 |
| 10*3d1 | E | E | ATA | D | A | 2 | 3 | >256 | 0.006 | 61.17 | 6 |
| N315ΔIP-rpoB621A | A | A | ATG | G | G | 0.25 | 0.25 | >256 | 0.006 | 38.22 | This study |
| N315ΔIP-rpoB621E | E | A | ATG | G | G | 1.5 | 3 | >256 | 0.006 | 39.31 | This study |
| N315ΔIP-SA182614A | A | A | ATG | G | G | 0.25 | 0.5 | >256 | 0.006 | 36.75 | This study |
| N315ΔIP-SA182614E | A | E | ATG | G | G | 0.25 | 0.5 | >256 | 0.006 | 36.83 | This study |
| 10*3d1-SA182614A | E | A | ATA | D | A | 1.5 | 3 | >256 | 0.006 | 66.46 | This study |
| 10*3d1-SA182614E | E | E | ATA | D | A | 1.5 | 3 | >256 | 0.006 | 70.54 | This study |
| 10*3d1-SA182614A-rpoB621A | A | A | ATA | D | A | 0.25 | 0.75 | >256 | 0.006 | 59.61 | This study |
| 10*3d1-SA182614A-rpoB621E | E | A | ATA | D | A | 1.5 | 3 | >256 | 0.006 | 72.12 | This study |
Substituted amino acids are given, except for rplV, for which the mutated initiation codon (first three nucleotides) is given. A, alanine; E, glutamic acid; G, glycine; D, aspartate.
Etest was performed with Mueller-Hinton agar, and the results were read after 24 h of incubation at 37°C.
MIC for the minority subpopulations is given in parentheses.
(ii) The effect of the rpoB mutation.
Next, we introduced the mutated rpoB into N315ΔIP, obtaining strains N315ΔIP-rpoB621E and N315ΔIP-rpoB621A (Table 2). To validate if the rpoB(A621E) mutation is a unique genetic alteration during the N315ΔIP-rpoB621E construction, we performed the whole-genome sequence comparison between N315ΔIP-rpoB621E and N315ΔIP using the NimbleGen microarray. We found no detectable mutation except for rpoB(A621E) in the N315ΔIP-rpoB621E in comparison to N315ΔIP (data not shown), supporting the hypothesis that phenotype changes of N315ΔIP- rpoB621E were caused by the rpoB(A621E) mutation. rpoB encodes the β subunit of the bacterial RNA polymerase. RpoB is a well-known target of action of rifampin, and its mutation is frequently associated with resistance to rifampin. The position of the mutation in 10*3d1, however, was found outside the previously identified rifampin resistance regions of RpoB known to confer rifampin resistance (2, 36). In fact, the mutation was not accompanied by rifampin resistance in either 10*3d1 or N315ΔIP-rpoB621E (Table 2). However, susceptibility tests showed that the rpoB(A621E) mutation caused a simultaneous increase of daptomycin and vancomycin MICs; the increment of daptomycin MIC was from 0.25 mg/liter to 1.5 mg/liter, and that of the vancomycin MIC was from 0.25 mg/liter to 3.0 mg/liter (Table 2). The extent of MIC changes were comparable to that between N315ΔIP and 10*3d1; N315ΔIP had a daptomycin MIC of 0.38 mg/liter and a vancomycin MIC of 0.5 mg/liter, whereas it was 2.0 mg/liter and 3.0 mg/liter, respectively, for 10*3d1 (Table 2 and Fig. 1).
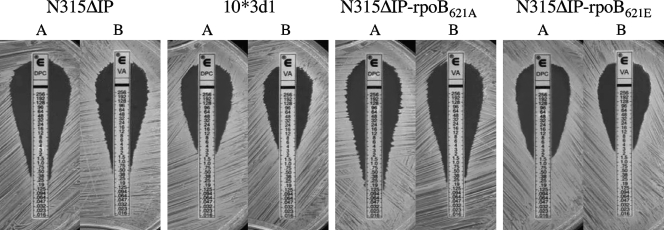
FIG. 1.
Daptomycin (A) and vancomycin (B) MICs were determined by Etest for N315ΔIP-rpoB621E and its relevant strains. Note that N315ΔIP-rpoB621E has reduced daptomycin and vancomycin susceptibilities similar to those of 10*3d1.
(iii) The effect of rplV and rplC mutations.
All our trials failed to introduce the mutations identified in rplV and rplC of 10*3d1 into N315ΔIP with unknown reason. To evaluate the effect of the two mutations in the ribosomal protein genes collectively, we changed the strategy of experiments. Instead of introducing mutations into N315ΔIP, we sequentially cured the SA1826 and rpoB mutations of 10*3d1, obtaining the strain 10*3d1-SA186214A-rpoB621A which harbors mutations only in the two rpl genes compared to N315ΔIP. The obtained mutant strain retained the slow growth phenotype of 10*3d1. Thus, it appears that the rplV and rplC mutations were the cause for the slow growth of 10*3d1. As shown in Table 2, restoration of the rpoB mutation but not the SA1862 mutation of 10*3d1 led to a significant decrease of both daptomycin and vancomycin resistance to the susceptible levels of N315ΔIP, while there was no susceptibility change in the control strain 10*3d1-SA186214A-rpoB621E. This clearly indicated that the mutations of rplV and rplC alone had no significant effect on vancomycin and daptomycin resistance. This experiment also confirmed that the rpoB(A621E) mutation alone is responsible for the dual heteroresistance to vancomycin and daptomycin.
Slow growth has been repeatedly reported for VISA strains and is considered one of the factors associated with VISA phenotype expression in S. aureus (10, 14, 19, 46). In our previous study, 10*3d1 showed a significantly decreased growth rate compared to its daptomycin- and vancomycin-susceptible parent strain N315ΔIP (6). We considered that the slow growth was associated with the expression of the resistance phenotype (6). However, as shown in Table 2 and Fig. 2, the rpoB-restored strain 10*3d1-SA186214A-rpoB621A retained the slow growth phenotype of 10*3d1 after having lost the vancomycin and daptomycin dual resistance. Thus, the slow growth rate itself did not influence the MIC values in any significant way.
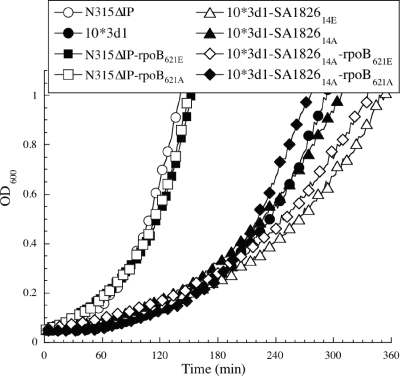
FIG. 2.
Growth curve characteristics of strains exhibiting two groups of different growth rates. Note that strains having rplC and rplV mutations (see Table 2) showed significantly slower growth than those without rplC and rplV mutations.
Although the slow growth phenotype did not raise MIC values, it may affect the bactericidal activities of daptomycin and vancomycin. To test this possibility, we evaluated the growth inhibition and time-kill effect of daptomycin against N315ΔIP and 10*3d1-SA186214A-rpoB621A. The two strains had the same susceptibility to bactericidal effects of daptomycin irrespective of the difference in the growth rates (Fig. 2 and Fig. 3). Thus, the slow growth rate was not a major contributor in the reduced susceptibility to daptomycin and vancomycin of 10*3d1.
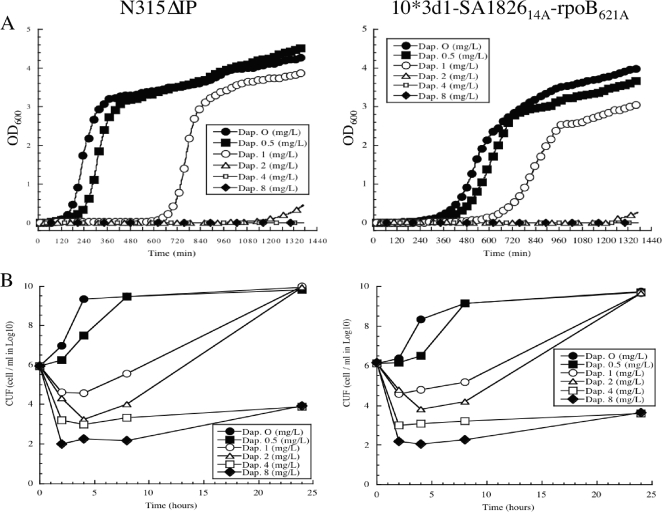
FIG. 3.
Growth inhibition (A) and time-killing effect (B) of daptomycin against strains N315ΔIP and 10*3d1-SA182614A-rpoB621A. Killing activity is presented as log10 CFU/ml.
The rpoB(A621E) mutation confers daptomycin and vancomycin heteroresistance to N315ΔIP.
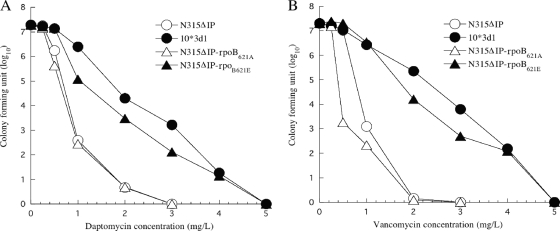
The introduction of the rpoB(A621E) mutation in N315ΔIP raised MICs for daptomycin and vancomycin as shown in Table 2. We previously showed that the types of daptomycin and vancomycin resistance expressed by strain 10*3d1 were heterogenous (6). In order to determine if the resistance pattern conferred by the rpoB(A621E) mutation was heterogeneous or not, we analyzed the resistant subpopulation profile of the rpoB(A621E)-carrying strain against vancomycin and daptomycin. Figure 4 shows the results. Population analysis showed that N315ΔIP-rpoB621E, compared to N315ΔIP-rpoB621A and parent strain N315ΔIP, had a significant increase in resistant subpopulations for both daptomycin and vancomycin, and the pattern of the population curves were similar to that of 10*3d1. Therefore, the rpoB(A621E) mutation alone could cause the heterogenous resistance phenotype of 10*3d1 against daptomycin and vancomycin.
FIG. 4.
Analysis of daptomycin- and vancomycin-resistant subpopulations of N315ΔIP-rpoB621E and its isogenic strains. The number of colonies on plates containing various concentrations of daptomycin (A) and vancomycin (B) was counted after 48 h of incubation at 37°C.
Friedman et al. (18) recently reported a finding of two strains with RpoB mutations, Ile953→Ser953 and Ala1086→Val1086, in a series of laboratory-derived S. aureus strains with decreased susceptibility to daptomycin. However, the two strains with the rpoB mutation also carried additional mutations in mprF and yycG. Furthermore, since they did not find the rpoB mutations in a collection of clinical isolates with decreased susceptibility to daptomycin, the role of the rpoB mutation was not explored any further. In this study, by using a gene replacement method, we unequivocally proved that an rpoB mutation can be a cause of dual heteroresistance to vancomycin and daptomycin.
rpoB(A621E) mutation causes thickening of the cell wall.
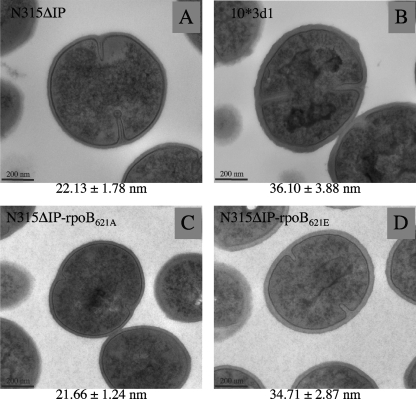
How does the rpoB mutation cause reduced susceptibility to daptomycin and vancomycin? The most significant feature of the VISA phenotype is cell wall thickness (11, 14, 22). The feature is directly associated with the “peptidoglycan-clogging mechanism” that prevents the passage of vancomycin across the thickened peptidoglycan layers (12). Unlike vancomycin, daptomycin does not have binding sites in the peptidoglycan, so the clogging effect may not apply to daptomycin resistance. However, daptomycin has a relatively bigger molecular size than vancomycin (molecular weight, 1,620.67 versus 1,485.7). Moreover, daptomycin forms oligomers with calcium ions, which dissociates only when the oligomers come close to the bacterial membrane (45). Therefore, the thickened peptidoglycan layer may serve as an obstacle for the access of daptomycin to the cell membrane. In agreement with our hypothesis (17), 10*3d1 showed a significantly thickened cell wall compared to that of its drug-susceptible parent strain N315ΔIP (6). Figure 5 shows the electronmicrograph showing that the strain N315ΔIP-rpoB621E harboring the rpoB(A621E) mutation had a much more thickened cell wall than that of the control strain N315ΔIP-rpoB621A. The extent of the cell wall thickening was 1.60 times, which was comparable to the case of 10*3d1 (whose cell wall was thicker by 63% than that of its parent strain N315ΔIP) (Fig. 5). Therefore, the rpoB mutation is considered to contribute to the heteroresistance to vancomycin and daptomycin by thickening the cell wall.
FIG. 5.
Transmission electron microscopic observation of N315ΔIP-rpoB621E and its isogenic strains. Transmission electron microscopy was carried out on N315ΔIP-rpoB621E (D), which was generated from N315ΔIP (A) by substitution of its rpoB gene with that of 10*3d1 (B), and experimental control strain N315ΔIP-rpoB621A (C), which retained the intact rpoB gene. Magnification is ×30,000. The values given under each picture are the means and standard deviations of each strain's cell wall thickness in nanometers. Note that the cell walls of N315ΔIP-rpoB621E and 10*3d1 were much thicker than those of the control and parent strains.
Decreased negative cell surface charge associated with the rpoB mutation.
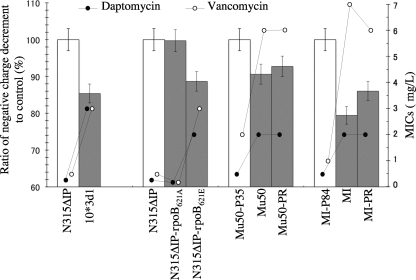
Decreased negative cell surface charge has been reported as one of the contributing factors for the increased daptomycin and vancomycin resistance in S. aureus (25, 33, 52), based on the charge repulsion mechanism assumption. Daptomycin and vancomycin are relatively positively charged at a neutral pH (4, 8, 20, 24). Therefore, if a bacterial cell generates a less negative cell surface, it would reduce the binding of vancomycin and daptomycin molecules to the cell (25, 33). In the case of daptomycin, however, there has been a discrepant observation that the increased negative cell surface charge is observed in in vitro-selected daptomycin-nonsusceptible mutants (34). Therefore, the charge repulsion mechanism might not be generally applicable or not in its simplest form. In any case, to test the contribution of the cell surface charge in our in vitro-selected strains, we determined the whole-cell surface charge of the constructed mutants with some control strains. The cell surface charge was measured by determining the bacterial zeta potential, which is an indicator of the electronic charge of colloidal suspensions (5, 51). A total of two groups of strains were used for the zeta potential determination. The first group consisted of N315ΔIP-rpoB621E, N315ΔIP-rpoB621A, 10*3d1, and N315ΔIP. The second group consisted of clinical VISA strains Mu50 and MI and their derivative substrains, passage-derived vancomycin-susceptible strains Mu50-P35 and MI-P84, and Mu50-PR and MI-PR, which were derived from the vancomycin-susceptible derivative strains by selection with 4 mg/liter of vancomycin (14). Their vancomycin and daptomycin MICs were as follows: the vancomycin MICs for Mu50, Mu50-P35, and Mu50-PR were 6, 2, and 6 mg/liter, respectively, and those for MI, MI-P84, and MI-PR were 7, 1, and 6 mg/liter, respectively. The daptomycin MICs for them were 2, 0.5, and 2, and 2, 0.5, and 2 mg/liter, respectively (14, 17). All the vancomycin-susceptible strains used in this experiment were susceptible to daptomycin, while the vancomycin-resistant strains were also resistant to daptomycin (17). Figure 6 shows the summary of decreased negative charge calculations of drug-resistant strains against their drug-susceptible isogenic control strains. Zeta potential values for control strain N315ΔIP, Mu50-P35, and MI-P84 were −28 ± 1.4, −30 ± 1.6, and −35 ± 1.5 mV, respectively. We calculated the ratio (percentage) of negative charge decrement in resistant strains to their respective daptomycin- and vancomycin-susceptible counterparts. It was noted that all the daptomycin- and vancomycin-resistant strains had a decreased negative charge compared to that of their susceptible controls (P < 0.01), and there was a positive correlation between the reduction of negative charge and the reduced susceptibility to daptomycin and vancomycin (Fig. 6). The results indicated that the decreased negative surface charge might contribute to the daptomycin and vancomycin resistance in 10*3d1 as well as in clinical VISA strains Mu50 and MI.
FIG. 6.
Decrease of whole-cell negative charge in daptomycin- and vancomycin-resistant strains. Whole-cell zeta potential, an indicator of the cell electronic charge, was determined at pH 7.4, and the zeta potential change ratio (percentage) (box bar) of the test strain (filled box) to its respective control (open box) was calculated as follows: {1 − [(Zt − Zc)/Zc] × 100}, where Zt and Zc represent the Zeta potential of tthe est and control, respectively. Values are means ± standard deviations.
Rifampin resistance associated with rpoB point mutations is a well-recognized phenomenon in S. aureus (2, 39, 50). The mutations are clustered predominantly in a conserved region of RpoB, designated rifampin resistance-determining region (RRDR), which spans amino acid residues ∼463 to 550. Mutations in the region result in a lessening of the hydrophobic interaction between RpoB and rifampin and a decrease in the binding of rifampin to RNA polymerase holoenzyme (40). The rpoB mutation identified in 10*3d1 is located outside the RRDR, and this coincides with the observation that the rpoB(A621E) mutation does not affect rifampin susceptibility of the strain (Table 2). However, the effect of this mutation on RNA polymerase activity and the detailed mechanism causing heteroresistance to both daptomycin and vancomycin remains to be elucidated.
rpoB mutations are also found in clinical VISA strain Mu50 (38) and JH9 (37). Our finding of the contribution of an rpoB mutation to daptomycin and vancomycin dual resistance in this study indicates that the rpoB mutations also contribute to the VISA phenotype of clinical MRSA strains.
Microarray transcriptional analysis.
In our previous attempt to understand the global regulatory mechanism for the expression of daptomycin resistance in 10*3d1, we performed microarray analysis on 10*3d1 and its parent strain N315ΔIP. The results, including raw signal data, were disclosed at the CIBEX site (http://cibex.nig.ac.jp/index.jsp) under the CIBEX accession number CBX67. By comparing the transcriptional profiles of 10*3d1and N315ΔIP, we identified dozens of specifically up- and downregulated genes in 10*3d1, including those involved in cell wall and cell membrane metabolism (6). However, since 10*3d1 had 5 single point mutations in 4 distinct genes compared to N315ΔIP (Table 1) and all of these mutations might have some influence on the transcriptional profiles of 10*3d1, the alterations of transcriptional profiles of 10*3d1 might not have directly reflected the effect of the rpoB(A621E) mutation. To better delineate the effect of the rpoB(A621E) mutation causing dual resistance to daptomycin and vancomycin, we performed microarray analysis of three combinations of strains: N315ΔIP-rpoB621E versus N315ΔIP, 10*3d1-SA182614A-rpoB621E versus 10*3d1-SA182614A-rpoB621A, and 10*3d1 versus N315ΔIP (Table 2). There is only one mutation difference [rpoB(A621E)] within each of the first two pairs. The last pair represents the cumulative effects of the five mutations (6).
All transcriptional profiles and comparison data are available on the CIBEX site under the accession number CBX135, and Table 3 shows the extracted representative data. The most significant alterations apparently caused by the rpoB(A621E) mutation were (i) repression of the genes in the purine and pyrimidine metabolic pathways; (ii) repression of urease genes; (iii) enhancement of the biosynthetic pathway genes for riboflavin (vitamin B2), phylloquinone (vitamin K1), and menaquinone (vitamin K2); and (iv) enhancement of several genes involved in the cell envelope metabolism and structure (Table 3). It is not clear yet how the rpoB(A621E)-mediated altered gene transcription profile mentioned above is correlated with the phenotypic changes such as cell wall thickening, decreased cell surface negative charge, and reduced susceptibility to vancomycin and daptomycin. The products of the dlt operon genes are reported to have a function of attaching positively charged d-alanine residues onto the negatively charged phosphate groups in the backbone of teichoic acids in S. aureus (9, 30, 41). An enhanced expression of the dlt operon associated with an increased cell surface positive charge was observed in daptomycin-resistant MRSA (34). The singly introduced rpoB(A621E) mutation in N315ΔIP did enhance the expression of dlt genes. However, curiously, the enhancement was not apparent in 10*3d1 (Table 3). The same was true with the sigB gene, whose overexpression is reported to thicken the cell wall (35). sigB was upregulated by the rpoB(A621E) mutation in N315ΔIP, but the effect was not apparent in the parent strain 10*3d1 (Table 3). Besides these genes, however, there were quite a number of genes involved in cell envelope metabolism enlisted as the genes up- or downregulated by the rpoB(A621E) mutation (Table 3). The altered expression of such genes as sle1, atl, stcA, SA1898 (sceD homolog), SA2097 (ssaA homolog), capABC, SA0126, SA0127, SA2284, SA2285, mgrA, fmtB, and sarUT may modify the cell envelope physiology and structure in a way that evades the attack of vancomycin and daptomycin.
TABLE 3.
Transcriptional profiles of RpoBA621E mutation-related strains
| ORF ID | Gene | Products | Ratio of signal intensitya |
||
|---|---|---|---|---|---|
| N315ΔIP-rpoB621E/N315ΔIP | 10*3d1-SA182614A-rpoB621E/10*3d1-SA182614A-rpoB621A | 10*3d1/ N315ΔIP | |||
| Purine metabolism | |||||
| SA0016 | purA | Adenylosuccinate synthase | 0.36 | 0.64 | 0.76 |
| SA0373 | xprT | Xanthine phosphoribosyltransferase | 0.64 | 0.40 | 0.12 |
| SA0374 | pbuX | Xanthine permease | 0.55 | 0.42 | 0.10 |
| SA0375 | guaB | Inositol-monophosphate dehydrogenase | 0.90 | 0.50 | 0.18 |
| SA0376 | guaA | GMP synthase (glutamine hydrolyzing) | 0.88 | 0.65 | 0.23 |
| SA0468 | Hypoxanthine-guanine phosphoribosyltransferase homologue | 1.51 | 1.18 | 1.53 | |
| SA0916 | Similar to phosphoribosylaminoimidazole carboxylase PurE | 0.84 | 0.80 | 0.72 | |
| SA0917 | purK | Phosphoribosylaminoimidazole carboxylase carbon dioxide-fixation chain PurK homolog | 0.93 | 0.54 | 0.43 |
| SA0918 | purC | Phosphoribosylaminoimidazolesuccinocarboxamide synthetase homolog | 1.00 | 0.57 | 0.43 |
| SA0919 | Conserved hypothetical protein | 0.96 | 0.69 | 0.45 | |
| SA0920 | purQ | Phosphoribosylformylglycinamidine synthase I PurQ | 1.10 | 0.52 | 0.32 |
| SA0922 | purF | Phosphoribosylpyrophosphate amidotransferase PurF | 0.81 | 0.52 | 0.26 |
| SA0923 | purM | Phosphoribosylformylglycinamidine cyclo-ligase PurM | 0.77 | 0.54 | 0.29 |
| SA0924 | purN | Phosphoribosylglycinamide formyltransferase | 0.79 | 0.56 | 0.30 |
| SA0925 | purH | Bifunctional purine biosynthesis protein PurH | 0.75 | 0.59 | 0.33 |
| SA0926 | purD | Phosphoribosylamine-glycine ligase PurD | 0.61 | 0.55 | 0.36 |
| SA1052 | gmk | Guanylate kinase homolog | 0.81 | 0.91 | 0.81 |
| SA1172 | Similar to GMP reductase | 0.34 | 0.37 | 0.57 | |
| SA1173 | Conserved hypothetical protein | 0.28 | 0.33 | 0.58 | |
| SA1461 | apt | Adenine phosphoribosyl transferase | 0.94 | 1.30 | 1.41 |
| Pyrimidine metabolism | |||||
| SA0440 | tmk | Thymidylate kinase homolog | 0.64 | 0.60 | 0.74 |
| SA0719 | trxB | Thioredoxine reductase | 0.45 | 0.46 | 0.68 |
| SA1041 | pyrR | Pyrimidine operon repressor chain A | 0.17 | 0.35 | 0.82 |
| SA1042 | pyrP | Uracil permease | 0.14 | 0.21 | 0.41 |
| SA1043 | pyrB | Aspartate transcarbamoylase chain A | 0.13 | 0.22 | 0.41 |
| SA1044 | pyrC | Dihydroorotase | 0.15 | 0.25 | 0.39 |
| SA1045 | pyrAA | Carbamoyl-phosphate synthase small chain | 0.18 | 0.28 | 0.40 |
| SA1046 | pyrAB | Carbamoyl-phosphate synthase large chain | 0.18 | 0.32 | 0.38 |
| SA1047 | pyrF | Orotidine-5-phosphate decarboxylase | 0.22 | 0.40 | 0.40 |
| SA1048 | pyrE | Orotate phosphoribosyltransferase | 0.20 | 0.43 | 0.41 |
| SA1049 | Hypothetical protein | 0.22 | 0.42 | 0.47 | |
| SA1439 | udk | Uridine kinase | 0.68 | 0.74 | 1.32 |
| SA2375 | Similar to dihydroorotate dehydrogenase | 0.62 | 0.75 | 0.56 | |
| Arginine metabolism and urea cycle | |||||
| SA0175 | Conserved hypothetical protein | 0.34 | 0.42 | 0.24 | |
| SA0176 | Similar to N-acetylglutamate 5-phosphotransferase | 0.63 | 0.76 | 0.64 | |
| SA0177 | argJ | Arginine biosynthesis bifunctional protein homolog | 0.61 | 0.76 | 0.64 |
| SA0178 | argC | N-acetylglutamate gamma-semialdehyde dehydrogenase | 0.59 | 0.73 | 0.65 |
| SA0179 | Ornithine aminotransferase | 0.64 | 0.80 | 0.82 | |
| SA0821 | argH | Argininosuccinate lyase | 0.44 | 0.54 | 0.27 |
| SA0822 | argG | Argininosuccinate synthase | 0.48 | 0.59 | 0.29 |
| SA2082 | ureA | Urease gamma subunit | 0.37 | 0.26 | 0.28 |
| SA2083 | ureB | Urease beta subunit | 0.23 | 0.19 | 0.24 |
| SA2084 | ureC | Urease alpha subunit | 0.30 | 0.21 | 0.24 |
| SA2085 | ureE | Urease accessory protein UreE | 0.26 | 0.19 | 0.24 |
| SA2086 | ureF | Urease accessory protein UreF | 0.30 | 0.27 | 0.29 |
| SA2087 | ureG | Urease accessory protein UreG | 0.44 | 0.33 | 0.32 |
| SA2088 | ureD | Urease accessory protein UreD | 0.39 | 0.30 | 0.32 |
| SA2428 | arcA | Arginine deiminase | 2.82 | 2.75 | 1.45 |
| SA2429 | Similar to arginine repressor | 3.62 | 6.20 | 2.27 | |
| Galactose metabolism | |||||
| SA0123 | Similar to UDP-glucose 4-epimerase (gale-1) | 0.34 | 0.45 | 0.41 | |
| SA0236 | Similar to PTS fructose-specific enzyme IIBC component | 0.68 | 0.83 | 0.82 | |
| SA0237 | Similar to PTS, galactitol-specific IIB component | 0.65 | 0.71 | 0.93 | |
| SA0238 | gatC | Probable PTS galactitol-specific enzyme IIC component | 0.64 | 0.81 | 1.28 |
| SA1521 | pfk | 6-Phosphofructokinase | 0.77 | 0.60 | 0.53 |
| SA1991 | lacG | 6-Phospho-beta-galactosidase | 0.33 | 0.73 | 1.75 |
| SA1992 | lacE | PTS system, lactose-specific IIBC component | 0.35 | 0.63 | 1.40 |
| SA1993 | lacF | PTS system, lactose-specific IIA component | 0.32 | 0.62 | 1.38 |
| SA1994 | lacD | Tagatose 1,6-diphosphate aldolase | 0.39 | 0.59 | 1.25 |
| SA1995 | lacC | Tagatose-6-phosphate kinase | 0.38 | 0.56 | 1.22 |
| SA1996 | lacB | Galactose-6-phosphate isomerase LacB subunit | 0.42 | 0.53 | 1.17 |
| SA1997 | lacA | Galactose-6-phosphate isomerase LacA subunit | 0.42 | 0.54 | 1.10 |
| SA2288 | gtaB | UTP-glucose-1-phosphate uridyltransferase | 0.63 | 0.69 | 0.53 |
| Nitrogen metabolism | |||||
| SA0008 | hutH | Histidine ammonia-lyase | 1.27 | 1.50 | 1.25 |
| SA0171 | fdh | NAD-dependent formate dehydrogenase | 0.81 | 0.78 | 1.18 |
| SA0774 | ABC transporter ATP-binding protein homolog | 2.85 | 2.95 | 3.28 | |
| SA0775 | FeS assembly protein SufD | 2.84 | 3.05 | 3.71 | |
| SA0776 | Aminotransferase NifS homolog | 2.76 | 3.12 | 3.41 | |
| SA0777 | Similar to nitrogen fixation protein NifU | 2.58 | 3.15 | 3.44 | |
| SA0778 | sufB | FeS assembly protein SufB | 2.27 | 2.91 | 3.23 |
| SA0779 | Hypothetical protein | 1.91 | 2.11 | 2.30 | |
| SA0819 | gudB | NAD-specific glutamate dehydrogenase | 1.78 | 2.14 | 1.75 |
| SA1241 | Similar to nitric-oxide reductase | 2.37 | 1.42 | 2.29 | |
| SA1310 | ansA | Probable l-asparaginase | 1.31 | 1.36 | 1.18 |
| dlt operon | |||||
| SA0792 | Hypothetical protein | 1.56 | 2.12 | 0.98 | |
| SA0793 | dltA | d-alanine-d-alanyl carrier protein ligase | 1.61 | 2.27 | 0.88 |
| SA0794 | dltB | DltB membrane protein | 1.60 | 2.54 | 0.81 |
| SA0795 | dltC | d-alanyl carrier protein | 1.55 | 2.60 | 0.83 |
| SA0796 | dltD | Poly(glycerophosphate chain) d-alanine transfer protein | 1.44 | 2.52 | 0.84 |
| sigB operon | |||||
| SA1869 | sigB | Sigma factor B | 2.96 | 2.05 | 1.19 |
| SA1870 | rsbW | Anti-sigmaB factor | 3.15 | 1.90 | 1.14 |
| SA1871 | rsbV | Anti-sigmaB factor antagonist | 2.64 | 1.74 | 1.22 |
| SA1872 | rsbU | SigmaB regulation protein RsbU | 2.08 | 1.62 | 1.22 |
| Riboflavin (vitamin B2) metabolism | |||||
| SA1586 | ribH | 6,7-Dimethyl-8-ribityllumazine synthase | 1.75 | 1.88 | 2.03 |
| SA1588 | ribB | Riboflavin synthase alpha chain | 1.92 | 1.94 | 2.20 |
| SA1589 | ribD | Riboflavin-specific deaminase | 2.03 | 2.04 | 2.08 |
| SA1587 | ribA | Riboflavin biosynthesis protein | 2.18 | 1.99 | 2.16 |
| Phylloquinone (vitamin K1) and menaquinone (vitamin K2) metabolism | |||||
| SA0898 | menB | Naphthoate synthase | 1.08 | 1.30 | 1.33 |
| SA1303 | gerCB | Menaquinone biosynthesis methyltransferase | 1.14 | 1.00 | 1.16 |
| SA0895 | Similar to menaquinone-specific isochorismate synthase | 1.79 | 1.44 | 2.66 | |
| SA1616 | Hypothetical protein | 1.85 | 2.12 | 2.37 | |
| SA0897 | Similar to prolyl aminopeptidase (EC 3.4.11.5) | 1.91 | 1.74 | 1.91 | |
| SA0896 | menD | Menaquinone biosynthesis protein | 1.93 | 1.65 | 2.33 |
| SA1615 | menE | O-succinylbenzoic acid-coenzyme A (CoA) ligase | 2.12 | 1.76 | 2.18 |
| SA1614 | menC | O-succinylbenzoic acid (OSB) synthetase | 2.14 | 1.65 | 1.78 |
| Citrate cycle (TCA cycle) | |||||
| SA0944 | phdB | Pyruvate dehydrogenase E1 component beta subunit | 1.25 | 0.97 | 0.90 |
| SA0945 | pdhC | Dihydrolipoamide S-acetyltransferase component of pyruvate dehydrogenase complex E2 | 1.28 | 1.02 | 0.97 |
| SA0946 | pdhD | Dihydrolipoamide dehydrogenase component of pyruvate dehydrogenase E3 | 0.90 | 1.04 | 1.11 |
| SA0994 | sdhC | Succinate dehydrogenase cytochrome b-558 | 1.72 | 1.13 | 1.50 |
| SA0995 | sdhA | Succinate dehydrogenase flavoprotein subunit | 1.65 | 1.31 | 1.50 |
| SA0996 | sdhB | Succinate dehydrogenase iron-sulfur protein subunit | 1.05 | 1.58 | 1.88 |
| SA1088 | sucC | Succinyl-CoA synthetase (beta subunit) | 1.46 | 1.54 | 1.59 |
| SA1089 | sucD | succinyl-CoA synthetase (alpha subunit) | 1.11 | 1.52 | 1.62 |
| SA1131 | Similar to 2-oxoacid ferredoxin oxidoreductase, alpha subunit | 1.49 | 1.25 | 1.77 | |
| SA1132 | Similar to 2-oxoacid ferredoxin oxidoreductase, beta subunit | 1.39 | 1.37 | 1.74 | |
| SA1184 | citB | Aconitate hydratase | 1.07 | 1.11 | 1.59 |
| SA1244 | odhB | Dihydrolipoamide succinyltransferase | 1.66 | 1.74 | 1.44 |
| SA1245 | odhA | 2-Oxoglutarate dehydrogenase E1 | 1.36 | 1.45 | 1.24 |
| SA1349 | Dihydrolipoamide dehydrogenase | 1.54 | 0.93 | 1.16 | |
| SA1517 | citC | Isocitrate dehyrogenase | 1.17 | 1.27 | 2.02 |
| SA1518 | citZ | Citrate synthase II | 1.03 | 1.18 | 2.11 |
| SA1609 | pckA | Phosphoenolpyruvate carboxykinase (ATP) | 1.27 | 0.82 | 1.10 |
| Cell envelope and cellular processes | |||||
| SA0111 | sirA | Lipoprotein | 2.32 | 2.19 | 0.65 |
| SA0126 | Similar to capsular polysaccharide synthesis protein 14H | 0.30 | 0.44 | 0.44 | |
| SA0127 | Similar to capsular polysaccharide synthesis protein 14H | 0.36 | 0.48 | 0.43 | |
| SA0265 | lytM | Peptidoglycan hydrolase | 2.15 | 2.06 | 1.72 |
| SA0295 | Similar to outer membrane protein precursor | 3.29 | 3.73 | 1.03 | |
| SA0423 | sle1 | Hypothetical protein, similar to autolysin (N-acetylmuramoyl-l-alanine amidase) | 2.76 | 2.75 | 2.89 |
| SA0457 | gcaD | UDP-N-acetylglucosamine pyrophosphorylase | 2.00 | 1.59 | 1.25 |
| SA0523 | Similar to poly(glycerol-phosphate) alpha-glucosyltransferase (teichoic acid biosynthesis) | 3.25 | 4.10 | 1.72 | |
| SA0620 | stcA | Secretory antigen SsaA homolog | 1.81 | 3.16 | 3.05 |
| SA0905 | atl | Autolysin (N-acetylmuramyl-l-alanine amidase and endo-β-N-acetylglucosaminidase) | 3.28 | 1.89 | 1.88 |
| SA1023 | ftsL | Cell division protein | 0.49 | 0.45 | 0.70 |
| SA1026 | murD | UDP-N-acetylmuramoylalanine-d-glutamate ligase | 2.16 | 1.37 | 1.29 |
| SA1027 | div1b | Cell division protein, FtsQ homolog | 2.28 | 1.32 | 1.19 |
| SA1212 | opp-2D | Oligopeptide transport ATPase | 1.90 | 2.31 | 2.01 |
| SA1267 | ebhA | Similar to streptococcal adhesin Emb | 0.30 | 0.12 | 0.64 |
| SA1898 | Similar to SceD precursor (lytic transglycosylase) | 2.35 | 2.24 | 4.50 | |
| SA2097 | ssaA homolog | Similar to secretory antigen precursor SsaA | 8.16 | 8.04 | 6.00 |
| SA2356 | isaA | Immunodominant antigen A | 0.38 | 0.60 | 1.22 |
| SA2442 | Preprotein translocase secA homolog | 2.57 | 1.88 | 1.23 | |
| SA2446 | Similar to preprotein translocase secY | 2.01 | 2.08 | 2.10 | |
| SA2455 | Capsular polysaccharide biosynthesis, capC | 0.37 | 0.41 | 0.44 | |
| SA2456 | Capsular polysaccharide biosynthesis, capB | 0.38 | 0.38 | 0.40 | |
| SA2457 | Capsular polysaccharide biosynthesis, capA | 0.40 | 0.36 | 0.41 | |
| Other functions | |||||
| SA0128 | sodM | Superoxide dismutase | 1.51 | 2.04 | 2.12 |
| SA0222 | coa | Staphylocoagulase precursor | 0.46 | 0.64 | 0.14 |
| SA0309 | geh | Glycerol ester hydrolase; lipase | 2.29 | 2.03 | 0.83 |
| SA0519 | sdrC | Ser-Asp rich fibrinogen-binding | 3.35 | 3.04 | 1.15 |
| SA0520 | sdrD | Ser-Asp rich fibrinogen-binding | 6.25 | 9.58 | 0.58 |
| SA0521 | sdrE | pfam04650, YSIRK-type signal peptide | 3.21 | 3.30 | 0.52 |
| SA0641 | mgrA | Transcriptional regulator, MarR family | 1.81 | 4.00 | 1.74 |
| SA0723 | clpP | ATP-dependent Clp protease proteolytic subunit homolog | 0.46 | 0.71 | 1.00 |
| SA0743 | graB | Similar to staphylocoagulase precursor | 0.23 | 0.22 | 0.31 |
| SA0746 | Staphylococcal nuclease | 2.35 | 4.54 | 0.74 | |
| SA1145 | Hfq, an abundant, ubiquitous RNA-binding protein | 0.30 | 0.41 | 0.70 | |
| SA1396 | bex | GTP-binding protein Era homolog | 2.02 | 1.39 | 1.48 |
| SA1452 | csbD | SigmaB-controlled gene product | 3.04 | 1.82 | 1.60 |
| SA1549 | Similar to serine proteinase Do, heat shock protein HtrA | 0.48 | 0.65 | 0.56 | |
| SA1559 | Similar to smooth muscle caldesmon | 2.79 | 1.44 | 0.93 | |
| SA1577 | sasC | Hypothetical protein, similar to FmtB protein | 0.41 | 0.38 | 0.83 |
| SA1617 | Similar to latent nuclear antigen | 4.69 | 3.16 | 1.90 | |
| SA1709 | Similar to ferritin | 1.53 | 2.24 | 2.84 | |
| SA1941 | dps | General stress protein 20U | 1.76 | 3.99 | 4.28 |
| SA1964 | fmtB | FmtB protein | 0.30 | 0.25 | 0.47 |
| SA1984 | asp23 | Alkaline shock protein 23, ASP23 | 3.26 | 2.00 | 1.25 |
| SA2284 | Similar to accumulation-associated protein | 0.05 | 0.45 | 0.02 | |
| SA2285 | Similar to accumulation-associated protein AAP | 0.05 | 0.50 | 0.02 | |
| SA2286 | sarT | Helix-turn-helix multiple antibiotic resistance protein | 0.45 | 1.03 | 0.36 |
| SA2287 | sarU | Helix-turn-helix multiple antibiotic resistance protein | 0.07 | 0.92 | 0.08 |
| SA2290 | fnbB | Fibronectin-binding protein homolog | 5.69 | 2.93 | 1.67 |
| SA2423 | clfB | Clumping factor B | 4.07 | 3.79 | 1.08 |
| SA2430 | aur | Zinc metalloproteinase aureolysin | 6.02 | 7.32 | 1.90 |
| SA2431 | isaB | Immunodominant antigen B | 2.28 | 4.16 | 1.07 |
Signal intensity ratios of strains with RpoB621E versus those with RpoB621A.
In conclusion, although detailed studies are needed to clarify the role of rpoB with an A621E mutation, the data presented in this study clearly show that the rpoB mutation does confer S. aureus cells heteroresistant to both vancomycin and daptomycin. Besides the rifampin resistance phenotype, rpoB mutation may have an important role in the multidrug-resistance phenotype of MRSA.
Acknowledgments
We thank Yiming Yang, Beckman Coulter K.K., for help with zeta potential determination.
This work was supported by a Grant-in-Aid for 21st Century COE Research and a Grant-in-Aid for Scientific Research (no. 18590438) to L. Cui from the Ministry of Education, Science, Sports, Culture, and Technology of Japan.
Footnotes
Published ahead of print on 13 September 2010.
REFERENCES
- 1.Akins, R. L., and M. J. Rybak. 2001. Bactericidal activities of two daptomycin regimens against clinical strains of glycopeptide intermediate-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus faecium, and methicillin-resistant Staphylococcus aureus isolates in an in vitro pharmacodynamic model with simulated endocardial vegetations. Antimicrob. Agents Chemother. 45:454-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubry-Damon, H., C. J. Soussy, and P. Courvalin. 1998. Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 42:2590-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, T., and O. Schneewind. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58-63. [DOI] [PubMed] [Google Scholar]
- 4.Best, G. K., and N. N. Durham. 1965. Vancomycin adsorption to Bacillus subtilis cell walls. Arch. Biochem. Biophys. 111:685-691. [DOI] [PubMed] [Google Scholar]
- 5.Bruinsma, G. M., M. Rustema-Abbing, H. C. van der Mei, C. Lakkis, and H. J. Busscher. 2006. Resistance to a polyquaternium-1 lens care solution and isoelectric points of Pseudomonas aeruginosa strains. J. Antimicrob. Chemother. 57:764-766. [DOI] [PubMed] [Google Scholar]
- 6.Camargo, I. L., H. M. Neoh, L. Cui, and K. Hiramatsu. 2008. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob. Agents Chemother. 52:4289-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; sixteenth informational supplement, vol. 26, no. 3. M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Coller, B. S., and H. R. Gralnick. 1977. Studies on the mechanism of ristocetin-induced platelet agglutination. Effects of structural modification of ristocetin and vancomycin. J. Clin. Invest. 60:302-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, L. V., S. A. Kristian, C. Weidenmaier, M. Faigle, K. P. Van Kessel, J. A. Van Strijp, F. Gotz, B. Neumeister, and A. Peschel. 2002. Staphylococcus aureus strains lacking d-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J. Infect. Dis. 186:214-219. [DOI] [PubMed] [Google Scholar]
- 10.Cosgrove, S. E., K. C. Carroll, and T. M. Perl. 2004. Staphylococcus aureus with reduced susceptibility to vancomycin. Clin. Infect. Dis. 39:539-545. [DOI] [PubMed] [Google Scholar]
- 11.Cui, L., and K. Hiramatsu. 2003. Vancomycin-resistant Staphylococcus aureus, p. 187-212. In A. C. Fluit and F. J. Schmitz (ed.), MRSA: current perspectives. Caister Academic Press, Norfolk, England.
- 12.Cui, L., A. Iwamoto, J. Q. Lian, H. M. Neoh, T. Maruyama, Y. Horikawa, and K. Hiramatsu. 2006. Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:428-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui, L., J. Lian, H. Neoh, R. Ethel, and K. Hiramatsu. 2005. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3404-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui, L., X. Ma, K. Sato, K. Okuma, F. C. Tenover, E. M. Mamizuka, C. G. Gemmell, M. N. Kim, M. C. Ploy, N. El-Solh, V. Ferraz, and K. Hiramatsu. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui, L., H. Murakami, K. Kuwahara-Arai, H. Hanaki, and K. Hiramatsu. 2000. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 44:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui, L., H. M. Neoh, M. Shoji, and K. Hiramatsu. 2009. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 53:1231-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui, L., E. Tominaga, H. M. Neoh, and K. Hiramatsu. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:1079-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman, L., J. D. Alder, and J. A. Silverman. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:2137-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardete, S., M. Aires-De-Sousa, A. Faustino, A. M. Ludovice, and H. de Lencastre. 2008. Identification of the first vancomycin intermediate-resistant Staphylococcus aureus (VISA) isolate from a hospital in Portugal. Microb. Drug Resist. 14:1-6. [DOI] [PubMed] [Google Scholar]
- 20.Hachmann, A. B., E. R. Angert, and J. D. Helmann. 2009. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob. Agents Chemother. 53:1598-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayden, M. K., K. Rezai, R. A. Hayes, K. Lolans, J. P. Quinn, and R. A. Weinstein. 2005. Development of daptomycin resistance in vivo in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5285-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147-155. [DOI] [PubMed] [Google Scholar]
- 23.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9(10):486-493. [DOI] [PubMed] [Google Scholar]
- 24.Ho, S. W., D. Jung, J. R. Calhoun, J. D. Lear, M. Okon, W. R. Scott, R. E. Hancock, and S. K. Straus. 2008. Effect of divalent cations on the structure of the antibiotic daptomycin. Eur. Biophys. J. 37:421-433. [DOI] [PubMed] [Google Scholar]
- 25.Jones, T., M. R. Yeaman, G. Sakoulas, S. J. Yang, R. A. Proctor, H. G. Sahl, J. Schrenzel, Y. Q. Xiong, and A. S. Bayer. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 52:269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Julian, K., K. Kosowska-Shick, C. Whitener, M. Roos, H. Labischinski, A. Rubio, L. Parent, L. Ednie, L. Koeth, T. Bogdanovich, and P. C. Appelbaum. 2007. Characterization of a daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus strain in a patient with endocarditis. Antimicrob. Agents Chemother. 51:3445-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung, D., J. P. Powers, S. K. Straus, and R. E. Hancock. 2008. Lipid-specific binding of the calcium-dependent antibiotic daptomycin leads to changes in lipid polymorphism of model membranes. Chem. Phys. Lipids 154:120-128. [DOI] [PubMed] [Google Scholar]
- 28.Katayama, Y., H. Murakami-Kuroda, L. Cui, and K. Hiramatsu. 2009. Selection of heterogeneous vancomycin-intermediate Staphylococcus aureus by imipenem. Antimicrob. Agents Chemother. 53:3190-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirkpatrick, P., A. Raja, J. LaBonte, and J. Lebbos. 2003. Daptomycin. Nat. Rev. Drug Discov. 2:943-944. [DOI] [PubMed] [Google Scholar]
- 30.Kraus, D., S. Herbert, S. A. Kristian, A. Khosravi, V. Nizet, F. Gotz, and A. Peschel. 2008. The GraRS regulatory system controls Staphylococcus aureus susceptibility to antimicrobial host defenses. BMC Microbiol. 8:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuwahara-Arai, K., N. Kondo, S. Hori, E. Tateda-Suzuki, and K. Hiramatsu. 1996. Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP2′ production. Antimicrob. Agents Chemother. 40:2680-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mariani, P. G., H. S. Sader, and R. N. Jones. 2006. Development of decreased susceptibility to daptomycin and vancomycin in a Staphylococcus aureus strain during prolonged therapy. J. Antimicrob. Chemother. 58:481-483. [DOI] [PubMed] [Google Scholar]
- 33.Meehl, M., S. Herbert, F. Gotz, and A. Cheung. 2007. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:2679-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishra, N. N., S. J. Yang, A. Sawa, A. Rubio, C. C. Nast, M. R. Yeaman, and A. S. Bayer. 2009. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:2312-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morikawa, K., A. Maruyama, Y. Inose, M. Higashide, H. Hayashi, and T. Ohta. 2001. Overexpression of sigma factor, sigma(B), urges Staphylococcus aureus to thicken the cell wall and to resist beta-lactams. Biochem. Biophys. Res. Commun. 288:385-389. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, C. K., S. Mullin, M. S. Osburne, J. van Duzer, J. Siedlecki, X. Yu, K. Kerstein, M. Cynamon, and D. M. Rothstein. 2006. In vitro activity of novel rifamycins against rifamycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 50:827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mwangi, M. M., S. W. Wu, Y. Zhou, K. Sieradzki, H. de Lencastre, P. Richardson, D. Bruce, E. Rubin, E. Myers, E. D. Siggia, and A. Tomasz. 2007. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 104:9451-9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neoh, H. M., L. Cui, H. Yuzawa, F. Takeuchi, M. Matsuo, and K. Hiramatsu. 2008. Mutated response regulator graR is responsible for phenotypic conversion of Staphylococcus aureus from heterogeneous vancomycin-intermediate resistance to vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 52:45-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Neill, A., B. Oliva, C. Storey, A. Hoyle, C. Fishwick, and I. Chopra. 2000. RNA polymerase inhibitors with activity against rifampin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 44:3163-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Neill, A. J., T. Huovinen, C. W. Fishwick, and I. Chopra. 2006. Molecular genetic and structural modeling studies of Staphylococcus aureus RNA polymerase and the fitness of rifampin resistance genotypes in relation to clinical prevalence. Antimicrob. Agents Chemother. 50:298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 42.Rose, W. E., S. N. Leonard, G. Sakoulas, G. W. Kaatz, M. J. Zervos, A. Sheth, C. F. Carpenter, and M. J. Rybak. 2008. Daptomycin activity against Staphylococcus aureus following vancomycin exposure in an in vitro pharmacodynamic model with simulated endocardial vegetations. Antimicrob. Agents Chemother. 52:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakoulas, G., J. Alder, C. Thauvin-Eliopoulos, R. C. Moellering, Jr., and G. M. Eliopoulos. 2006. Induction of daptomycin heterogeneous susceptibility in Staphylococcus aureus by exposure to vancomycin. Antimicrob. Agents Chemother. 50:1581-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauermann, R., M. Rothenburger, W. Graninger, and C. Joukhadar. 2008. Daptomycin: a review 4 years after first approval. Pharmacology 81:79-91. [DOI] [PubMed] [Google Scholar]
- 45.Scott, W. R., S. B. Baek, D. Jung, R. E. Hancock, and S. K. Straus. 2007. NMR structural studies of the antibiotic lipopeptide daptomycin in DHPC micelles. Biochim. Biophys. Acta 1768:3116-3126. [DOI] [PubMed] [Google Scholar]
- 46.Sieradzki, K., T. Leski, J. Dick, L. Borio, and A. Tomasz. 2003. Evolution of a vancomycin-intermediate Staphylococcus aureus strain in vivo: multiple changes in the antibiotic resistance phenotypes of a single lineage of methicillin-resistant S. aureus under the impact of antibiotics administered for chemotherapy. J. Clin. Microbiol. 41:1687-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silverman, J. A., N. G. Perlmutter, and H. M. Shapiro. 2003. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tally, F., M. Zeckel, M. Wasilewski, C. Carini, C. Berman, G. Drusano, and F. B. J. Oleson. 1999. Daptomycin: a novel agent for Gram-positive infections. Expert Opin. Investig. Drugs 8:1223-1238. [DOI] [PubMed] [Google Scholar]
- 49.Toivonen, M. 2007. How bacteraemia is reviewed by medicines licensing authorities in Europe. Int. J. Antimicrob. Agents 30(Suppl. 1):S103-S107. [DOI] [PubMed] [Google Scholar]
- 50.Wichelhaus, T., V. Schafer, V. Brade, and B. Boddinghaus. 2001. Differential effect of rpoB mutations on antibacterial activities of rifampicin and KRM-1648 against Staphylococcus aureus. J. Antimicrob. Chemother. 47:153-156. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, W. W., M. M. Wade, S. C. Holman, and F. R. Champlin. 2001. Status of methods for assessing bacterial cell surface charge properties based on zeta potential measurements. J. Microbiol. Methods 43:153-164. [DOI] [PubMed] [Google Scholar]
- 52.Yang, S. J., C. C. Nast, N. N. Mishra, M. R. Yeaman, P. D. Fey, and A. S. Bayer. 2010. Cell wall thickening is not a universal accompaniment of the daptomycin nonsusceptibility phenotype in Staphylococcus aureus: evidence for multiple resistance mechanisms. Antimicrob. Agents Chemother. 54:3079-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]