Abstract
Acinetobacter lwoffii, a species whose natural habitat is the human skin, intrinsically possesses a chromosomal gene encoding a carbapenem-hydrolyzing class D β-lactamase, OXA-134. This species may therefore constitute a reservoir for carbapenemase genes that may spread among other Acinetobacter species.
Acinetobacter baumannii, the most common Acinetobacter species isolated from humans, is an opportunistic pathogen for which resistance to carbapenems is increasing worldwide (13-15). Carbapenem resistance in A. baumannii is associated mostly with acquired carbapenem-hydrolyzing class D β-lactamases (CHDLs) (19). Four groups of acquired CHDLs in A. baumannii, OXA-23, OXA-40, OXA-58, and OXA-143, have been identified (9, 18). In addition, A. baumannii possesses a naturally occurring blaOXA-51 or blaOXA-69 CHDL-encoding gene that reduces the efficacy of carbapenems when it is overexpressed (4, 5, 7, 22). Identification of the sources of acquired and clinically relevant CHDLs is important to better understand the way and the reason why these resistance determinants are spreading. Acinetobacter radioresistens has recently been identified as the natural carrier of blaOXA-23, a gene encoding one of the most commonly acquired CHDLs in A. baumannii (16). However, the progenitors of the other acquired CHDLs identified in Acinetobacter species remain unknown. Our study aimed to evaluate whether other Acinetobacter species may represent additional reservoirs of CHDL-encoding genes.
The screening panel included strains belonging to 23 Acinetobacter species, including A. junii, A. johnsonii, A. haemolyticus, A. baylyi, A. lwoffii, A. radioresistens, A. schindleri, A. ursingii, A. calcoaceticus, A. gerneri, A. tjernbergiae, A. bouvetii, A. tandoii, A. grimontii, A. towneri, A. parvus, and Acinetobacter genomospecies 3, 6, 9, 10, 13, 15, 16, and 17. Acinetobacter genomospecies 9 is now classified as A. lwoffii (15). Strains were identified at the species level by using 16S rRNA sequencing (3). Susceptibility testing was analyzed by the disk diffusion method in accordance with the guidelines of the Clinical and Laboratory Standards Institute (1), and MICs were determined by using Etest strips (AB bioMérieux, Solna, Sweden).
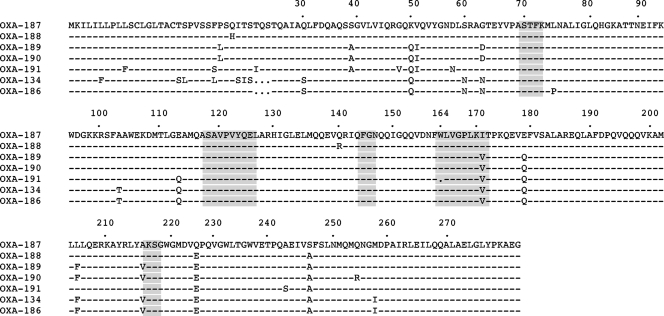
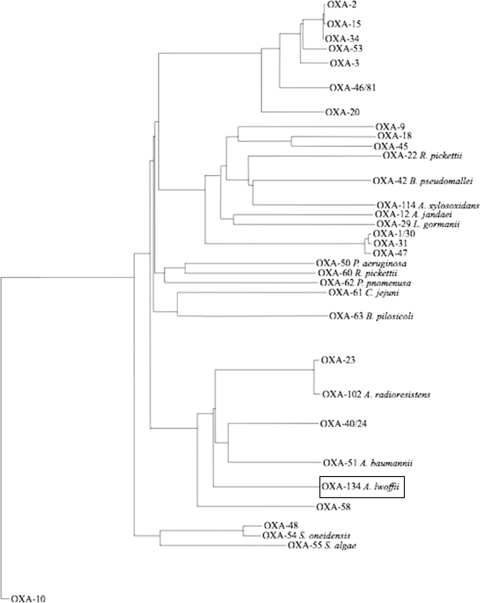
Screening for the known CHDL-encoding blaOXA-23, blaOXA-40, blaOXA-58, and blaOXA-143 genes was performed by PCR using internal primers (8, 9). This screening was positive only for the blaOXA-23 gene and only for A. radioresistens strain 3 and A. lwoffii strain 1. After sequencing, A. radioresistens strain 3 was found to possess the blaOXA-23 gene, in accordance with previous results (16). Sequencing of the amplicon obtained from A. lwoffii strain 1 identified a gene encoding a novel OXA-type β-lactamase. Thermal asymmetric interlaced (TAIL) PCR experiments were performed in order to obtain the entire sequence of this gene (11, 12). It encoded a 273-amino-acid protein named OXA-134 that shared 63, 58, 57, and 53% amino acid identity with OXA-23, OXA-40, OXA-51, and OXA-58, respectively. OXA-134 possessed the typical features of a class D β-lactamase, including the STFK tetrad at positions 70 to 73 according to class D β-lactamase (DBL) numbering (Fig. 1) (2). Also, as observed for other CHDLs (except for OXA-58), an FGN motif at DBL positions 144 to 146 replaced the usual YGN motif of classical class D β-lactamases (18). Finally, a KSG element was identified at DBL positions 216 to 218, as observed in the CHDLs OXA-40 and OXA-51, whereas a KTG motif is present in most class D β-lactamases, including the CHDLs OXA-23 and OXA-58 (18). A phylogenetic analysis showed that OXA-134-like β-lactamases were constituting a separate subgroup of CHDLs but that this subgroup was more closely related to the identified class D β-lactamases from Acinetobacter spp. than to other known CHDLs (Fig. 2) .
FIG. 1.
Amino acid alignment of the seven OXA-134-like class D β-lactamases identified in this study. Dashes indicate amino acids identical to those in the OXA-187 sequence. Amino acid motifs which are well conserved (even if possibly variable) among class D β-lactamases are shaded in gray. Numbering is according to DBL numbering (2).
FIG. 2.
Dendrogram obtained for 32 class D β-lactamases by neighbor-joining analysis. The alignment used for tree calculation was performed with ClustalX. Branch lengths are drawn to scale and are proportional to the number of amino acid changes. The distance along the vertical axis has no significance. The different clusters identified allowed the identification of nine main groups, considering that proteins from the same group have more than 80% amino acid identity. The class D β-lactamases which are considered to be naturally occurring are indicated together with the names of the corresponding species. R. pickettii, Ralstonia pickettii; B. pseudomallei, Burkholderia pseudomallei; A. xylosoxidans, Alcaligenes xylosoxidans; A. jandaei, Aeromonas jandaei; L. gormanii, Legionella gormanii; P. aeruginosa, Pseudomonas aeruginosa; P. pnomenusa, Pandoraea pnomenusa; C. jejuni, Campylobacter jejuni; B. pilosicoli, Brachyspira pilosicoli; S. oneidensis, Shewanella oneidensis; S. algae, Shewanella algae.
A. lwoffii is a commensal organism found on human skin, the perineum, and the oropharynx. It has been associated with catheter-related bloodstream infections in immunocompromised patients and with bacteremia associated with community-acquired gastroenteritis and gastritis (20, 21). All the Acinetobacter genomospecies 9/A. lwoffii isolates we included in our study were fully susceptible to all antibiotics tested, including penicillins, imipenem, and meropenem. It is therefore likely that the blaOXA-134-like genes were not expressed (or were expressed at an insignificant level) in these hosts.
In order to study the biochemical properties of OXA-134, cloning of the blaOXA-134 gene into the kanamycin-resistant plasmid pCR-BluntII-TOPO (Invitrogen, Life Technologies, Cergy-Pontoise, France) was performed using PCR products generated with primers PreOXA-134A (5′-GAAAAATGACCAAAATTTGATCG-3′) and PreOXA-134B (5′-TATTTGCATCATCCTTCAGC-3′) as described previously (16). Escherichia coli TOP10(pOXA-134) showed reduced susceptibility to imipenem and meropenem and resistance to most penicillins that was not inhibited by β-lactamase inhibitors (Table 1).
TABLE 1.
MICs of β-lactams for the different A. lwoffii isolates, E. coli TOP10 harboring recombinant plasmid pOXA-134, and the E. coli TOP10 reference strain
| β-Lactam(s)a | MIC (μg/ml) for: |
||
|---|---|---|---|
| A. lwoffii isolates | E. coli TOP10(pOXA-134) | E. coli TOP10 | |
| Amoxicillin | 0.5-1 | >512 | 4 |
| Amoxicillin + CLA | 0.5-1 | 128 | 4 |
| Ticarcillin | 0.5-1 | >512 | 4 |
| Ticarcillin + CLA | 0.5-1 | 256 | 4 |
| Piperacillin | 1.5-4 | 8 | 1 |
| Piperacillin + TZB | 1.5-4 | 8 | 1 |
| Cephalothin | 4-8 | 8 | 2 |
| Cefuroxime | 4-8 | 4 | 2 |
| Ceftazidime | 1-4 | 0.12 | 0.06 |
| Cefotaxime | 0.75-3 | 0.12 | 0.12 |
| Cefepime | 0.25-1 | 0.12 | 0.06 |
| Moxalactam | 1-4 | 0.12 | 0.06 |
| Aztreonam | 4-8 | 0.12 | 0.12 |
| Imipenem | 0.12-0.5 | 0.5 | 0.06 |
| Meropenem | 0.12-0.5 | 0.5 | 0.06 |
CLA, clavulanic acid at a fixed concentration of 4 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
To further characterize the activity of OXA-134, the enzyme was purified from E. coli cultures containing recombinant plasmid pOXA-134 as described previously (17). After DNase treatment and ultracentrifugation at 40,000 × g for 1 h, the extract was loaded successively onto two Q-Sepharose columns with 20 mM diethanolamine (pH 8.5) and 20 mM diethanolamine (pH 9.5) buffers. The specific activity of the purified β-lactamase OXA-134, measured with 100 μM imipenem as the substrate, was 116 U·mg of protein−1, with a 20-fold purification factor. The kinetic measurements of the purified enzymes were carried out at 25°C in 50 mM sodium phosphate (pH 7.0), and Km and kcat values were determined as described previously (6). β-Lactamase OXA-134 showed a narrow-spectrum hydrolysis profile, including mostly penicillins (Table 2). The rates of imipenem and meropenem hydrolysis were low, whereas the MICs of both carbapenems for E. coli TOP10 expressing OXA-134 were increased by 3-fold (Table 1). Overall, the catalytic activities obtained for OXA-134 were similar to those for OXA-58 and OXA-40, taken as references for CHDL activity (17).
TABLE 2.
Kinetic parameters for purified β-lactamase OXA-134a
| Substrate | kcat (s−1) | Km (μM) | kcat/Km ratio (s−1·mM−1) |
|---|---|---|---|
| Benzylpenicillin | 70 | 50 | 1,400 |
| Ampicillin | 150 | 250 | 600 |
| Ticarcillin | 0.2 | 200 | 1 |
| Piperacillin | 30 | 200 | 150 |
| Ceftazidime | <0.01 | NDb | |
| Cefotaxime | <0.01 | ND | |
| Cefepime | <0.01 | ND | |
| Cefoxitin | <0.01 | ND | |
| Aztreonam | <0.01 | ND | |
| Imipenem | 0.1 | 10 | 10 |
| Meropenem | 0.05 | 250 | 0.2 |
Data are means of results from three independent experiments. Standard deviations were within 10% of the means.
ND, no detectable hydrolysis (<0.01 s−1).
In order to assess whether the blaOXA-134-like gene was naturally present in A. lwoffii, a blaOXA-134-specific PCR was performed using whole-cell DNA samples from a collection of 10 A. lwoffii isolates recovered from clinical specimens, including blood cultures, urine samples, cerebrospinal fluids, and central venous catheter tips, from Bicêtre and Cologne hospitals. PCR results showed that all strains possessed a blaOXA-134-like gene. Sequencing of the amplicons allowed the identification of six additional OXA-134 derivatives (named OXA-186 to OXA-191) (see www.lahey.org/Studies) differing by 3 to 18 amino acid substitutions (Fig. 1). Noteworthily, OXA-134 and OXA-186 each possessed 273 amino acids whereas the five other variants each possessed an additional 3-amino-acid stretch (Fig. 1). In three isolates, the blaOXA-134-like gene was disrupted by nucleotide substitutions located in the center of the gene and likely leading to interruption of the open reading frame (data not shown). In order to evaluate whether an OXA-134-like variant possessing additional amino acids may confer a different β-lactam resistance pattern, the blaOXA-187 gene chosen as a representative was cloned and expressed in the same manner as the blaOXA-134 gene. MICs of β-lactams for E. coli(pOXA-187) were similar to those for E. coli(pOXA-134) (data not shown), showing that those additional amino acids did not play any significant role in hydrolysis. The chromosomal locations of the blaOXA-134-like genes in these A. lwoffii isolates were confirmed by using the endonuclease I-CeuI technique, as described previously (10).
In order to evaluate whether blaOXA-134-like genes might have disseminated among A. baumannii strains, a collection of 100 A. baumannii isolates (with variable susceptibilities to imipenem, including 50 carbapenem-resistant isolates) were screened by PCR. None of the screened A. baumannii isolates harbored a blaOXA-134-like gene.
A. lwoffii was found to be a reservoir of a novel type of CHDL-encoding gene. Detection of that β-lactamase gene might be used as a tool for rapid and accurate identification of the A. lwoffii species.
Nucleotide sequence accession number.
The nucleotide sequence of the blaOXA-134 gene described in this work is available in the GenBank nucleotide database under accession number HQ122933.
Acknowledgments
This work was funded by the INSERM (U914), by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, Paris, France, and mostly by a grant from the European Community (TROCAR HEALTH-F3-2008-223031). S.F. was funded by a grant-in-aid from the Fond d'Etudes et de Recherche du Corps Médical des Hôpitaux de Paris. The contribution of H.S. was supported by a grant from the Bundesministerium für Bildung und Forschung (BMBF), Germany, Klinische Forschergruppe Infektiologie (BMBF grant 01KI0771).
We thank G. Jacoby, who provided us with the OXA numbering, and T. Naas for precious advice.
Footnotes
Published ahead of print on 13 September 2010.
REFERENCES
- 1.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing. CLSI M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA.
- 2.Couture, F., J. Lachapelle, and R. C. Levesque. 1992. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol. Microbiol. 6:1693-1705. [DOI] [PubMed] [Google Scholar]
- 3.Dortet, L., P. Legrand, C. J. Soussy, and V. Cattoir. 2006. Bacterial identification, clinical significance, and antimicrobial susceptibilities of Acinetobacter ursingii and Acinetobacter schindleri, two frequently misidentified opportunistic pathogens. J. Clin. Microbiol. 44:4471-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figueiredo, S., L. Poirel, J. Croizé, C. Recule, and P. Nordmann. 2009. In vivo selection of reduced susceptibility to carbapenems in Acinetobacter baumannii related to ISAba1-mediated overexpression of the natural blaOXA-66 oxacillinase gene. Antimicrob. Agents Chemother. 53:2657-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Figueiredo, S., L. Poirel, A. Papa, V. Koulourida, and P. Nordmann. 2009. Overexpression of the naturally occurring blaOXA-51 gene in Acinetobacter baumannii mediated by novel insertion sequence ISAba9. Antimicrob. Agents Chemother. 53:4045-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giuliani, F., J. D. Docquier, M. L. Riccio, L. Pagani, and G. M. Rossolini. 2005. OXA-46, a new class D β-lactamase of narrow substrate specificity encoded by a blaVIM-1-containing integron from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 49:1973-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Héritier, C., L. Poirel, P. E. Fournier, J. M. Claverie, D. Raoult, and P. Nordmann. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4174-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Héritier, C., L. Poirel, T. Lambert, and P. Nordmann. 2005. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:3198-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins, P. G., L. Poirel, M. Lehmann, P. Nordmann, and H. Seifert. 2009. OXA-143, a novel carbapenem-hydrolyzing class D β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:5035-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, Y. G., N. Mitsukawa, T. Oosumi, and R. F. Whittier. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8:457-463. [DOI] [PubMed] [Google Scholar]
- 12.Liu, Y. G., and R. F. Whittier. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674-681. [DOI] [PubMed] [Google Scholar]
- 13.Maragakis, L. L., and T. M. Perl. 2008. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 46:1254-1263. [DOI] [PubMed] [Google Scholar]
- 14.Munoz-Price, L. S., and R. A. Weinstein. 2008. Acinetobacter infection. N. Engl. J. Med. 358:1271-1281. [DOI] [PubMed] [Google Scholar]
- 15.Peleg, A. Y., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel, L., S. Figueiredo, V. Cattoir, A. Carattoli, and P. Nordmann. 2008. Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob. Agents Chemother. 52:1252-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel, L., S. Marqué, C. Héritier, C. Segonds, G. Chabanon, and P. Nordmann. 2005. OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirel, L., T. Naas, and P. Nordmann. 2010. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob. Agents Chemother. 54:24-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel, L., and P. Nordmann. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826-836. [DOI] [PubMed] [Google Scholar]
- 20.Seifert, H., A. Strate, A. Schulze, and G. Pulverer. 1994. Bacteremia due to Acinetobacter species other than Acinetobacter baumannii. Infection 22:379-385. [DOI] [PubMed] [Google Scholar]
- 21.Rathinavelu, S., Y. Zavros, and J. L. Merchant. 2003. Acinetobacter lwoffii infection and gastritis. Microbes Infect. 5:651-657. [DOI] [PubMed] [Google Scholar]
- 22.Turton, J. F., M. E. Ward, N. Woodford, M. E. Kaufmann, R. Pike, D. M. Livermore, and T. L. Pitt. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72-77. [DOI] [PubMed] [Google Scholar]