Abstract
While genome sequencing projects are advancing rapidly, EST sequencing and analysis remains a primary research tool for the identification and categorization of gene sequences in a wide variety of species and an important resource for annotation of genomic sequence. The TIGR Gene Indices (http://www.tigr.org/tdb/tgi.shtml) are a collection of species-specific databases that use a highly refined protocol to analyze EST sequences in an attempt to identify the genes represented by that data and to provide additional information regarding those genes. Gene Indices are constructed by first clustering, then assembling EST and annotated gene sequences from GenBank for the targeted species. This process produces a set of unique, high-fidelity virtual transcripts, or Tentative Consensus (TC) sequences. The TC sequences can be used to provide putative genes with functional annotation, to link the transcripts to mapping and genomic sequence data, to provide links between orthologous and paralogous genes and as a resource for comparative sequence analysis.
INTRODUCTION
The sequencing of eukaryotic genomes is progressing at an astonishing rate. The fruit fly, Drosophila melanogaster, was published in the spring of 2000, Arabidopsis thaliana, a plant model organism, has recently been completed, a draft-quality human sequence is now available, and the sequencing of mouse, rat and rice are well under way. However, for many organisms of scientific, economic or agricultural interest, complete genomic sequencing is unlikely to be completed in the foreseeable future and the sequencing of Expressed Sequence Tags (ESTs) (1) remains the primary tool for genomic exploration and for functional genomics projects. There are nearly 5 000 000 ESTs in GenBank (nearly half of which are human), and the number of species represented by 50 000 or more ESTs has increased dramatically in the past year (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html).
Even for completed genomes, EST data remains a crucial tool for gene identification, genomic annotation and comparative genomics. Regardless of how ESTs are ultimately used, their value can be significantly enhanced if the data are used to reconstruct a high-fidelity set of non-redundant transcripts. There are a number of publicly available databases that attempt to provide such analysis for some species, including UniGene (2) and STACK (3). However, the TIGR Gene Indices (4) are unique in the number of species surveyed, in the approach used to construct the individual species-specific databases and in the manner in which they can be used.
The TIGR Gene Indices provide an analysis for humans, experimental models of human disease such as mouse and rat, valuable crop plants and other important experimental organisms sampled extensively by EST sequencing. TIGR Gene Indices are maintained for 21 species, including the 15 most heavily sampled organisms, potato and five parasitic eukaryotes that are currently the subject of genomic sequencing projects. The current state of EST sequencing and a summary of the currently available TIGR Gene Indices can be found in Table 1.
Table 1. EST sequence entries from dbEST as of November 29, 2000 for the most heavily sampled organisms with the corresponding TIGR Gene Indices and their most recent release dates.
| Species |
Entries |
TIGR Gene Index |
Release date |
| Homo sapiens (human) | 2 454 447 | HGI 6.0 | June 30, 2000 |
| Mus musculus + domesticus (mouse) | 1 661 949 | MGI 5.0 | October 11, 2000 |
| Rattus sp. (rat) | 188 736 | RGI 4.1 | August 11, 2000 |
| Bos taurus (cattle) | 126 879 | BtGI 2.0 | September 26, 2000 |
| Glycine max (soybean) | 121 051 | GmGI 3.0 | July 12, 2000 |
| Arabidopsis thaliana (thale cress) | 112 467 | AtGI 4.0 | July 8, 2000 |
| Caenorhabditis elegans (nematode) | 101 252 | CeGI 2.0 | August 13, 2000 |
| Drosophila melanogaster (fruit fly) | 91 055 | DGI 3.0 | August 2, 2000 |
| Lycopersicon esculentum (tomato) | 87 680 | LGI 5.0 | August 4, 2000 |
| Danio rerio (zebrafish) | 73 703 | ZGI 5.0 | August 2, 2000 |
| Zea mays (maize) | 73 698 | ZmGI 3.0 | July 13, 2000 |
| Medicago truncatula (barrel medic) | 72 828 | MtGI 1.0 | July 11, 2000 |
| Oryza sativa (rice) | 62 126 | OsGI 4.0 | August 3, 2000 |
| Sorghum bicolor (sorghum) | 45 265 | SbGI 1.0 | September 18, 2000 |
| Triticum aestivum (wheat) | 44 132 | TaGI 1.0 | October 3, 2000 |
| Schistosoma mansoni (blood fluke) | 12 959 | SmGI 1.0 | July 26, 2000 |
| Trypanosoma cruzi | 9919 | TcGI 1.0 | March 24, 2000 |
| Solanum tuberosum (potato) | 8582 | StGI 1.0 | July 19, 2000 |
| Trypanosoma brucei rhodesiense | 4821 | TbGI 1.0 | March 28, 2000 |
| Plasmodium falciparum (malaria parasite) | 2871 | PfGI 1.0 | September 18, 2000 |
| Leishmania major | 2191 | LshGI 1.0 | April 18, 2000 |
| Total number of sequences | 5 358 611 |
Together, these species represent more than 91% of the 5 879 539 ESTs available in dbEST.
To create the TIGR Gene Indices, we have developed a highly refined, rigorously tested protocol for cleaning, clustering and assembling ESTs and gene sequences to produce high-fidelity consensus sequences for the represented genes while eliminating low quality, misclustered or chimaeric sequences (5). This has several advantages over competing approaches: it separates closely related genes into distinct consensus sequences, it separates splice variants and it produces longer representations of the underlying gene sequences. The resulting Tentative Consensus (TC) sequences can be used for eukaryotic genome sequence annotation (6,7), integration of complex mapping data and identification of orthologous genes.
CONSTRUCTION OF THE GENE INDICES
Each Gene Index is assembled using an identical process. For each species, EST sequences are downloaded from dbEST and trimmed to remove remove vector, polyA/T tails, adaptor sequences and contaminating bacterial sequences. Gene sequences (NP sequences) are parsed through Entrez from CDS and CDS-join features in GenBank records; additional Expressed Transcript (ET) sequences are obtained from the TIGR EGAD database (http://www.tigr.org/tdb/egad/egad.html).
EST and gene sequences are then compared using FLAST, a rapid sequence comparison program based on DDS (8), in which query sequences are first concatenated and then searched against a nucleotide database. Sequences sharing a minimum of 95% identity over a 40 nt or longer region with <20 bases of mismatched sequence at either end are grouped into a cluster. Each cluster is then assembled separately. For each cluster, component EST, NP and ET sequences are downloaded and these sequences are then assembled using CAP3 (9) to produce TCs. Assembly produces one or more consensus sequence for each cluster and rejects any chimeric, low-quality and non-overlapping sequences. Each cluster is assembled in the same fashion until the entire set of clusters has been exhausted. The resulting set of TCs is loaded into the appropriate species-specific Gene Index database for annotation.
Following assembly, TCs are annotated to provide a provisional functional assignment. A TC containing a known gene is assigned the function of that gene; TCs without assigned functions are searched using DPS (8) against a non-redundant protein database; high-scoring hits are assigned a putative function. For the Human, Mouse and Rat Gene Indices, mapping locations are assigned by using e-PCR (10). The resulting Gene Index is released through the TIGR web site (http://www.tigr.org/tbd/tdb.html); an example THC from the Human Gene Index is shown in Figure 1.
Figure 1.

An example THC from the Human Gene Index. The consensus sequence is presented in FASTA format below which the locations of the gene sequences (red) and ESTs that comprise the assembly are shown with their respective locations within the assembly. Links are provided to GenBank records, internal data for all ESTs sequenced at TIGR and to clones available through the ATCC. This THC has been assigned a putative ID of ‘insulin receptor inhibitor, muscle’ as it contains a HT853 (as well as gene sequences from GenBank).
Gene Indices can be searched by TC number, the GenBank Accession number of any EST contained within the dataset or any ET used to build the Index. Users can perform a tissue-based search in which the library information in EST records is used to generate an ‘electronic northern blot’, identifying the tissue-specificity of expression based on the relative EST abundance. DNA and protein sequences can also be used to search the Gene Indices using WU-BLAST (http://www.tigr.org/cgi-bin/BlastSearch/blast_tgi.cgi), a gapped BLAST program developed by Warren Gish (Washington University, St Louis, MO).
The TIGR Gene Indices and the component TC assemblies are maintained within Sybase relational databases that allow versioning and heritability to be maintained. Each time a new version of the database is created, novel assemblies, caused by either the joining or splitting of previous TCs, are assigned a new, unique TC identifier. Previously-used identifiers are never reused and information regarding previous assemblies is never lost. Database queries using a TC identifier from a previous build return the most current version of that assembly. This allows assemblies to evolve as more data are available while providing tracking from build to build and maintaining functional assignments across multiple releases.
IDENTIFICATION OF ORTHOLOGS AND PARALOGS
The pending completion of the sequence of human and Arabidopsis genomes represent significant scientific achievements and sets the stage for the sequencing of other plant and animal genomes, including mouse, rat and rice. These data promise unprecedented opportunities for functional and evolutionary studies, including the identification and functional annotation of genes and non-coding regulatory regions. The utility of such analysis depends on the identification of homologous genes across species and the integration of data from a wide range of organisms. Homologous genes can be separated into two classes, orthologs and paralogs (11). Orthologs are homologous genes that perform the same biological function in different species but that have diverged in sequence due to evolutionary separation; paralogs are homologous genes within a species that are the result of a gene duplication event within the lineage. The study of orthologs is of particular importance because it is assumed that these genes play similar developmental or physiological roles and, consequently, should share conserved functional and regulatory domains.
While genome sequencing will provide a significant quantity of data, for many species, ESTs provide the primary source of gene sequence data. We have developed two separate approaches to the identification and representation of orthologous gene sequence data: the TIGR Orthologous Gene Alignment (TOGA) database and sequence-based genome alignments.
The TOGA database was introduced in January 2000 and represents the first attempt to identify orthologs using the gene and EST sequence resources. At present, TOGA is divided into separate sections for mammals and plants; the mammalian section consists of orthologs from human, mouse, rat and cattle while the plant section includes Arabidopsis, rice, tomato, potato, Medicago, soybean and maize. While the comparison of millions of ESTs from these species represents a significant computational challenge, this task is vastly simplified through the use of the TCs that comprise the TIGR Gene Indices.
For each species to be included in TOGA, the TCs contained within the respective Gene Indices are compared pairwise. Tentative Ortholog Groups (TOGs) are identified by requiring reciprocal best hits across three or more species with a minimum of 75% identity over a length of 400 bp or more for any single sequence match. High-scoring hits that did not meet the reciprocal best hit criteria, but which matched members of existing TOGs using the same criteria, were classified as Tentative Paralogs. Using these criteria, 8300 TOGs were identified containing TCs from three or more of the four mammalian species and 3074 from among the eight plant species surveyed. The distribution of species represented in TOGA is summarized in Table 2. An example mammalian TOG can be seen in Figure 2.
Table 2. Statistics for the most recent release of the TOGA database showing the relative numbers of TOGs of various sizes (not including paralogs).
| TOG sizea |
Mammals |
Plants |
Total |
| 3 | 6111 | 1948 | 8059 |
| 4 | 2189 | 689 | 2878 |
| 5 | 279 | 279 | |
| 6 | 109 | 109 | |
| 7 | 39 | 39 | |
| 8 | 10 | 10 | |
| Total TOGs | 8300 | 3074 | 11 374 |
In building the mammalian TOGs, only four species were considered while eight plant species contributed to the plant analysis.
aNumber of sequences.
Figure 2.

An example TOG from the TOGA database. The human, mouse and rat TCs all contain annotated genes; those in mouse and rat have been identified as ‘bithoraxoid-like protein’ while the human gene is simply annotated as ‘HSPC162’ and the cattle TC consists only of ESTs. The stringent overlap criteria used to construct the TOGs makes it unlikely that these matches are spurious and provides putative functional annotation for the previously unclassified human and bovine gene and EST sequences.
Like the TIGR Gene Indices, TOGA is a relational database that maintains the TOGs as accessionable objects that can be tracked across subsequent releases. TOGs can be searched using either a name-based search that allows users to enter a gene name and look for approximate matches or using a WU-BLAST (12) to search the dataset. TOGA can be found at http://www.tigr.org/tdb/toga/toga.shtml. More information regarding WU-BLAST can be found at http://blast.wustl.edu.
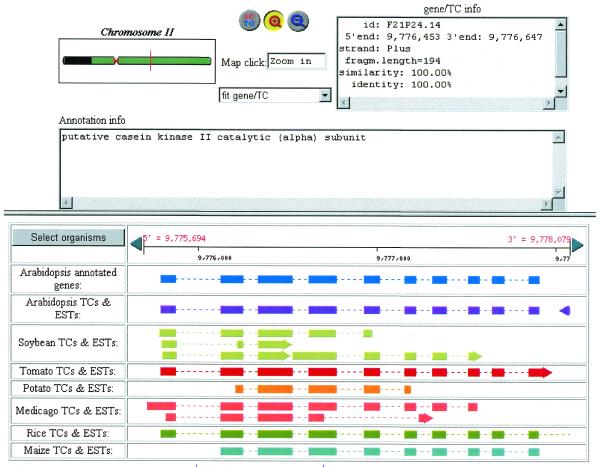
Additional interspecies information can be gained by examining the alignment of the EST and gene sequence data in the TIGR Gene Indices with reference to plant and animal genomes. Using the completed genome of Arabidopsis we tabulated the alignment of the TCs from the various TIGR Plant Gene Indices with the chromosomal sequence (http://www.tigr.org/tdb/at/alignTC.html). An example alignment in the region of an annotated gene on Chromosome II can be seen in Figure 3. We have completed a similar analysis using the recently published sequences of the long arms of human chromosomes 21 and 22.
Figure 3.
Alignment of TCs from the TIGR Plant Gene Indices with the sequence of Arabidopsis thaliana Chromsome II. The coding sequence of a putative casein kinase II catalytic subunit shows significant homology to the same gene in other plants as is evident from an alignment between the Arabidopsis genomic sequence and the various plant TCs. This gene is well conserved across both monocots and dicots. The multiple hits seen in some species may represent paralogs, gene families, alternative splice forms or partial TC assemblies.
USING THE TIGR GENE INDICES
Effective use of genomic resources for functional, comparative and evolutionary studies will rely on developing an accurate catalog of the genes encoded within each species as well as tools for cross-referencing various genomes of interest. The TIGR Gene Indices and the TOGA database represent an effort to provide such a resource by first attempting to identify and annotate the genes in a variety of organisms and then providing mechanisms to link to candidate orthologs in other species.
There are a variety of means by which a user might gain entry to the TIGR Gene Indices. For example, the radiation hybrid mapping data allows users to search for TC sequences that map to a candidate genomic region. Other users may search for TCs that appear to be expressed in a tissue-specific fashion or that contain ESTs from a particular disease state. However, the most common entry point for most users is the sequence search page (http://www.tigr.org/cgi-bin/BlastSearch/blast_tgi.cgi). Both BLASTN and TBLASTN versions of the WU-BLAST package have been implemented allowing DNA and protein queries to be used. Alignments to high scoring TCs and singleton ESTs in the organism searched are returned and users can view the appropriate sequence by clicking on the TC number or EST ID brings the user to an appropriate display of the sequence, similar to that in Figure 1. These TCs can be used to identify TOGs in the TOGA database or to search the genomic sequence alignments.
In addition to the Web interface, the TIGR Gene Indices are available as flat files. The TC consensus sequences are provided in a FASTA format file; the ESTs comprising each TC are specified in a separate file. Many users involved in the annotation of genomic sequence and in analysis of cDNA microarray data have found these to be particularly useful.
CONCLUSIONS
An increasing number of species are being subjected to genomic analysis, rapidly increasing the pace of gene discovery and accelerating functional genomics applications. For most species, EST sequencing remains the primary method of genomic sequence analysis. The TIGR Gene Indices, which represent the most comprehensive, publicly available analysis of EST sequences, have expanded significantly in the past year, adding 10 additional species-specific databases. In addition, we have expanded both the scope and utility of the resources we provide for cross-species comparisons through the TOGA database and genomic sequence alignments.
The TIGR Gene Indices have proven invaluable for annotation of genomic sequence and for functional analysis of ESTs. They are available via a free license for academic and non-profit use; commercial licenses are available for a fee. Parties interested in obtaining a license should visit http://www.tigr.org/tdb/license.html or email license@tigr.org.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are indebted to A. Glodek for database development. The authors also wish to thank M. Heaney and S. Lo for database support, and V. Sapiro, B. Lee, S. Gregory, R. Kramchedu, C. Irwin, M. Sengamalay and E. Arnold for computer system support. This work was supported by the US Department of Energy, grant DE-FG02-99ER62852 and the US National Science Foundation, grant DBI-9983070. Additional support was provided by the US National Science Foundation through grants DBI-9813392 and DBI-9975866. J.Q. was supported in part by NSF grant KDI-9980088.
References
- 1.Adams M.D., Kelley,J.M., Gocayne,J.D., Dubnick,M., Polymeropoulos,M.H.M., Xiao,H., Merril,C.R., Wu,A., Olde,B., Moreno,R.F. et al. (1991) Complementary DNA sequencing: expressed sequence tags and human genome project. Science, 252, 1651–1656. [DOI] [PubMed] [Google Scholar]
- 2.Boguski M.S. and Schuler,G.D. (1995) ESTablishing a human transcript map. Nature Genet., 10, 369–371. [DOI] [PubMed] [Google Scholar]
- 3.Burke J., Wang,H., Hide,W. and Davison,D.B. (1998) Alternative gene form discovery and candidate gene selection from gene indexing projects. Genome Res., 8, 276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quackenbush J., Liang,F., Holt,I., Pertea,G. and Upton,J. (2000). The TIGR Gene Indices: reconstruction and representation of expressed gene sequences. Nucleic Acids Res., 28, 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang F., Holt,I., Pertea,G., Karamycheva,S., Salzberg,S.L. and Quackenbush,J. (2000) An optimized protocol for analysis of EST sequences. Nucleic Acids Res., 28, 3657–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin X., Kaul,S., Rounsley,S., Shea,T.P., Benito,M.I., Town,C.D., Fujii,C.Y., Mason,T., Bowman,C.L., Barnstead,M. et al. (1999) Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature, 402, 761–768. [DOI] [PubMed] [Google Scholar]
- 7.Liang F., Holt,I., Pertea,G., Karamycheva,S., Salzberg,S.L. and Quackenbush,J. (2000) Gene index analysis of the human genome estimates approximately 120, 000 genes. Nature Genet., 25, 239–240. [DOI] [PubMed] [Google Scholar]
- 8.Huang X., Adams,M.D., Zhou,H. and Kerlavage,A.R. (1997) A Tool for Analyzing and Annotating Genomic Sequence. Genomics, 46, 37–45. [DOI] [PubMed] [Google Scholar]
- 9.Huang X. and Madan,A. (1999) CAP3: A DNA sequence assembly program. Genome Res., 9, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuler G.D. (1997) Sequence mapping by electronic PCR. Genome Res., 7, 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitch W.M. (1970) Distinguishing homologous from analogous proteins. Syst. Zool., 19, 99–113. [PubMed] [Google Scholar]
- 12.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]