Abstract
Treponema pallidum reacts poorly with the antibodies present in rabbit and human syphilitic sera, a property attributed to the paucity of proteins in its outer membrane. To better understand the basis for the syphilis spirochete's “stealth pathogenicity,” we used a dual-label, 3-step amplified assay in which treponemes encapsulated in gel microdroplets were probed with syphilitic sera in parallel with anti-FlaA antibodies. A small (approximately 5 to 10%) but reproducible fraction of intact treponemes bound IgG and/or IgM antibodies. Three lines of evidence supported the notion that the surface antigens were likely β-barrel-forming outer membrane proteins (OMPs): (i) surface labeling with anti-lipoidal (VDRL) antibodies was not observed, (ii) immunoblot analysis confirmed prior results showing that T. pallidum glycolipids are not immunoreactive, and (iii) labeling of intact organisms was not appreciably affected by proteinase K (PK) treatment. With this method, we also demonstrate that TprK (TP0897), an extensively studied candidate OMP, and TP0136, a lipoprotein recently reported to be surface exposed, are both periplasmic. Consistent with the immunolabeling studies, TprK was also found to lack amphiphilicity, a characteristic property of β-barrel-forming proteins. Using a consensus computational framework that combined subcellular localization and β-barrel structural prediction tools, we generated ranked groups of candidate rare OMPs, the predicted T. pallidum outer membrane proteome (OMPeome), which we postulate includes the surface-exposed molecules detected by our enhanced gel microdroplet assay. In addition to underscoring the syphilis spirochete's remarkably poor surface antigenicity, our findings help to explain the complex and shifting balance between pathogen and host defenses that characterizes syphilitic infection.
Syphilis is a multistage, sexually transmitted illness whose protean clinical manifestations and protracted natural history reflect the extraordinary invasiveness and immunoevasiveness of its etiologic agent, the spirochetal bacterium Treponema pallidum subsp. pallidum (72, 79, 104). The syphilis spirochete's capacity to replicate in tissues and disseminate hematogenously in the face of the robust cellular and humoral immune responses it elicits has earned it the designation “stealth pathogen” (72, 104). Secondary syphilis represents a prime example of treponemal immunoevasiveness; during this stage of the acute disease, patients can be spirochetemic for prolonged periods and develop new metastatic lesions despite extremely high titers of T. pallidum-specific antibodies (8, 35). Even so, the natural course of untreated syphilis clearly indicates that host defenses eventually gain the upper hand; over several years, infectious relapses become infrequent, and patients enter the prolonged, asymptomatic state known as late latency (28, 79). Containment, however, is often incomplete, as approximately one-third of untreated patients develop one of the potentially devastating forms of recrudescent disease called tertiary syphilis (28). The factors that influence the complex and shifting balance between this persistent bacterium and host clearance mechanisms are among the least understood aspects of syphilis pathogenesis (72, 104).
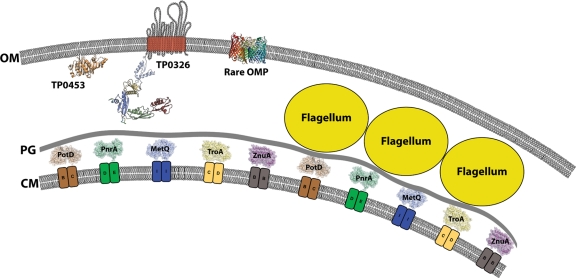
Efforts dating back almost 3 decades to identify T. pallidum outer membrane proteins (OMPs) using recombinant DNA techniques led to the discovery of the spirochete's distinctive molecular architecture, the ultrastructural basis for its stealth pathogenicity (20, 102). Our current working model of the T. pallidum cell envelope is shown in Fig. 1. The spirochete's outer membrane (OM) lacks lipopolysaccharide (9, 47) and hence is poorly inflammatory (87). The OM also contains a paucity of integral membrane proteins, still largely undefined at the molecular level, that present few surface antigenic targets to the host immune system (20, 102). Situated below the OM, where they are inaccessible to antibodies, are the organism's major protein immunogens, many of which are lipoproteins tethered by N-terminal lipids to the periplasmic face of the cytoplasmic membrane (CM) (15, 20, 25, 47, 85). There are three principal reasons why, more than 10 years after the publication of the genomic sequence of the Nichols type strain (47), the quest for T. pallidum OMPs remains largely unfulfilled. First, direct localization of surface-exposed proteins has been hindered by the fragility of the treponemal OM, coupled with limits to the sensitivity of techniques for detecting the low density of antigenic determinants on the T. pallidum surface (20, 32, 102). Second, even with the development of cell fractionation techniques tailored to the distinctive physical properties of the T. pallidum outer membrane (13, 106), rare OMPs have been exceedingly difficult to distinguish from periplasmic contaminants (1, 115). Lastly, with one notable exception, TP0326/Tp92 (22), the spirochete's genome does not encode any orthologs for well-characterized Gram-negative OMPs (47).
FIG. 1.
Current model for the molecular architecture of the T. pallidum cell envelope. Embedded within the OM are TP0326 (TP92), a BamA ortholog (22; D. C. Desrosiers, M. A. D. Cummings, C. E. Cameron, and J. D. Radolf, unpublished), and other, as yet unidentified membrane-spanning proteins predicted to form β-barrels, represented schematically by a prototypical OMP, the E. coli porin OmpC (7). Anchored to the inner leaflet of the OM is the pore-forming lipoprotein TP0453 (56), the crystal structure of which was recently solved (A. Luthra, G. Zhu, D. C. Desrosiers, V. Mulay, A. P. Heuck, F. B. Romano, I. Barcena-Uribarri, R. Benz, M. G. Malkowski, and J. D. Radolf, unpublished data). Within the periplasmic space, but below the peptidoglycan (PG) (64), are shown some structurally characterized ABC transporter substrate-binding proteins, TP0655 (PotD) (82), TP0319 (PnrA) (38), MetQ (38a), TP0163 (TroA) (75, 76), and TP0034 (ZnuA) (40), associated with their corresponding inner membrane (IM) permeases. The locations of the CM and the CM proteins, the positioning of the flagellar filaments in relation to the PG, and the scale of the diagram are based upon data obtained by cryoelectron tomography of vitreously frozen treponemes (64, 77a).
Two fundamental questions emanating from the discovery of rare OMPs are whether they induce an antibody response during syphilitic infection and, if so, the extent to which such responses contribute to containment of the bacterium. Not surprisingly, efforts to address these and related issues are greatly impeded by our lack of knowledge concerning the identities of T. pallidum OMPs. The notion that antibodies are induced against rare OMPs is supported by ex vivo studies demonstrating that syphilitic serum inhibits cytadherence (45), promotes opsonophagocytosis (34, 72, 78, 80, 81, 87), and mediates complement-dependent treponemicidal activity (12). Moreover, passive-transfer experiments have demonstrated that immune rabbit serum partially protects against intradermal challenge (97, 125). On the other hand, investigators have experienced considerable difficulty in directly detecting antibodies bound to the surface of intact treponemes using a variety of techniques, including agglutination (55), radioimmunoassay (32), immunofluorescence (31, 96), immunoelectron microscopy (61, 103), and cell surface radioimmunoprecipitation (118). A number of years ago, we devised the gel microdroplet technique to probe the antigenic structure of T. pallidum under conditions that minimize damage to its fragile outer membrane during the immunolabeling procedure (31). In its original format, this assay was unable to detect labeling of intact treponemes with syphilitic sera. To bring the results of the method more in line with those obtained from the ex vivo biological assays mentioned above, particularly opsonophagocytosis, we modified the microdroplet procedure to strengthen the signal generated by surface-bound antibodies in parallel with enhanced anti-flagellar antibody staining in order to assess the structural integrity of labeled organisms. Using this amplified, dual-labeling approach, we have been able to demonstrate, for the first time, unequivocal surface immunostaining of T. pallidum using both human and rabbit syphilitic sera; the finding that only small percentages of undisrupted organisms could be surface labeled serves as further evidence for the spirochete's ultrastructurally based strategy for immune evasion. We also used this method to examine the surface exposure of VDRL (i.e., cardiolipin) antigen, which has been reported to be an opsonic target (6); TprK, an extensively investigated, sequence-variable candidate OMP (23, 37, 72, 78); and TP0136, a lipoprotein recently reported to be surface exposed (14). All three were determined to be subsurface antigens and therefore unlikely contributors to the surface labeling observed with syphilitic sera. Using a consensus computational framework that combined subcellular localization and protein structural prediction tools, we generated ranked groups of candidate rare OMPs that comprise the putative T. pallidum outer membrane proteome (OMPeome), among which, we postulate, are the surface-exposed molecules detected with our enhanced microdroplet assay.
MATERIALS AND METHODS
Propagation and harvesting of T. pallidum.
Male New Zealand White rabbits (approximately 3.5 kg) maintained on antibiotic-free food and water and housed at 16°C were inoculated by injection of each testis with 1 × 108 T. pallidum organisms. Ten days later, the animals were euthanized, and the testes were removed aseptically. Several lengthwise cuts were made in each testis, following which the treponemes were extracted on a rotary shaker in CMRL-1066 (Invitrogen, Carlsbad, CA) or T. pallidum cultivation medium (TpCM) (30). Gross testicular contaminants were removed from the extract by centrifugation at 400 × g for 15 min. The animal protocols for this work were approved by the University of Connecticut Health Center (UCHC) Animal Care Committee and the Animal Care Committee of the Centers for Disease Control and Prevention (CDC) under the auspices of Animal Welfare Assurance numbers A3471-01 and A4365-01, respectively.
Immunologic reagents.
Immune rabbit serum (IRS) with high-titer reactivity in opsonophagocytosis assays with rabbit peritoneal macrophages was described previously (58, 116). Immunolabeling experiments with human syphilitic serum (HSS) were performed using a pool of sera from 5 HIV-seronegative secondary-syphilis patients that was previously shown to promote opsonophagocytosis of treponemes by human peripheral blood monocytes (34, 87). Normal human serum was obtained from volunteers without a history of syphilis and confirmed to be negative by rapid plasma reagin test in the UCHC clinical laboratory. Antisera from rabbits immunized with VDRL antigen (6) were generously provided by Sheila Lukehart (University of Washington); the VDRL titer was measured in the Division of Sexually Transmitted Diseases, Laboratory Reference and Research Branch, at the CDC using standard methodology (74). Rabbit antiserum generated against recombinant TP0136 (14) was the gift of Timothy Palzkill (University of Texas, Houston, Medical School). Rat antisera directed against T. pallidum FlaA (TP0249) (58), TroA (TP0163) (57), and thioredoxin (TP0919) (94) and rabbit antiserum produced using purified T. pallidum flagella (63) have been described previously. Normal rabbit serum was obtained from healthy, uninfected rabbits comparable in size and age to those inoculated with T. pallidum.
Purified N-terminally His-tagged recombinant TprK (TP0897; Nichols-Seattle) corresponding to the full-length protein without the predicted signal sequence (58) was kindly provided by Sheila Lukehart (University of Washington, Seattle, WA). To generate monospecific rat antisera directed against TprK and TP0326 (see cloning and purification of TP0326 periplasmic polypeptide transport-associated [POTRA] domains below), female Sprague-Dawley rats were immunized by intraperitoneal injection with a 1:1 mixture of Freund's complete adjuvant and 40 μg of purified recombinant protein in 250 μl of phosphate-buffered saline (PBS). Three and 5 weeks postimmunization, the rats were boosted by intraperitoneal injection of a 1:1 mixture of Freund's incomplete adjuvant and 40 μg of protein in 250 μl of PBS. Two weeks following the final boost, the animals were euthanized and exsanguinated.
Preparation of gel microdroplets and immunolabeling of treponemes.
T. pallidum organisms freshly harvested from rabbit testes were encapsulated in gel microdroplets composed of type VII low-melting-temperature agarose (Sigma Aldrich, St. Louis, MO) as previously reported (31). The beads were washed three times with TpCM and collected by centrifugation at 500 × g. The volume of the packed beads was noted after the final centrifugation, following which the beads were resuspended at a final concentration of 0.25 ml of beads per ml of TpCM.
The general immunolabeling scheme is shown in Fig. S1 in the supplemental material. In experiments using syphilitic serum, 20 μl of IRS or pooled HSS and 20 μl of rat anti-FlaA antiserum were added to 1-ml portions of the resuspended beads; Triton X-100 (TX100) (final concentration, 0.1%) was added to some samples immediately after the addition of the primary antibody. Following incubation for 2 h in a 34°C water bath with gentle mixing, the beads were washed three times with TpCM and resuspended in 4 ml of medium. In the second step, the samples were incubated with 2 μg each of biotinylated goat anti-IgG and anti-IgM (both from Sigma Aldrich) with the appropriate species specificity and 2 μg of chicken anti-rat IgG conjugated to Alexa 488 (Invitrogen, Carlsbad, CA) for 2 h at 34°C with gentle mixing. For experiments in which binding by IgG and IgM antibodies in syphilitic sera was compared, samples were incubated separately with biotinylated probes for IgG or IgM. The beads were washed three times with 4 ml of TpCM and suspended in 4 ml of TpCM as described above. In the third step, samples were incubated for 2 h at 34°C with 0.5 μg each of streptavidin conjugated to Alexa 546 (Invitrogen) and goat anti-chicken IgG conjugated to Alexa 488 (Invitrogen), washed three times with 4 ml of TpCM, resuspended in 4 ml of TpCM, and then allowed to sediment by gravity prior to examination by microscopy.
For the proteinase K (PK) experiments, encapsulated treponemes were first washed three times with PBS to remove serum proteins and then incubated with 0, 50, or 100 μg/ml of PK for 2 h at 34°C. PK activity was stopped by the addition of 20% fetal bovine serum (FBS), and the beads were immediately washed 3 times with TpCM containing 10% serum before being probed with HSS as described above. For localization of VDRL antigen and TP0136, 20 μl of the corresponding rabbit antiserum (described above) was used in place of IRS. For localization of TprK, 20 μl of rat anti-TprK antiserum was used as the primary antibody, and the beads were coincubated with 20 μl of rabbit anti-flagellar antiserum. In the second step, the beads were incubated with 2 μg of biotinylated goat anti-rat IgG (Invitrogen) and chicken anti-rabbit IgG-Alexa 488 conjugate (Invitrogen). In the third step, the beads were incubated with 0.5 μg each of Streptavidin-Alexa 546 and goat anti-chicken IgG-Alexa 488.
Scoring, photomicroscopy, and quantitative analysis of labeling.
Twenty microliters of gravity-sedimented beads from each sample was placed on a glass slide, and a coverslip was gently applied. Organisms were scored using a Nikon E-400 fluorescence microscope equipped with a dark-field condenser and with fluorescein and rhodamine filters. The integrity of each organism was determined by the presence or absence of discernible green fluorescence (Alexa 488), indicative of labeling with anti-FlaA or anti-flagellar antibodies; the OM of an organism was deemed to be disrupted if labeling (green) was observed on any portion of the cell. Labeling with syphilitic serum or antigen-specific antiserum was determined by the presence or absence of orange fluorescence (Alexa 546); an organism with orange labeling on any portion of the cell was deemed labeled. Consequently, in each experimental sample, four classifications were possible: (i) intact, unlabeled; (ii) intact, labeled; (iii) OM disrupted, labeled; and (iv) OM disrupted, unlabeled. The last category was not used because organisms with disrupted OMs were always labeled with primary antibodies (see Results). Except where noted, a minimum of three independent experiments were conducted. Photomicrographs were taken using a Spot RT Slider digital imager (Spot Diagnostics, Sterling Heights, MI) and processed using ImageJ software (http://rsb.info.nih.gov/ij/). To produce digital images representative of lightly labeled organisms observed microscopically, the signal gain and/or the exposure time was increased. The adjustments for the gain and the exposure required to produce these images were no more than 5-fold and 8-fold, respectively, over the acquisition settings used to visualize heavily labeled organisms.
To quantitatively compare the labeling of intact and TX100-treated treponemes with HSS, the exposure time, gain, brightness, and contrast of images for the two conditions were held constant. Briefly, images were converted into 8-bit gray-scale micrographs, giving a signal intensity from 0 (darkest pixel) to 255 (brightest pixel). The background was determined from 20 random samples and subtracted from all quantified images. The relative fluorescence intensity (RFI) values were quantified using ImageJ.
SDS-PAGE and immunoblot analysis of proteinase K-treated T. pallidum lysates.
Aliquots of T. pallidum (5 × 109 treponemes), freshly isolated from rabbit testes, were centrifuged at 8,000 × g for 20 min at 4°C and then gently washed twice with cold PBS. The resulting pellets were resuspended in 0.1 ml PBS. For proteinase digestion, 5 × 109 treponemes were treated for 1 h at 56°C with freshly prepared PK (0.1-mg/ml final concentration). Treated and untreated whole-cell lysates were boiled in Laemmli sample buffer, separated on a 12.5% SDS-PAGE gel, and then either stained with silver or transferred to nylon-supported nitrocellulose membranes (Micro Separations Inc., Westborough, MA). For immunoblotting, membranes were incubated overnight with the same pool of HSS described above (1:500), followed by incubation with horseradish peroxidase-conjugated goat anti-human IgG plus IgM secondary antibody (1:40,000) (Southern Biotechnology Associates, Birmingham, AL). Chemiluminescent detection was performed using the SuperSignal West Pico chemiluminescence substrate (Pierce, Rockford, IL) according to the manufacturer's instructions.
Cloning.
The portion of tp0326 encoding all five of its predicted POTRA domains (D. C. Desrosiers, et al., unpublished) was PCR amplified from T. pallidum DNA using primers 5′ tp0326 (GCCCATATGCAGGCAAACGACAATTGGTACG) and 3′ tp0326 POTRA 1-5 (GGCGAATTCTTACTAGTTTGCCGTCGACTGCTCCTC). The resulting amplicon was cloned into the expression vector pET28a (Novagen, San Diego, CA) using NdeI (5′ end) and EcoRI (3′ end) restriction sites. Nucleotide sequencing was performed to confirm that the sequence of the protein construct was correct.
Expression and purification of the POTRA domains of TP0326.
A 1-liter culture of LB was inoculated with 50 ml of overnight culture grown at 37°C. At an optical density (600 nm) of 0.6, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to the culture to a final concentration of 1 mM. The cells were grown for an additional 3 h and then harvested by centrifugation at 6,000 × g for 15 min at 4°C. The pellets were resuspended with 20 ml of 50 mM Tris (pH 7.5), 10% glycerol, 100 μg of lysozyme (Sigma-Aldrich, St. Louis, MO), and 100 μl of protease inhibitor cocktail (PIC) (Sigma-Aldrich). After the cells were thawed on ice, the bacterial suspension was lysed by sonication for three 30-s pulses interspersed with 30 s of incubation on ice. Following sonication, NaCl and β-mercaptoethanol were added to the lysates to give final concentrations of 0.5 M and 0.1%, respectively. The supernatants were then cleared of cellular debris by centrifugation at 18,000 × g for 20 min at 4°C and applied to a superflow Ni-nitrilotriacetic acid (NTA) (Qiagen, Valencia, CA) immobilized metal affinity chromatography (IMAC) column, which had been equilibrated with 25 mM Tris (pH 7.5), 0.5 M NaCl, 10% glycerol, and 0.1% β-mercaptoethanol (POTRA IMAC buffer). The protein was eluted with POTRA IMAC buffer supplemented with 250 mM imidazole. To excise the N-terminal His tag, 100 U of thrombin (Sigma-Aldrich) was added to the pooled protein fractions, dialyzed for 10 to 12 h at 4°C against POTRA IMAC buffer, and subjected to another round of IMAC purification to remove uncleaved His-tagged protein. Fractions containing the protein were concentrated using an Amicon-Ultra concentrator (Millipore, Billerica, MA) with a nominal molecular mass cutoff of 10 kDa and dialyzed into 50 mM potassium phosphate (pH 6.5) and 1 mM dithiothreitol (DTT) (POTRA CE buffer). Following dialysis, the purified POTRA domain construct was loaded onto a column with Macro-Prep High S cation-exchange resin (Bio-Rad, Hercules, CA), which had been equilibrated with POTRA CE buffer, and was eluted with a 0 to 1 M NaCl gradient. The final purification step consisted of size exclusion chromatography using a Superdex 75 preparation grade HiLoad 16/60 column (GE Healthcare BioSciences, Piscataway, NJ) equilibrated with POTRA CE buffer.
Expression, purification, and refolding of E. coli OmpG and BamA.
The expression vectors containing the Escherichia coli ompGm2 (pET29A:OmpGm2) and bamA (pET15b::Ec-yaeT) genes were generous gifts from Jörg Kleinschmidt (Universität Konstanz). OmpGm2 and BamA were expressed in the BL21(DE3) and BL21(DE3)/pLysS strains (Agilent Technologies, Santa Clara, CA), respectively, and purified as previously described (101). Purified OmpGm2 in 20 mM Tris (pH 7.0), 8 M urea, and 0.1% β-mercaptoethanol was refolded by dilution into 20 mM Tris (pH 7.0), 0.1% β-mercaptoethanol, and 4% β-octylglucopyranoside (Anatrace, Maumee, OH) (OmpGm2 buffer) to yield a final urea concentration of 1.5 M, followed by incubation at 37°C for 5 h. After being refolded, the urea was removed by dialysis against OmpGm2 buffer. To refold BamA, inclusion bodies were dissolved in 50 mM Tris (pH 7.0) and 8 M urea, followed by rapid 1:10 dilution into 50 mM Tris (pH 7.0) and 0.07% dodecylmaltopyranoside (Anatrace) (BamA buffer). The sample was refolded during incubation at 4°C for 24 h and then at room temperature for 8 h. Proper folding of reconstituted BamA and OmpG was confirmed by heat modifiability upon SDS-PAGE (29, 119).
Triton X-114 phase partitioning and immunoblot analysis.
Phase partitioning of T. pallidum proteins with Triton X-114 has been described previously (16). Briefly, approximately 4 × 109 freshly harvested T. pallidum organisms were pelleted by centrifugation at 10,000 × g for 20 min at 4°C; resuspended in 250 μl of PBS, 2% Triton X-114, and 0.005% PIC (Sigma-Aldrich, St. Louis, MO); and diluted with 990 μl of PBS. After incubation for 1 hour at 4°C, the solubilized T. pallidum organisms were dialyzed overnight in PBS at 4°C. Insoluble material was removed by centrifugation at 20,000 × g for 20 min at 4°C and stored at −80°C. The supernatants were phase separated, and the resulting detergent and aqueous fractions were washed five times. Samples were then precipitated with 10 volumes of acetone overnight at −80°C. Each fraction (aqueous, detergent, and insoluble) was resuspended with 50 μl of 1× Laemmli sample buffer (Bio-Rad, Hercules, CA) and subsequently boiled for SDS-PAGE and immunoblotting analysis using antisera to TrpK, TP0326, TroA, and thioredoxin (described above).
Bioinformatics prediction of the T. pallidum OMPeome.
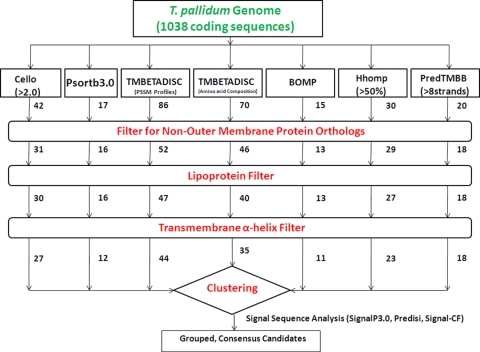
A flow diagram depicting the consensus computational framework used to predict the putative T. pallidum OMPeome is shown in Fig. 2, while Table 1 contains a summary of the various computational tools used. Initially, we analyzed 1,030 sequences in the T. pallidum genome (http://cmr.jcvi.org) for proteins predicted to be OM located or to form β-barrels using the following seven computational tools: CELLO (128), PSORTb 3.0 (129), TMBETADISC-Position-Specific Scoring Matrix (PSSM) profiles (93), TMBETADISC amino acid composition (AAC) (93), BOMP (11), Hhomp (109), and PRED-TMBB (5), using 8 strands, based on the structure of E. coli OmpX (124), as the minimum number required to form a β-barrel. In addition, using the same seven programs, we analyzed 8 frame-shifted sequences with identifiable translational start sites that encode polypeptides of 100 amino acids or greater; these included TP0009 (TprA) and TP0316 (TprF). The outputs from each program were then filtered sequentially for (i) annotated orthologs of non-OMPs; (ii) lipoproteins, using the database created by Setubal et al. (114); and (iii) α-helical transmembrane segments (other than potential N-terminal signal sequences), using TMHMM (69) and PHOBIUS (66). The remaining proteins were then clustered based on the number of programs predicting OM location or β-barrel formation. Proteins identified by four or more programs were analyzed for cleaved signal sequences by SignalP 3.0 (10), PrediSi (59), and Signal-CF (26). The 5′ sequences of selected proteins were manually inspected for alternative start codons.
FIG. 2.
Computational framework for prediction of the T. pallidum OMPeome. The numerals indicate the numbers of proteins identified by each OMP prediction tool and subsequent filters. Following the filter steps, the remaining candidates were grouped based on the number of OM localization/β-barrel topology programs that identified them and then analyzed for the presence of a cleaved signal sequence.
TABLE 1.
Computational tools used to generate the consensus T. pallidum OMPeome
| Program | Category | Description | Genome mininga | Reference | URL |
|---|---|---|---|---|---|
| CELLO | Subcellular localization | A two-level support vector machine system based on distinctive sets of multiple n-peptide compositions generated by primary sequence data | + | 128 | http://cello.life.nctu.edu.tw/ |
| PSORTb 3.0 | Subcellular localization | A Bayesian network that combines six analytical modules to generate one final localization result based on the performance of each module | + | 129 | http://www.psort.org/psortb/ |
| TMBETADISC(PSSM) | OMPs | Position-specific scoring matrix profiles generated by PSI-BLAST and nonredundant database search | + | 93 | http://rbf.bioinfo.tw/∼sachen/OMP.html/ |
| TMBETADISC(AAC) | OMPs | Utilizes amino acid composition based on radial basis function network (RBFN) generated by PSI-BLAST and nonredundant database | + | 93 | http://rbf.bioinfo.tw/∼sachen/OMP.html/ |
| BOMP | β-Barrel OMPs | C-terminal pattern recognition combined with analysis of amino acid frequencies in alternating positions | + | 11 | http://services.cbu.uib.no/tools/bomp/ |
| Hhomp | β-Barrel OMPs | Detects relationships to known OMPs using profile and pairwise hidden Markov models | − | 109 | http://toolkit.tuebingen.mpg.de/hhomp/ |
| PRED-TMBB | β-Barrel topology | A Hidden Markov model trained according to conditional maximum likelihood methods, also capable of predicting transmembrane β-barrel topology | − | 5 | http://biophysics.biol.uoa.gr/PRED-TMBB/ |
| TMHMM | Transmembrane α-helix | A hidden Markov model that determines seven states or parameters related to membrane protein topology and the probability of each over 20-amino-acid stretches | + | 69 | http://www.cbs.dtu.dk/services/TMHMM/ |
| PHOBIUS | Transmembrane α-helix | Based on a hidden Markov model trained to make an optimal choice between transmembrane segments and signal peptides | + | 66 | http://phobius.sbc.su.se/ |
| SignalP 3.0 | Signal sequence | Based on two different neural networks (NN), one for determining the probability of an amino acid in a signal peptide and the other for recognition of the cleavage site | + | 10 | http://www.cbs.dtu.dk/services/SignalP/ |
| PrediSi | Signal sequence | Position wweight matrices (PWM) based on amino acid frequencies in known signal sequences | + | 59 | http://www.predisi.de/ |
| Signal-CF | Signal sequence | A two-layer predictor in which the first layer identifies the secretory character of the query protein and, if a signal sequence is identified, the second program predicts the cleavage site | − | 26 | http://www.csbio.sjtu.edu.cn/bioinf/Signal-CF/ |
+ and − indicate, respectively, that the program does or does not have a genome-mining feature.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism 4.0 (GraphPad Software, San Diego, CA). For each condition studied, both the standard deviation and the standard error of the mean were calculated. The percentages of intact spirochetes labeled with IRS versus HSS and the RFI values for intact and TX100-treated organisms were compared using an unpaired Student's t test, with a P value of ≤0.05 considered to be significant.
RESULTS
Surface immunolabeling of T. pallidum in gel microdroplets.
As discussed in Materials and Methods and depicted in Fig. S1 in the supplemental material, we made two major modifications to our original gel microdroplet procedure (31). First, following the primary antibody incubation step, samples were incubated with biotinylated probes for rabbit or human immunoglobulins. Streptavidin-Alexa 546 (orange) was then used to amplify the signal generated by the small amounts of antibodies bound to the surfaces of intact treponemes. Second, to assess the integrity of labeled organisms, samples were coincubated with rat anti-FlaA antibodies during the primary incubation step and then probed with chicken anti-rat IgG, followed by goat anti-chicken IgG, both conjugated to Alexa 488 (green). The double conjugate was used to enhance identification of organisms with disrupted OMs. Parallel staining with anti-flagellar antibodies was an important modification, inasmuch as small percentages of organisms showed some degree of OM disruption in each experiment, and these might otherwise have been scored as surface labeled. Table 2 presents a summary of all of the labeling experiments described below.
TABLE 2.
Summary of gel microdroplet data
| Expta,b | No. of expts | Without TX100 [mean % ± SEM (total per category)] |
With TX100 (mean %) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of T. pallidum organisms counted | Intact, unlabeled | Intact, labeled | OM disrupted, labeled | No. of T. pallidum organisms counted | Unlabeled | Labeled | |||||||
| IRS (IgG + IgM)c | |||||||||||||
| IRS | 7 | 700 | 83.3 ± 1.71 (583) | 9.8 ± 1.78 (69) | 6.8 ± 0.70 (48) | 700 | 0 | 100 | |||||
| NRS | 7 | 700 | 99.7 ± 0.49 (698) | 0 | 0.2 ± 0.49 (2) | 700 | 100 | 0 | |||||
| HSS (IgG + IgM)d | |||||||||||||
| HSS | 7 | 709 | 89.0 ± 2.19 (622) | 5.8 ± 1.84 (41) | 5.1 ± 1.94 (37) | 700 | 0 | 100 | |||||
| NHS | 7 | 700 | 99.8 ± 0.41 (699) | 0 | 0.1 ± 0.41 (1) | 700 | 100 | 0 | |||||
| IRS (Ig vs. IgM) | |||||||||||||
| IgG + IgM | 2 | 200 | 81.5 ± 0.71 (163) | 11.5 ± 0.71 (23) | 7.0 ± 0.0 (14) | 200 | 0 | 100 | |||||
| IgG only | 2 | 200 | 85.0 ± 0.71 (170) | 10.0 ± 1.42 (20) | 5.0 ± 1.42 (10) | 200 | 0 | 100 | |||||
| IgM only | 2 | 200 | 89.5 ± 0.71 (179) | 4.5 ± 2.12 (9) | 6.0 ± 1.42 (12) | 200 | 0 | 100 | |||||
| NRS (IgG + IgM) | 2 | 200 | 100 | 0 | 0 | 200 | 100 | 0 | |||||
| HSS (IgG vs. IgM) | |||||||||||||
| IgG + IgM | 2 | 200 | 88.0 ± 2.83 (176) | 6.5 ± 3.54 (13) | 5.5 ± 0.71 (11) | 200 | 0 | 100 | |||||
| IgG only | 2 | 200 | 87.5 ± 6.36 (175) | 6.5 ± 6.0 (13) | 6.0 ± 0 (12) | 200 | 0 | 100 | |||||
| IgM only | 2 | 200 | 92.5 ± 0.71 (185) | 1.0 ± 0.0 (2) | 6.5 ± 0.71 (13) | 200 | 0 | 100 | |||||
| NHS (IgG + IgM) | 2 | 200 | 100 (200) | 0 | 0 | 200 | 100 | 0 | |||||
| VDRL | |||||||||||||
| VDRL antiserum (IgG + IgM) | 3 | 300 | 92.3 ± 1.16 (277) | 0.6 ± 0.58 (2) | 7.0 ± 1.0 (21) | 300 | 0 | 100 | |||||
| NRS (IgG + IgM) | 3 | 300 | 99.6 ± 0.58 (299) | 0 | 0.3 ± 0.58 (1) | 300 | 100 | 0 | |||||
| IRS (Ig-G + IgM) PK | |||||||||||||
| IRS no PK | 2 | 200 | 84.0 ± 0 (168) | 8.5 ± 0.71 (17) | 7.5 ± 0.71 (15) | 200 | 0 | 100 | |||||
| IRS 50μg PK | 2 | 200 | 70.5 ± 2.12 (141) | 7.5 ± 0.71 (15) | 22.0 ± 2.83 (44) | 200 | NDe | ND | |||||
| IRS 100μg PK | 2 | 200 | 67.5 ± 0.71 (135) | 8.0 ± 2.83 (16) | 24.5 ± 2.13 (49) | 200 | 0 | 100 | |||||
| NRS (IgG + IgM) | 2 | 200 | 100 | 0 | 0 | 200 | 100 | 0 | |||||
| Anti-TprK | |||||||||||||
| Anti-TprK (IgG) | 3 | 292 | 96.2 ± 0.25 (281) | 0 | 3.7 ± 0.25 (11) | 300 | 0 | 100 | |||||
| NRatS (IgG) | 3 | 300 | 100 | 0 | 0 | 300 | 100 | 0 | |||||
| HSS (IgG + IgM) | 3 | 300 | 90.1 ± 1.83 (270) | 5.9 ± 0.15 (18) | 3.9 ± 1.98 (12) | 300 | 100 | 0 | |||||
| Anti-Tp0136 | |||||||||||||
| Anti-Tp0136 (IgG + IgM) | 3 | 300 | 93.0 ± 1.0 (279) | 0.6 ± 0.58 (2) | 6.3 ± 1.53 (19) | 300 | 0 | 100 | |||||
| NRS (IgG + IgM) | 3 | 277 | 100 | 0 | 0 | 300 | 100 | 0 | |||||
NRS, normal rabbit serum; NHS, normal human serum; NRatS, normal rat serum.
The mean percentages for NRS, NHS, and NRatS represent labeling with Alexa 546 only.
Includes IRS and NRS controls used in the IgG/IgM and PK experiments.
Includes HSS and NHS controls used in the IgG/IgM and TprK experiments.
ND, not determined.
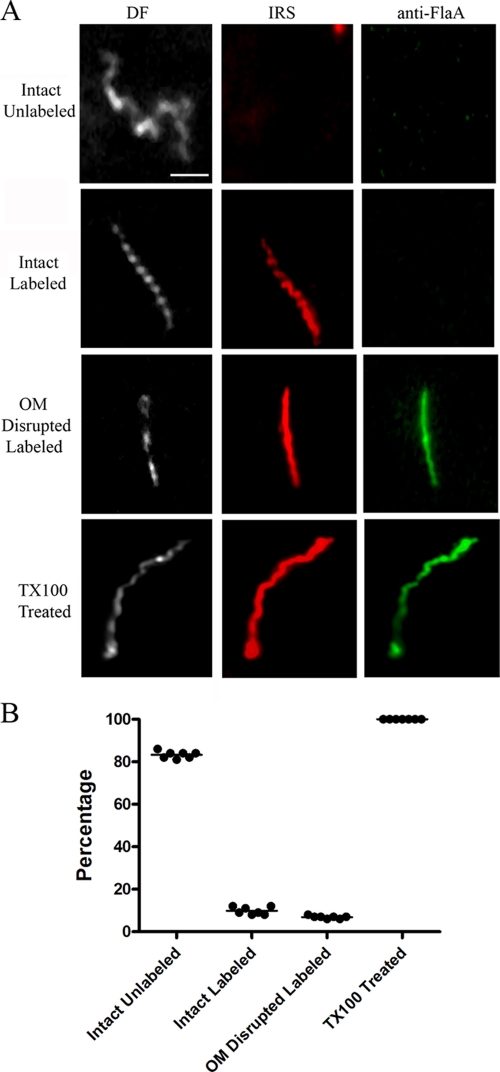
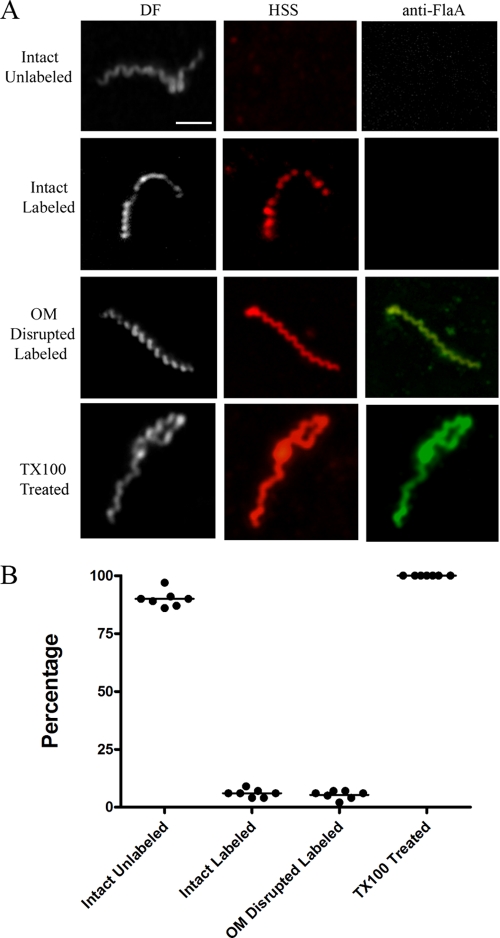
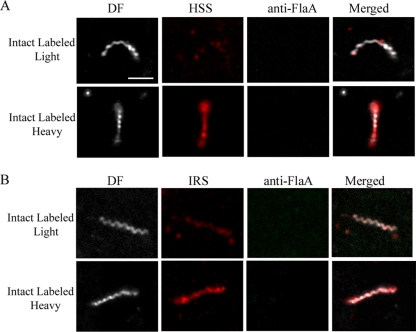
As shown in Fig. 3 and 4, with both IRS and HSS, we observed treponemes labeled with Alexa 546, but not with Alexa 488, a result indicative of true surface labeling. With both syphilitic sera, however, the percentages of intact, labeled treponemes were low, averaging 9.8% (IRS) and 5.8% (HSS) of organisms in 7 independent experiments performed with each primary antibody; the difference between these percentages was highly significant (P = 0.001). Also noteworthy was the fact that the patterns and intensities of labeling of intact treponemes with both IRS and HSS also varied. The labeling of some organisms was light and sporadic (confirmed by merging the dark-field and immunostaining images), whereas others fluoresced uniformly (Fig. 5). Five to 7% of organisms displayed various degrees of disruption of their outer membranes, as determined by labeling them with anti-FlaA antibodies; all of them were also labeled with syphilitic sera. Whereas the overwhelming majority of intact organisms failed to label with either IRS or HSS, 100% of TX100-treated spirochetes were uniformly labeled with both anti-flagellar antibodies and syphilitic sera. Quantitative image analysis confirmed that the labeling of detergent-treated spirochetes with HSS was significantly greater than that of intact organisms (mean RFI ± standard error of the mean [SEM] = 23.04 ± 3.78 and 13.8 ± 1.10, respectively; P = 0.034). There was no labeling of intact treponemes with normal rabbit or human sera and only rare, weak labeling of disrupted organisms, confirming the specificity of the staining with syphilitic sera (Table 2 and data not shown).
FIG. 3.
Surface labeling of T. pallidum with IRS. (A) Representative micrographs from seven independent experiments. DF, dark field. Scale bar = 2.5 μm. (B) Dot plot of key results. In this and subsequent plots, each dot presents the percentage of organisms in a particular labeling category from one experiment; the lines represent mean percentages. Complete results for all immunolabeling experiments are presented in Table 2.
FIG. 4.
Surface labeling of T. pallidum with pooled HSS. (A) Representative micrographs from seven independent experiments. Scale bar = 2.5 μm. (B) Dot plot of key results.
FIG. 5.
Variability of surface labeling of T. pallidum by IRS (A) and HSS (B). Scale bar = 2.5 μm.
In the above-mentioned experiments, samples were incubated with probes for both IgG and IgM to maximize the detection of surface-bound antibodies. To distinguish surface labeling by these two classes of immunoglobulins, we also performed experiments in which samples were incubated separately with probes directed against IgG or IgM. With IRS, approximately twice as many intact organisms were labeled with IgG than with IgM antibodies (10% versus 4.5% in two independent experiments); the intensity of labeling was also discernibly greater for IgG than for IgM (not shown). The disparity between IgG and IgM labeling was much greater for HSS in that light labeling for IgM was observed only rarely (6.5% versus 1% for IgG and IgM, respectively, in two experiments).
T. pallidum surface antigens are nonlipoidal but proteinase K resistant.
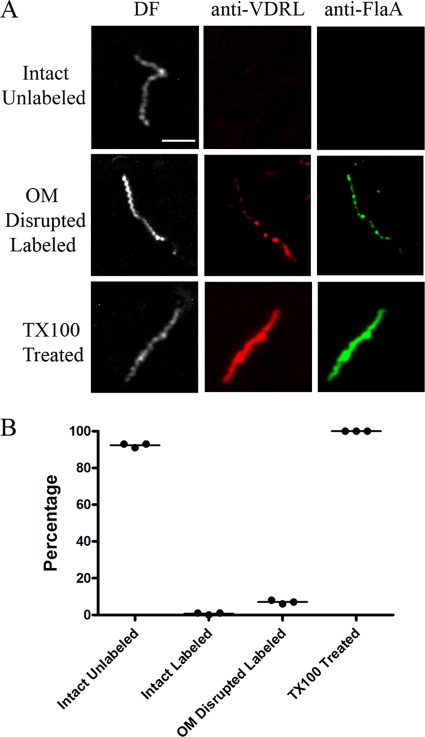
Syphilitic infection elicits two distinctly different types of antibody responses, traditionally designated “treponemal” and “nontreponemal” (79, 105). The former are directed against polypeptide antigens of T. pallidum. Nontreponemal antibodies, on the other hand, recognize lipoidal antigens and are detected using a defined mixture of phosphatidylcholine, cholesterol, and cardiolipin in which cardiolipin is the major antigenic component (73, 74). The term “nontreponemal” derives from the longstanding belief that such antibodies are elicited by host lipids liberated as a consequence of the inflammatory response evoked by spirochetes in tissues (79, 105). We previously showed, however, that T. pallidum contains cardiolipin (9, 106), while Baker-Zander et al. (6) reported that VDRL antibodies promote opsonophagocytosis of treponemes by rabbit peritoneal macrophages. Thus, it was important to determine whether the nontreponemal antibodies in syphilitic sera contribute to the surface immunolabeling of virulent T. pallidum. To address this question, we assessed the reactivity of encapsulated spirochetes with an opsonic rabbit serum generated against VDRL antigen (6); prior to its use, we confirmed that the antiserum contained high-titer (1:1,024) VDRL reactivity. Of the 300 organisms examined in three independent experiments, only 2 of 279 intact organisms were faintly labeled by VDRL antibodies (i.e., 277 intact organisms were unlabeled), while all 21 of the organisms scored as having disrupted OMs were labeled to various extents (Fig. 6). Particularly striking was the fact that all detergent-treated treponemes fluoresced intensely and uniformly following incubation with the VDRL antiserum (Fig. 6). Collectively, these findings indicate that the contribution of VDRL antibodies to the surface labeling observed with syphilitic sera is negligible. These results are in accord with our previously reported fractionation results (106), demonstrating that, as with Gram-negative bacteria (84, 117), cardiolipin is also a CM constituent in T. pallidum.
FIG. 6.
Cytoplasmic membrane association of T. pallidum lipoidal antigens recognized by anti-VDRL antiserum. (A) Representative micrographs from three independent experiments. Scale bar = 2.5 μm. (B) Dot plot of key results.
In addition to phospholipids, T. pallidum outer membranes contain one or more uncharacterized glycolipids, which we previously showed by a combination of thin-layer chromatography and immunoblot analysis do not react with syphilitic serum (106). Here, we used the highly sensitive chemiluminescence technique for immunoblot analysis to further examine the possibility that T. pallidum contains glycolipids that could be surface antigenic targets; antibodies in HSS did not recognize any protease-resistant, low-molecular-mass molecules in T. pallidum lysates digested extensively with proteinase K (data not shown). Because OMPs of Gram-negative bacteria form β-barrels that are highly resistant to surface proteolysis (48, 60, 126), it was of interest to assess the effect of PK treatment on the surface immunolabeling of T. pallidum. Encapsulated treponemes were pretreated with 50 μg or 100 μg of PK prior to incubation with IRS, as described in Materials and Methods. PK treatment (which included additional washing steps) increased the percentage of damaged, labeled organisms but had no discernible effect on the labeling of intact treponemes.
Periplasmic location of both TprK and TP0136.
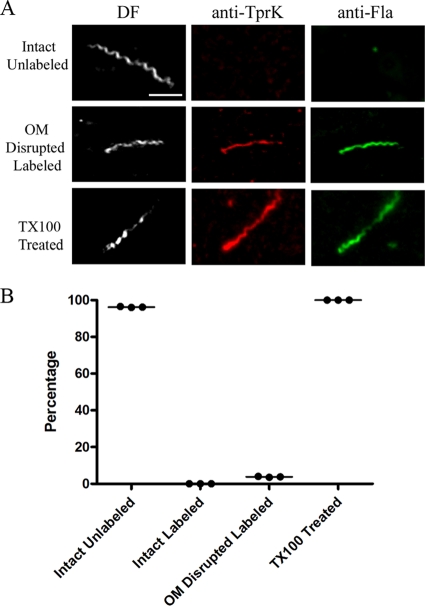
In the postgenomic era, TprK has been the most extensively studied candidate rare OMP. While Centurion-Lara et al. (23) reported that TprK is an opsonic target, we were unable to confirm this finding or to obtain other corroborating evidence for an outer membrane location by other localization methodologies, including immunofluorescence analysis of encapsulated treponemes (58). To address these discrepant results, we reexamined the cellular location of TprK using our improved microdroplet assay, along with a newly generated rat antiserum directed against full-length recombinant protein. Immunoblot analysis revealed that the antiserum was monospecific, recognizing a polypeptide in T. pallidum whole-cell lysates with an apparent molecular mass (46 kDa) close to that (48.8 kDa) predicted for the processed (i.e., with signal peptide removed) form of TprK (see Fig. 9 and data not shown). In three independent experiments, we failed to observe labeling of any intact treponemes with this antiserum (Fig. 7). Disrupted organisms, however, labeled with anti-TprK antibodies, as did all detergent-treated treponemes (Fig. 7).
FIG. 7.
Periplasmic location of TprK. (A) Representative micrographs from three independent experiments. Scale bar = 2.5 μm. (B) Dot plot of key results.
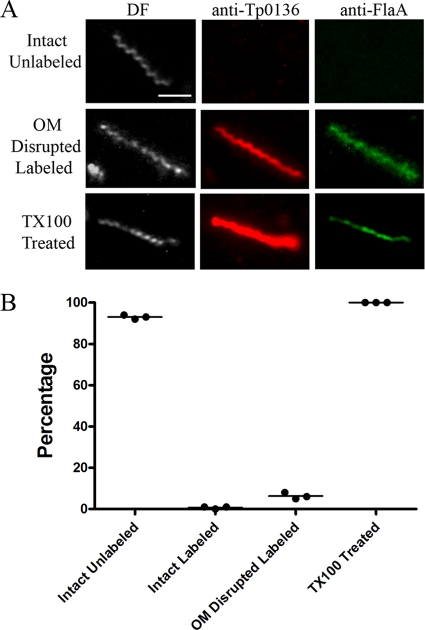
TP0136 is a fibronectin-binding lipoprotein recently reported to be surface exposed. In three independent experiments using the same anti-TP0136 antiserum as Brinkman et al. (14), we observed light labeling of only 2 of 281 intact treponemes; in contrast, all disrupted organisms (total, 19) identified were labeled, as were all detergent-treated organisms (Fig. 8).
FIG. 8.
TP0136 is cytoplasmic membrane associated. (A) Representative micrographs from three independent experiments. Scale bar = 2.5 μm. (B) Dot plot of key results.
A consensus computational framework to predict the T. pallidum OMPeome.
The T. pallidum genome encodes only one polypeptide, TP0326/Tp92 (22), with sequence relatedness to families of known outer-membrane-spanning proteins; TP0326 is an ortholog of BamA (β-barrel assembly machinery A) (Desrosiers et al., unpublished), the principal component of the outer membrane biogenesis complex (117). Consequently, alternative bioinformatics approaches must be employed to analyze the spirochete's genomic sequence for candidate rare OMPs. In recent years, numerous predictive tools have been devised for subcellular localization and structural analysis of actual and translated proteins from prokaryotic genomic sequences. We reasoned that a strategy using diverse computational approaches to achieve a consensus would yield the most robust prediction for the T. pallidum OMPeome. As described in Materials and Methods and depicted in Fig. 2, we used seven computational tools to analyze 1,038 coding sequences for predicted OM localization or β-barrel topology. Interestingly, the outputs of each program (summarized in Table S1 in the supplemental material) differed considerably with respect to both the number of potential OMPs and the specific proteins identified. For some programs (e.g., TMBETADISC [PSSM and AAC]), the number of predicted OMPs considerably exceeded the percentage (approximately 2 to 3%) of the total proteome considered to be typical for a eubacterial OMPeome (127) and included a number of annotated non-OMPs and lipoproteins. By comparison, BOMP and Hhomp identified relatively few annotated non-OMPs but still showed only limited overlap (six proteins). To eliminate as many false positives as possible, the outputs were filtered to remove (i) annotated orthologs of non-OMPs, (ii) lipoproteins, and (iii) proteins with α-helical transmembrane sequences other than potential signal sequences. The remaining proteins, grouped according to the number of OM localization/topology programs that identified them, were examined for the presence of cleaved signal sequences. Table 3 contains a summary of the results for the total of 19 proteins identified by four or more programs, while the predicted signal sequences and cleavage sites are shown in Fig. S2 in the supplemental material.
TABLE 3.
Predicted candidate T. pallidum rare outer membrane proteins
| Protein tag | Name | OM localization/β-barrel predictionsa |
Signal sequence predictionsd |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cello | Psort | TMBETADISC (PSSM profileb) | TMBETADISC (AACc) | Bomp | Hhomp | Predtmbb | Ymaxe (SignalP 3.0) | Sf (PrediSI) | Signal-CFg | ||
| Group 1 | |||||||||||
| TP0326 | BamA | + | + | + | + | + | + | + | 0.831 | 1 | + |
| TP0865 | HPh | + | + | + | + | + | + | + | 0.125 | 0.70 | + |
| Group 2 | |||||||||||
| TP0117 | TprC/D | + | − | + | + | + | + | + | 0.531 | 0.65 | + |
| TP0620 | TprI | + | − | + | + | + | + | + | 0.531 | 0.65 | + |
| TP0621 | TprJ | + | + | + | + | − | + | + | 0.691 | 0.72 | + |
| TP0969 | HP | + | + | + | + | − | + | + | 0.205 | 0.37 | + |
| Group 3 | |||||||||||
| TP0316 | TprF | + | − | + | + | − | + | + | 0.518 | 0.65 | + |
| TP1031 | TprL | + | − | + | − | + | + | + | 0.065 | 0 | − |
| Group 4 | |||||||||||
| TP0009 | TprA | + | − | + | + | + | − | − | 0.234 | 0.22 | − |
| TP0011 | TprB | + | − | + | − | − | + | + | 0.324 | 0.477 | + |
| TP0155 | CHPi | + | + | + | + | − | − | − | 0.13 | 0.7 | + |
| TP0313 | TprE | + | − | + | − | − | + | + | 0.691 | 0.72 | + |
| TP0325 | CHP | + | − | − | + | + | + | − | 0.218 | 0.47 | + |
| TP0421 | CHP | + | + | + | − | − | − | + | 0.443 | 0.75 | + |
| TP0548 | HP | + | − | + | − | − | + | + | 0.226 | 0.64 | + |
| TP0729 | HP | + | + | + | + | − | − | − | 0.27 | 0.80 | + |
| TP0855 | HP | + | + | + | + | − | − | − | 0.432 | 0.54 | + |
| TP0858 | HP | + | − | + | − | − | + | + | 0.249 | 1 | + |
| TP0897 | TprK | + | − | + | − | − | + | + | 0.75 | 0 | − |
Proteins are grouped according to the number of programs that predict OM localization or β-barrel formation.
Position-specific scoring matrix profiles.
Amino acid composition.
Predicted N-terminal sequences and cleavage sites are shown in Fig. S1 in the supplemental material.
Ymax is a derivative of the C score combined with the S score, where the C score is the cleavage site score and the S score is the amino acid position score in the signal sequence (10). A Ymax score of >0.4 indicates a cleaved signal sequence.
The score was calculated using the equation 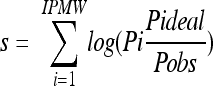 (59). A score of >0.5 indicates a cleaved signal sequence.
(59). A score of >0.5 indicates a cleaved signal sequence.
Presence (+) or absence (−) of a cleavable signal sequence.
HP, hypothetical protein.
CHP, conserved hypothetical protein.
Only two proteins, TP0326/Tp92 and TP0865, were identified by all seven algorithms (group 1); of note, TP0326/Tp92 was originally identified as an opsonic target of syphilitic sera, supporting its OM location and surface exposure (22). SignalP 3.0, PrediSi, and Signal-CF identified the same canonical cleaved signal sequence for TP0326. On the other hand, SignalP was unable to identify a cleavage site for the signal sequence of TP0865, while two different cleavage sites were predicted by PrediSi and Signal-CF (residues 55 and 17, respectively), suggesting the need for experimental analysis of the TP0865 N terminus for a functional, cleaved export signal. Group 2 contains four proteins, three of which, TP0117/TP0131, TP0620, and TP0621, are members of the T. pallidum repeat (Tpr) family (TprC/D, TprI, and TprJ, respectively). All three Tprs in group 2 were predicted to have cleaved signal sequences by all three programs. Group 2 also contains the hypothetical protein TP0969, which was predicted only by Signal-CF to possess a cleaved signal sequence. Given the divergence in signal sequence predictions for TP0969, we manually inspected the DNA sequence in the vicinity of the annotated translational start site for an alternative translational start codon but found none.
The two proteins in group 3, TP1031 and TP0316, both members of the Tpr family (TprL and TprF, respectively), were identified by five OM localization/topology programs. Interestingly, none of the programs identified a signal sequence in TprL. We considered the possibility that the translational start site of TprL was incorrectly annotated. Even with careful manual inspection of the DNA sequence near the predicted translational start site, however, we were unable to identify an alternative translational start site. TP0316 is virtually identical to TP0117/TP0131 and TP0620 for the first 39 kDa of its sequence; its lower ranking results from the fact that it is truncated due to a frameshift. Lastly, the 11 proteins in group 4 consisted of seven hypothetical proteins (TP0155, TP0325, TP0421, TP0548, TP0729, TP0855, and TP0858) and four Tprs (TP0009 [TprA], TP0011 [TprB], TP0313 [TprE], and TP0897 [TprK]). All three signal sequence algorithms predicted cleaved signal peptides for only TP0313, TP0421, and TP0855 and failed to identify any export signal for TP0009. TP0155, a fibronectin-binding protein (21), was predicted by PrediSi and Signal-CF to have a cleaved signal peptide, although the cleavage sites predicted by the two programs differed; SignalP, on the other hand, predicted an uncleaved signal peptide. Our analysis of the TprK N terminus was based upon our prior experimental demonstration that it is translated from an alternative start codon and has an N-terminal export signal (58). Surprisingly, neither PrediSi nor Signal-CF identified an N-terminal export signal from this start site, most likely because of the noncanonical arginine residues at positions 23 and 26, whereas SignalP 3.0 predicted a signal peptide cleaved at residue 30.
TprK lacks the amphiphilic character expected of OM-spanning proteins.
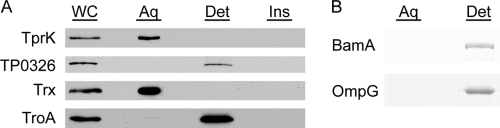
The localization results for TprK (Fig. 7) are clearly discordant with the OM predictions of some algorithms (Table 3). To help assess the accuracy of the bioinformatics predictions, we conducted experiments to determine whether TprK possesses the amphiphilic character expected of an outer-membrane-spanning protein. Triton X-114 phase partitioning of freshly harvested T. pallidum, shown in Fig. 9A, revealed that native TprK partitions exclusively into the aqueous phase; this finding, obtained with the new anti-TprK antiserum, confirms previously reported phase-partitioning results showing that TprK lacks amphiphilicity (58). In silico analysis of TP0326 predicts that it has a bipartite structure consisting of 5 periplasmic POTRA domains and a C-terminal β-barrel containing 18 membrane-spanning segments (Desrosiers et al., unpublished). In contrast to TprK, native TP0326 partitioned exclusively into the detergent-enriched phase (Fig. 9A). Figure 9A also reveals that TroA (TP0163), a periplasmic protein with an uncleaved leader sequence, partitions into the detergent-enriched phase, as previously reported (1), whereas the cytoplasmic enzyme thioredoxin (TP0919), as expected, is hydrophilic. Recombinant TroA without a signal sequence partitions into the aqueous phase and will not integrate into liposomes (1). The amphiphilic behavior of native TroA, therefore, supports the prediction of SignalP 3.0 that TprK has a cleaved signal sequence and, more generally, demonstrates that a single transmembrane domain is sufficient to completely alter a protein's phase-partitioning behavior.
FIG. 9.
TprK lacks the amphiphilic character of β-barrel-forming proteins. (A) Freshly harvested T. pallidum, solubilized in 2% Triton X-114, was phase partitioned and subjected to immunoblot analysis with monospecific antisera directed against TrpK, TP0326, thioredoxin (Trx), and TroA. Lanes: whole cells (WC), aqueous phase (Aq), detergent-enriched phase (Det), and insoluble material (Ins). (B) Purified recombinant E. coli BamA and OmpG were phase partitioned and stained with Gel-code Blue following SDS-PAGE.
Based on work with Leptospira, the concern has been raised that OMPs do not always partition as expected into the TX114 detergent-enriched phase (98). A number of years ago, we reported that native, trimeric porins (i.e., OmpF/C) solubilized from E. coli cell envelopes are amphiphilic by the TX114 method (121). To obtain further evidence that amphiphilicity is a general property of β-barrel-forming proteins, we also examined the phase-partitioning behavior of reconstituted, recombinant E. coli BamA and OmpG. E. coli BamA is proposed to have 16 membrane-spanning domains (49); OmpG is a monomeric β-barrel with 14 membrane-spanning segments (29, 120). As shown in Fig. 9B, both proteins partitioned exclusively into the detergent-enriched phase.
DISCUSSION
Although rare examples of T. pallidum residing within nonphagocytic eukaryotic cells in both tissue culture (46, 68) and lesional biopsy specimens from patients (122) have been documented, the spirochete is widely considered to be an extracellular parasite (36, 104, 113). As such, one might anticipate that the appearance of specific antibodies would herald the eradication of the invader and the control of syphilitic infection, yet as discussed above, clinical and experimental evidence to support this assumption has not been easy to garner. More than 5 decades ago, investigators began to note the poor surface reactivity of motile T. pallidum in serologic tests (55, 86, 89). Their studies fostered the notion that the spirochete is surrounded by a protective layer comprised of serum proteins and/or mucopolysaccharides (3, 27, 44). Over the years, we and others have presented multiple lines of evidence to disprove the so-called “outer coat hypothesis,” most recently by using cryoelectron tomography to show that treponemes preserved in a near-native state do not possess a discernible layer external to the OM (32, 64, 77a, 102). A contemporary understanding of the unusual interactions between live spirochetes and anti-treponemal antibodies dates from the discovery of T. pallidum's unique protein-deficient outer membrane and the description of its major lipoprotein antigens as proteins anchored by N-terminal lipids to the periplasmic leaflet of the CM (20, 25, 102). Here, we present additional immunofluorescence data in support of our ultrastructural scheme (Fig. 1), including findings contrary to the claim by Brinkman et al. (14) that TP0136 is an exception to the generalization that T. pallidum lacks surface-exposed lipoproteins.
Assays for examining the interactions of antibodies in syphilitic sera with motile T. pallidum fall into two broad categories: those that measure the effects of syphilitic sera on a biological function or property of live treponemes (sometimes referred to as “functional” assays) and those that directly assess surface antibody binding. Whereas studies involving direct detection of surface antibody binding to intact organisms generally have yielded negative results (31, 32, 95, 118), biological/functional assays have shown unequivocally that syphilitic serum contains antibodies that bind to the treponemal surface, albeit inefficiently and/or with delayed kinetics (2, 45, 72, 80, 89, 110). We previously speculated that these discordant results reflect differences in the sensitivities of the two methodologies (102), and we modified our gel microdroplet technique to help resolve this dichotomy. These modifications enabled us to directly detect antibodies bound to the surfaces of intact treponemes, although, notably, on only a small proportion of intact organisms, and they have yielded indirect evidence that the molecules recognized by treponemal antibodies are rare OMPs. Also noteworthy was the small but significantly greater surface binding of antibodies in IRS than in HSS. Whether this difference merely reflects the heterologous nature of the HSS with respect to the Nichols strain or is a biological correlate of the protected status of immune rabbits is an important question. In recent years, opsonophagocytosis has become the biological/functional assay of choice, supplanting complement-dependent immobilization (78). It is not widely appreciated, however, that the readout for opsonophagocytosis is the percentage of cells with internalized treponemes (80), not the percentage of input treponemes ingested. In our assays using human monocytes and HSS, we found that at least half of the input treponemes were recoverable at 4 h, even at low (i.e., 10:1) multiplicities of infection (34). Thus, while opsonophagocytosis still appears to be the more sensitive indicator of surface binding of antibodies in syphilitic serum, in our hands, both methods seem to be in agreement on two key, interrelated points: (i) treponemal populations are heterogeneous with respect to the density of surface antigenic targets on individual organisms and (ii) the surface antigenicity of a substantial percentage of treponemes falls below the threshold for detection of antibody binding.
The prediction that rare OMPs in T. pallidum form β-barrels (19, 36, 102) is well grounded in three assumptions derived from innumerable investigations of the composition, structure, function, and biogenesis of bacterial cell envelopes (90, 92, 100, 117): (i) even with the spirochete's remarkably low rate of multiplication (83) and comparatively fluid OM (20, 33), fixed channels or pores are needed to facilitate influx across the OM permeability barrier of the numerous hydrophilic nutrients the parasite must acquire from its obligate mammalian host (47); (ii) α-helical transmembrane domains serve as stop-transfer sequences during translocation of polypeptides across the CM by the highly conserved Sec export machinery; and (iii) OMP precursors containing multiple short amphipathic segments circumvent the constraints on Sec-dependent export associated with hydrophobic α-helices yet can form closed channels once properly folded into the OM bilayer. Evidence accumulated in the postgenomic era has further solidified the concept of the β-barrel as a universal topology of prokaryotic OMPs (112, 127). Mycobacteria, long classified as Gram positive, possess OMs that contain pore-forming β-barrels (91), though they are structurally quite different from the trimeric porins of Gram-negative organisms (42). β-Barrels have also been identified in the OMs of mitochondria and chloroplasts, eukaryotic organelles originating, respectively, from alphaproteobacterial and cyanobacterial endosymbiotic precursors (62, 130). Perhaps most important has been the discovery of the conserved assembly machinery that chaperones newly exported OMP precursors into the OMs of virtually all diderms, including Borrelia burgdorferi (77) and oral treponemes (65), as well as mitochondria and chloroplasts (67, 111). Indeed, the presence in T. pallidum of a protein, TP0326, that appears to possess the characteristic BamA bipartite structure (i.e., N-terminal POTRA domains and a C-terminal β-barrel [Fig. 1]) (Desrosiers et al., unpublished) is prima facie evidence that T. pallidum synthesizes as yet unidentified β-barrel precursors as substrates for an OM assembly machinery.
Collectively, the above considerations have transformed our quest for rare OMPs, heretofore conducted largely at the protein level, into a bioinformatics-based investigation of the spirochete's genome for sequences predicted to assume a β-barrel conformation. Whereas the α-helical transmembrane domains found in CM proteins are readily predictable, the diversity of computational approaches developed to identify β-barrels reflects the difficulties inherent in distinguishing hydrophilic proteins from proteins whose amphiphilic behavior stems from multiple short (approximately 10-amino-acid) segments of variable alternating polar and nonpolar residues interspersed throughout the polypeptide sequence (4, 53, 112). An additional confounder is the fact that all extant OMP prediction tools have been devised and evaluated using training and test sets derived from Gram-negative organisms and have yet to be validated for phylogenetically distant diderms, including spirochetes, using structural methodologies. An unintended bias in the algorithms toward known OMPs may explain why TP0326 was one of only two proteins identified by all seven computational tools. To avoid the pitfalls associated with any single prediction method, we adopted a consensus computational framework based on the straightforward assumption that a protein's candidacy would be strengthened in proportion to the number of algorithms that identified it. Indeed, the necessity for a consensus-based approach was evident from the marked disparities in outputs, including numerous false-positive annotated proteins, generated by the seven predictive tools. Because structure-based sequence analytical tools often do not heavily weigh whether a signal sequence is present, we also analyzed the clustered outputs for cleavable signal sequences. This additional step proved useful, demoting two candidates (TP0009 [TprA] and TP1031 [TprL]) and further suggesting that the β-barrel predictions for both were false positives despite the concordance of multiple algorithms (5 for TP0131 and 4 for TP0009). It must be emphasized, however, that the bioinformatics predictions in Table 3 indicate only that a subset of proteins has achieved “candidate” status. Confirmation that a candidate is a bona fide OMP ultimately rests on obtaining empirical evidence that it possesses commensurate physicochemical properties in concert with surface localization. Also to be noted is the important caveat that our bioinformatics approach was designed to identify proteins predicted to form monomeric β-barrels and likely would have missed proteins that, like TolC of E. coli (99) and MspA of Mycobacterium smegmatis (42), form closed cylinders only as multimeric complexes.
The 17 candidates identified by consensus predictions for both OM localization/β-barrel topology and cleaved signal sequences consisted, in addition to TP0326, of 7 members of the Tpr family (23, 72) and 9 hypothetical or conserved hypothetical proteins (“non-Tprs”) for which there is no other information available in the databases. In a report published in 2003, Cameron (19) used an earlier version of PSORT (v1.0), in concert with structural algorithms, to screen the T. pallidum genome for OMPs. Of the 10 candidates she described, TP0155 (21), TP0326, TP0316 (TprF), and TP0620 (TprI) were identified in the present study. A protein that is noteworthy because it was not identified by our consensus framework is TP0262 (TpN50). Originally reported as an ortholog for E. coli OmpA (54), we demonstrated using the original bead method that it is periplasmic and proposed that its sequence homology to OmpA was due to the presence of a C-terminal peptidoglycan binding domain (31, 39). Based on cryoelectron tomography findings, we suggested that TpN50 is part of a scaffold that stabilizes the peptidoglycan layer within the periplasmic space, creating a partition between CM-associated proteins and a less viscous zone containing the rotating flagellar filaments (Fig. 1) (64). Our current bioinformatics results are consistent with these earlier localization data and our more recent functional speculations.
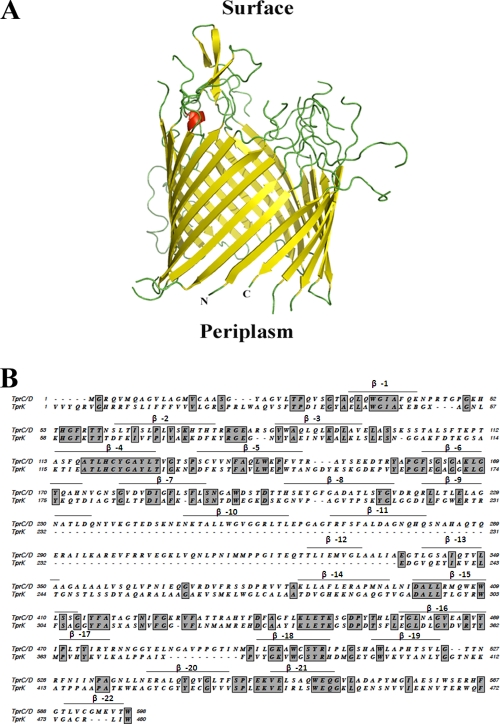
The discovery of the 12-member polymorphic family of Tpr proteins was arguably the greatest immediate dividend of the T. pallidum Nichols genomic sequencing effort (23, 47, 107). The OM location of at least some of these polypeptides initially was supported by their various degrees of sequence relatedness to the amphiphilic, surface-exposed major sheath protein of Treponema denticola (18, 41, 43), as well as the rudimentary cellular localization bioinformatics tools of the early genomics era (19, 23, 47, 107). The computational framework described here, the most extensive yet performed on this unique family, revealed that these proteins have a broad distribution of OM localization/β-barrel predictions, a result that is not surprising given their complex phylogenetic relationships and wide degree of sequence heterogeneity (23, 52); it is noteworthy, in fact, that two family members, TP0317 (TprG) and TP0610 (TprH), did not even meet our minimal criteria to be designated candidates. These analyses imply that differences in the amino acid compositions and sequences of individual Tprs could engender varying propensities to form OM-spanning proteins and that lower-ranked Tpr candidates have a greater likelihood of being false positives. While physical characterization of the top-ranked Tprs (e.g., those in group 2) has only just begun in our laboratory, our results to date indicating that TprK, which resides in group 4, is periplasmic and lacks the amphiphilic character of typical OMPs is in line with this hypothesis. Molecular modeling of TprC and alignments of TprC and TprK reveal further why this supposition is plausible. As shown in Fig. 10A, TprC is predicted to form a 22-stranded β-barrel. TprK has low sequence homology with TprC (23% identity) and lacks three of its predicted β-strands (Fig. 10B). Moreover, many of the regions in TprK that align with predicted β-strands of TprC have significant amino acid substitutions from the homologous TprC segments (Fig. 10B). We caution, however, that given the well-known caveats associated with modeling of proteins as β-barrels, this analysis needs detailed experimental examination.
FIG. 10.
(A) β-Barrel structure of TprC (TP0117) predicted by TMBpro (108). Depicted is a cartoon representation of the model in which β-strands, loops, and α-helices are shown in yellow, green, and red, respectively. (B) Amino acid sequence alignment of TprC and TprK generated by MacVector using a PAM matrix. Identical residues are shown in boxes; above the aligned sequences are the β-sheets predicted for TprC by TMBPro.
How, then, does one reconcile our findings and this entire line of thinking with the large amount of published work purportedly consistent with an OM location of TprK (37, 72, 78)? With the exception of opsonophagocytosis assay results (23), which have not been independently verified (58), no direct evidence for the molecule's OM location has been forthcoming, nor have empirical data showing that TprK has structural features of an OMP been reported. Indeed, most of the evidence for surface exposure is indirect, consisting of small degrees of regional sequence heterogeneity in TprKs from different T. pallidum strains and/or correlations of TprK sequence variants generated by selected treponemal strains with concomitant antibody responses (24, 50, 51, 70, 71, 88). A recent attempt to drive TprK antigenic diversity by artificial immunization was successful for the V6 variable region at a single time point during early infection, but not for V5, while comparable data for the molecule's five other variable domains was not presented (51). Whether sequence variation of TprK is a requirement for the establishment of chronic syphilitic infection also remains unsettled. In our hands, the Nichols strain has extremely limited capacity for generating sequence variants of TprK yet is fully capable of persisting in rabbits (58, 123). Given the current state of the evidence, the contention that TprK is a porin whose variable regions reside on surface-exposed loops seems unfounded (51).
Molecular characterization of rare OMPs is universally regarded as a linchpin of strategies to elucidate host-pathogen interactions during syphilitic infection (20, 36, 105). As the present study attests, the development of robust methodologies for probing the surfaces of individual treponemes is no less important for unraveling the twin enigmas of immunity and immune evasion during syphilis. In addition to providing new evidence that lack of antibody binding is a primary mechanism for evading clearance, our gel microdroplet results with syphilitic sera point to a complex dynamic in which spirochetes in a population undergo different fates determined by heterogeneous levels of antibody binding. One of the most perplexing features of early syphilis is that disease progression and regression occur concomitantly (8, 72, 104); simultaneous clearance and immune escape of organisms at a given site or within the bloodstream could help to explain this phenomenon. Conceivably, the balance would shift in favor of the host as infection progresses and the antibody repertoire both broadens and intensifies, narrowing the window for immune escape and suppressing spirochetal loads. Modulation of the total density of surface antigens, the specific molecules that are expressed, and the antigenic specificities of individual molecules all could be intrinsic components of the bacterium's countermeasures, contributing to the waxing-and-waning character of the disease, as well as promoting infectious relapses during early latency. Indeed, this conjecture is in accord with results from an elegant 1992 study by Lukehart and coworkers (81) demonstrating that treponemes recovered from rabbit testes consist of opsonization-susceptible and -resistant subpopulations and that the latter, potentially representing organisms with the capacity to persist, increases with the duration of infection. The changes in OM composition necessitated by this strategy, of course, would also have to be exquisitely calibrated to the bacterium's physiological and nutritional requirements in a given microenvironment. Interpretation of immunolabeling data obtained with syphilitic sera is difficult because the results likely represent the collective binding of antibodies directed against multiple surface molecules. The methodological and bioinformatics advances reported here will enable investigators to move beyond the broad picture painted with syphilitic serum to a detailed dissection of antibody responses to individual rare OMPs, coupled with structure-function analyses of these molecules and delineation of their in situ expression profiles during infection.
Supplementary Material
Acknowledgments
This work was supported by NIH grants AI-26756 (J.D.R.) and 5R03TW008023 (J.C.S.) and by the Connecticut Children's Medical Center (CCMC) Arrison and Burr Curtis Research Funds (J.C.S.). S. D.-E. is the recepient of a career development award from the Northeastern Research Center for Excellence (NIH grant U54 AI057159).
We acknowledge the excellent technical assistance of Morgan LeDoyt.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 27 September 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Akins, D. R., E. Robinson, D. Shevchenko, C. Elkins, D. L. Cox, and J. D. Radolf. 1997. Tromp1, a putative rare outer membrane protein, is anchored by an uncleaved signal sequence to the Treponema pallidum cytoplasmic membrane. J. Bacteriol. 179:5076-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alder, J. D., L. Friess, M. Tengowski, and R. F. Schell. 1990. Phagocytosis of opsonized Treponema pallidum subsp. pallidum proceeds slowly. Infect. Immun. 58:1167-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alderete, J. F., and J. B. Baseman. 1979. Surface-associated host proteins on virulent Treponema pallidum. Infect. Immun. 26:1048-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagos, P. G., T. D. Liakopoulos, and S. J. Hamodrakas. 2005. Evaluation of methods for predicting the topology of Β-barrel outer membrane proteins and a consensus prediction method. BMC Bioinformatics 6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagos, P. G., T. D. Liakopoulos, I. C. Spyropoulos, and S. J. Hamodrakas. 2004. PRED-TMBB: a web server for predicting the topology of Β-barrel outer membrane proteins. Nucleic Acids Res. 32:W400-W404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker-Zander, S. A., J. M. Shaffer, and S. A. Lukehart. 1993. VDRL antibodies enhance phagocytosis of Treponema pallidum by macrophages. J. Infect. Dis. 167:1100-1105. [DOI] [PubMed] [Google Scholar]
- 7.Baslé, A., G. Rummel, P. Storici, J. P. Rosenbusch, and T. Schirmer. 2006. Crystal structure of osmoporin OmpC from E. coli at 2.0 A. J. Mol. Biol. 362:933-942. [DOI] [PubMed] [Google Scholar]
- 8.Baughn, R. E., and D. M. Musher. 2005. Secondary syphilitic lesions. Clin. Microbiol. Rev. 18:205-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belisle, J. T., M. E. Brandt, J. D. Radolf, and M. V. Norgard. 1994. Fatty acids of Treponema pallidum and Borrelia burgdorferi lipoproteins. J. Bacteriol. 176:2151-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 11.Berven, F. S., K. Flikka, H. B. Jensen, and I. Eidhammer. 2004. BOMP: a program to predict integral beta-barrel outer membrane proteins encoded within genomes of Gram-negative bacteria. Nucleic Acids Res. 32:W394-W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bishop, N. H., and J. N. Miller. 1983. Humoral immune mechanisms in experimental syphilis, p. 241-270. In R. F. Schell and D. M. Musher (ed.), Pathogenesis and immunology of treponemal infection, vol. 1. Marcel Dekker, Inc., New York, NY. [Google Scholar]
- 13.Blanco, D. R., K. Reimann, J. Skare, C. I. Champion, D. Foley, M. M. Exner, R. E. W. Hancock, J. N. Miller, and J. N. Lovett. 1994. Isolation of the outer membranes from Treponema pallidum and Treponema vincentii. J. Bacteriol. 176:6088-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brinkman, M. B., M. A. McGill, J. Pettersson, A. Rogers, P. Matejkova, D. Smajs, G. M. Weinstock, S. J. Norris, and T. Palzkill. 2008. A novel Treponema pallidum antigen, TP0136, is an outer membrane protein that binds human fibronectin. Infect. Immun. 76:1848-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brinkman, M. B., M. McKevitt, M. McLoughlin, C. Perez, J. Howell, G. M. Weinstock, S. J. Norris, and T. Palzkill. 2006. Reactivity of antibodies from syphilis patients to a protein array representing the Treponema pallidum proteome. J. Clin. Microbiol. 44:888-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brusca, J. S., and J. D. Radolf. 1994. Isolation of integral membrane proteins by phase partitioning with Triton X-114. Methods Enzymol. 228:182-193. [DOI] [PubMed] [Google Scholar]
- 17.Reference deleted.
- 18.Caimano, M. J., K. W. Bourell, T. D. Bannister, D. L. Cox, and J. D. Radolf. 1999. The Treponema denticola major sheath protein is predominantly periplasmic and has only limited surface exposure. Infect. Immun. 67:4072-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cameron, C. E. 2003. Identification of a Treponema pallidum laminin-binding protein. Infect. Immun. 71:2525-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cameron, C. E. 2006. The T. pallidum outer membrane and outer membrane proteins, p. 237-266. In J. D. Radolf and S. A. Lukehart (ed.), Pathogenic treponema: molecular and cellular biology. Caister Academic Press, Norwich, United Kingdom.
- 21.Cameron, C. E., E. L. Brown, J. M. Kuroiwa, L. M. Schnapp, and N. L. Brouwer. 2004. Treponema pallidum fibronectin-binding proteins. J. Bacteriol. 186:7019-7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cameron, C. E., S. A. Lukehart, C. Castro, B. Molini, C. Godornes, and W. C. Van Voorhis. 2000. Opsonic potential, protective capacity, and sequence conservation of the Treponema pallidum subspecies pallidum Tp92. J. Infect. Dis. 181:1401-1413. [DOI] [PubMed] [Google Scholar]
- 23.Centurion-Lara, A., C. Castro, L. Barrett, C. Cameron, M. Mostowfi, W. C. Van Voorhis, and S. A. Lukehart. 1999. Treponema pallidum major sheath protein homologue TprK is a target of opsonic antibody and the protective immune response. J. Exp. Med. 189:647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Centurion-Lara, A., C. Godornes, C. Castro, W. C. Van Voorhis, and S. A. Lukehart. 2000. The tprK gene is heterogeneous among Treponema pallidum strains and has multiple alleles. Infect. Immun. 68:824-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chamberlain, N. R., M. E. Brandt, A. L. Erwin, J. D. Radolf, and M. V. Norgard. 1989. Major integral membrane protein immunogens of Treponema pallidum are proteolipids. Infect. Immun. 57:2872-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou, K. C., and H. B. Shen. 2007. Signal-CF: a subsite-coupled and window-fusing approach for predicting signal peptides. Biochem. Biophys. Res. Commun. 357:633-640. [DOI] [PubMed] [Google Scholar]
- 27.Christiansen, S. 1963. Protective layer covering pathogenic treponemata. Lancet i:423-425. [DOI] [PubMed] [Google Scholar]
- 28.Clark, E. G., and N. Danbolt. 1955. The Oslo study of the natural history of untreated syphilis. An epidemiologic investigation based on a restudy of the Boeck-Bruusgaard material. J. Chronic Dis. 2:311-344. [DOI] [PubMed] [Google Scholar]
- 29.Conlan, S., and H. Bayley. 2003. Folding of a monomeric porin, OmpG, in detergent solution. Biochemistry (Moscow) 42:9453-9465. [DOI] [PubMed] [Google Scholar]
- 30.Cox, D. L. 1994. Culture of Treponema pallidum. Methods Enzymol. 236:390-405. [DOI] [PubMed] [Google Scholar]
- 31.Cox, D. L., D. R. Akins, S. F. Porcella, M. V. Norgard, and J. D. Radolf. 1995. Treponema pallidum in gel microdroplets: a novel strategy for investigation of treponemal molecular architecture. Mol. Microbiol. 15:1151-1164. [DOI] [PubMed] [Google Scholar]
- 32.Cox, D. L., P. Chang, A. W. McDowall, and J. D. Radolf. 1992. The outer membrane, not a coat of host proteins, limits antigenicity of virulent Treponema pallidum. Infect. Immun. 60:1076-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox, D. L., and J. D. Radolf. 2001. Insertion of fluorescent fatty acid probes into the outer membranes of the pathogenic spirochaetes Treponema pallidum and Borrelia burgdorferi. Microbiology 147:1161-1169. [DOI] [PubMed] [Google Scholar]
- 34.Cruz, A. R., M. W. Moore, C. J. La Vake, C. H. Eggers, J. C. Salazar, and J. D. Radolf. 2008. Phagocytosis of Borrelia burgdorferi, the Lyme disease spirochete, potentiates innate immune activation and induces apoptosis in human monocytes. Infect. Immun. 76:56-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cruz, A. R., A. Pillay, A. V. Zuluaga, L. G. Ramirez, J. E. Duque, G. E. Aristizabal, M. D. Fiel-Gan, R. Jaramillo, R. Trujillo, C. Valencia, L. Jagodzinski, D. L. Cox, J. D. Radolf, and J. C. Salazar. 2010. Secondary syphilis in Cali, Colombia: new concepts in disease pathogenesis. PLoS Negl. Trop. Dis. 4:e690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cullen, P. A., and C. E. Cameron. 2006. Progress towards an effective syphilis vaccine: the past, present and future. Expert Rev. Vaccines 5:67-80. [DOI] [PubMed] [Google Scholar]
- 37.Deitsch, K. W., S. A. Lukehart, and J. R. Stringer. 2009. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat. Rev. Microbiol. 7:493-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deka, R. K., C. A. Brautigam, X. F. Yang, J. S. Blevins, M. Machius, D. R. Tomchick, and M. V. Norgard. 2006. The PnrA (Tp0319; TmpC) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an ATP-binding cassette (ABC)-like operon in Treponema pallidum. J. Biol. Chem. 281:8072-8081. [DOI] [PubMed] [Google Scholar]
- 38a.Deka, R. K., L. Neil, K. E. Hagman, M. Machius, D. R. Tomchick, C. A. Brautigam, and M. V. Norgard. 2004. Structural evidence that the 32-kilodalton lipoprotein (Tp32) of Treponema pallidum is an l-methionine-binding protein. J. Biol. Chem. 279:55644-55650. [DOI] [PubMed] [Google Scholar]
- 39.De Mot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333-334. [DOI] [PubMed] [Google Scholar]
- 40.Desrosiers, D. C., Y. C. Sun, A. A. Zaidi, C. H. Eggers, D. L. Cox, and J. D. Radolf. 2007. The general transition metal (Tro) and Zn2+ (Znu) transporters in Treponema pallidum: analysis of metal specificities and expression profiles. Mol. Microbiol. 65:137-152. [DOI] [PubMed] [Google Scholar]
- 41.Edwards, A. M., H. F. Jenkinson, M. J. Woodward, and D. Dymock. 2005. Binding properties and adhesion-mediating regions of the major sheath protein of Treponema denticola ATCC 35405. Infect. Immun. 73:2891-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faller, M., M. Niederweis, and G. E. Schulz. 2004. The structure of a mycobacterial outer-membrane channel. Science 303:1189-1192. [DOI] [PubMed] [Google Scholar]
- 43.Fenno, J. C., K. H. Muller, and B. C. McBride. 1996. Sequence analysis, expression, and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J. Bacteriol. 178:2489-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fitzgerald, T. J., P. Cleveland, R. C. Johnson, J. N. Miller, and J. A. Sykes. 1977. Scanning electron microscopy of Treponema pallidum (Nichols strain) attached to cultured mammalian cells. J. Bacteriol. 130:1333-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fitzgerald, T. J., R. C. Johnson, J. N. Miller, and J. A. Sykes. 1977. Characterization of the attachment of Treponema pallidum (Nichols strain) to cultured mammalian cells and the potential relationship of attachment to pathogenicity. Infect. Immun. 18:467-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fitzgerald, T. J., J. N. Miller, and J. A. Sykes. 1975. Treponema pallidum (Nichols strain) in tissue cultures: cellular attachment, entry, and survival. Infect. Immun. 11:1133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. D. Cotton, and J. C. Venter. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 48.Freudl, R., S. MacIntyre, M. Degen, and U. Henning. 1986. Cell surface exposure of the outer membrane protein OmpA of Escherichia coli K-12. J. Mol. Biol. 188:491-494. [DOI] [PubMed] [Google Scholar]
- 49.Gentle, I. E., L. Burri, and T. Lithgow. 2005. Molecular architecture and function of the Omp85 family of proteins. Mol. Microbiol. 58:1216-1225. [DOI] [PubMed] [Google Scholar]
- 50.Giacani, L., B. Molini, C. Godornes, L. Barrett, V. W. Van, A. Centurion-Lara, and S. A. Lukehart. 2007. Quantitative analysis of tpr gene expression in Treponema pallidum isolates: differences among isolates and correlation with T-cell responsiveness in experimental syphilis. Infect. Immun. 75:104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giacani, L., B. J. Molini, E. Y. Kim, B. C. Godornes, B. T. Leader, L. C. Tantalo, A. Centurion-Lara, and S. A. Lukehart. 2010. Antigenic variation in Treponema pallidum: TprK sequence diversity accumulates in response to immune pressure during experimental syphilis. J. Immunol. 184:3822-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gray, R. R., C. J. Mulligan, B. J. Molini, E. S. Sun, L. Giacani, C. Godornes, A. Kitchen, S. A. Lukehart, and A. Centurion-Lara. 2006. Molecular evolution of the tpr C, D, I, K, G, and J. genes in the pathogenic genus Treponema. Mol. Biol. Evol. 23:2220-2233. [DOI] [PubMed] [Google Scholar]
- 53.Gromiha, M. M., and M. Suwa. 2007. Current developments on Β-barrel membrane proteins: sequence and structure analysis, discrimination and prediction. Curr. Protein Pept. Sci. 8:580-599. [DOI] [PubMed] [Google Scholar]
- 54.Hardham, J. M., and L. V. Stamm. 1994. Identification and characterization of the Treponema pallidum tpn50 gene, an ompA homolog. Infect. Immun. 62:1015-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hardy, P. H., Jr., and E. E. Nell. 1957. Study of the antigenic structure of Treponema pallidum by specific agglutination. Am. J. Hyg. 66:160-172. [DOI] [PubMed] [Google Scholar]
- 56.Hazlett, K. R., D. L. Cox, M. Decaffmeyer, M. P. Bennett, D. C. Desrosiers, C. J. La Vake, M. E. La Vake, K. W. Bourell, E. J. Robinson, R. Brasseur, and J. D. Radolf. 2005. TP0453, a concealed outer membrane protein of Treponema pallidum, enhances membrane permeability. J. Bacteriol. 187:6499-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hazlett, K. R., F. Rusnak, D. G. Kehres, S. W. Bearden, C. J. LaVake, M. E. LaVake, M. E. Maquire, R. D. Perry, and J. D. Radolf. 2003. The Treponema pallidum tro operon encodes a multiple metal transporter, a Zn-dependent transcriptional repressor, and a semi-autonomously expressed phosphoglycerate mutase. J. Biol. Chem. 278:20687-20694. [DOI] [PubMed] [Google Scholar]
- 58.Hazlett, K. R., T. J. Sellati, T. T. Nguyen, D. L. Cox, M. L. Clawson, M. J. Caimano, and J. D. Radolf. 2001. The TprK protein of Treponema pallidum is periplasmic and is not a target of opsonic antibody or protective immunity. J. Exp. Med. 193:1015-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hiller, K., A. Grote, M. Scheer, R. Munch, and D. Jahn. 2004. PrediSi: prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 32:W375-W379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoenger, A., J. M. Pages, D. Fourel, and A. Engel. 1993. The orientation of porin OmpF in the outer membrane of Escherichia coli. J. Mol. Biol. 233:400-413. [DOI] [PubMed] [Google Scholar]
- 61.Hovind-Hougen, K., A. Birch Andersen, and H. A. Nielsen. 1979. Electron microscopy of treponemes subjected to the Treponema pallidum immobilization (TPI) test. II. Immunoelectron microscopy. Acta Pathol. Microbiol. Scand. 87C:263-268. [PubMed] [Google Scholar]
- 62.Hsu, S. C., and K. Inoue. 2009. Two evolutionarily conserved essential beta-barrel proteins in the chloroplast outer envelope membrane. Biosci. Trends 3:168-178. [PubMed] [Google Scholar]
- 63.Isaacs, R. D., J. H. Hanke, L. M. Guzman-Verduzco, G. Newport, N. Agabian, M. V. Norgard, S. A. Lukehart, and J. D. Radolf. 1989. Molecular cloning and DNA sequence analysis of the 37-kilodalton endoflagellar sheath protein gene of Treponema pallidum. Infect. Immun. 57:3403-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Izard, J., C. Renken, C. E. Hsieh, D. C. Desrosiers, S. Dunham-Ems, V. C. La, L. L. Gebhardt, R. J. Limberger, D. L. Cox, M. Marko, and J. D. Radolf. 2009. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J. Bacteriol. 191:7566-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jun, H. K., Y. M. Kang, H. R. Lee, S. H. Lee, and B. K. Choi. 2008. Highly conserved surface proteins of oral spirochetes as adhesins and potent inducers of proinflammatory and osteoclastogenic factors. Infect. Immun. 76:2428-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Käll, L., A. Krogh, and E. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027-1036. [DOI] [PubMed] [Google Scholar]
- 67.Knowles, T. J., A. Scott-Tucker, M. Overduin, and I. R. Henderson. 2009. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7:206-214. [DOI] [PubMed] [Google Scholar]
- 68.Konishi, H., Z. Yoshii, and D. L. Cox. 1986. Electron microscopy of Treponema pallidum (Nichols) cultivated in tissue cultures of Sf1Ep cells. Infect. Immun. 53:32-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 70.LaFond, R. E., A. Centurion-Lara, C. Godornes, A. M. Rompalo, W. C. Van Voorhis, and S. A. Lukehart. 2003. Sequence diversity of Treponema pallidum subsp. pallidum tprK in human syphilis lesions and rabbit-propagated isolates. J. Bacteriol. 185:6262-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.LaFond, R. E., A. Centurion-Lara, C. Godornes, W. C. Van Voorhis, and S. A. Lukehart. 2006. TprK sequence diversity accumulates during infection of rabbits with Treponema pallidum subsp. pallidum Nichols strain. Infect. Immun. 74:1896-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lafond, R. E., and S. A. Lukehart. 2006. Biological basis for syphilis. Clin. Microbiol. Rev. 19:29-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Larsen, S. A., B. M. Steiner, and A. H. Rudolph. 1995. Laboratory diagnosis and interpretation of tests for syphilis. Clin. Microbiol. Rev. 8:1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Larsen, S. A., V. Pope, and E. J. K. Re Johnson. 1998. A manual of tests for syphilis, vol. 9. American Public Health Association, Washington, DC.
- 75.Lee, Y. H., R. K. Deka, M. V. Norgard, J. D. Radolf, and C. A. Hasemann. 1999. Treponema pallidum TroA is a periplasmic zinc-binding protein with a helical backbone. Nat. Struct. Biol. 6:628-633. [DOI] [PubMed] [Google Scholar]
- 76.Lee, Y. H., M. R. Dorwart, K. R. O. Hazlett, R. K. Deka, M. V. Norgard, J. D. Radolf, and C. A. Hasemann. 2002. The crystal structure of Zn(II)-free Treponema pallidum TroA, a periplasmic metal-binding protein, reveals a closed conformation. J. Bacteriol. 184:2300-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lenhart, T. R., and D. R. Akins. 2010. Borrelia burgdorferi locus BB0795 encodes a BamA orthologue required for growth and efficient localization of outer membrane proteins. Mol. Microbiol. 75:692-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77a.Lui, J., J. K. Howell, S. D. Bradley, Y. Zheng, Z. H. Zhou, and S. J. Norris. 2010. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J. Mol. Biol. 403:546-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lukehart, S. A. 2008. Scientific monogamy: thirty years dancing with the same bug. 2007 Thomas Parran Award Lecture. Sex. Transm. Dis. 35:2-7. [DOI] [PubMed] [Google Scholar]
- 79.Lukehart, S. A. 2008. Syphilis, p. 1038-1046. In A. S. Fauci, E. Braunwald, D. L. Kasper, S. J. Hauser, D. L. Longo, and J. L. Jameson (ed.), Harrison's principles of internal medicine, vol. 17. McGraw Hill, New York, NY. [Google Scholar]
- 80.Lukehart, S. A., and J. N. Miller. 1978. Demonstration of the in vitro phagocytosis of Treponema pallidum by rabbit peritoneal macrophages. J. Immunol. 121:2014-2024. [PubMed] [Google Scholar]
- 81.Lukehart, S. A., J. M. Shaffer, and S. A. Baker-Zander. 1992. A subpopulation of Treponema pallidum is resistant to phagocytosis: possible mechanisms of persistence. J. Infect. Dis. 166:1449-1453. [DOI] [PubMed] [Google Scholar]
- 82.Machius, M., C. A. Brautigam, D. R. Tomchick, P. Ward, Z. Otwinowski, J. S. Blevins, R. K. Deka, and M. V. Norgard. 2007. Structural and biochemical basis for polyamine binding to the Tp0655 lipoprotein of Treponema pallidum: putative role for Tp0655 (TpPotD) as a polyamine receptor. J. Mol. Biol. 373:681-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Magnuson, H. J., H. Eagle, and R. Fleischman. 1948. The minimal infectious inoculum of Spirochaeta pallida (Nichols Strain), and a consideration of its rate of multiplication in vivo. Am. J. Syph. Gonorrhea Vener. Dis. 32:1-19. [PubMed] [Google Scholar]
- 84.McAuley, K. E., P. K. Fyfe, J. P. Ridge, N. W. Isaacs, R. J. Cogdell, and M. R. Jones. 1999. Structural details of an interaction between cardiolipin and an integral membrane protein. Proc. Natl. Acad. Sci. U. S. A. 96:14706-14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McGill, M. A., D. G. Edmondson, J. A. Carroll, R. G. Cook, R. S. Orkiszewski, and S. J. Norris. 2010. Characterization and serologic analysis of the Treponema pallidum proteome. Infect. Immun. 78:2631-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Metzger, M., P. H. Hardy, Jr., and E. E. Nell. 1961. Influence of lysozyme upon the treponeme immobilization reaction. Am. J. Hyg. 73:236-244. [DOI] [PubMed] [Google Scholar]
- 87.Moore, M. W., A. R. Cruz, C. J. LaVake, A. L. Marzo, C. H. Eggers, J. C. Salazar, and J. D. Radolf. 2007. Phagocytosis of Borrelia burgdorferi and Treponema pallidum potentiates innate immune activation and induces gamma interferon production. Infect. Immun. 75:2046-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morgan, C. A., S. A. Lukehart, and W. C. Van Voorhis. 2003. Protection against syphilis correlates with specificity of antibodies to the variable regions of Treponema pallidum repeat protein K. Infect. Immun. 71:5605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nelson, R. A., Jr., and M. M. Mayer. 1949. Immobilization of Treponema pallidum in vitro by antibody produced in syphilitic infection. J. Exp. Med. 89:369-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Niederweis, M. 2008. Nutrient acquisition by mycobacteria. Microbiology 154:679-692. [DOI] [PubMed] [Google Scholar]
- 91.Niederweis, M., O. Danilchanka, J. Huff, C. Hoffmann, and H. Engelhardt. 2010. Mycobacterial outer membranes: in search of proteins. Trends Microbiol. 18:109-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ou, Y. Y., M. M. Gromiha, S. A. Chen, and M. Suwa. 2008. TMBETADISC-RBF: discrimination of Β-barrel membrane proteins using RBF networks and PSSM profiles. Comput. Biol. Chem. 32:227-231. [DOI] [PubMed] [Google Scholar]
- 94.Parsonage, D., D. C. Desrosiers, K. R. Hazlett, Y. Sun, K. J. Nelson, D. L. Cox, J. D. Radolf, and L. B. Poole. 2010. Broad specificity AhpC-like peroxiredoxin and its thioredoxin reductant in the sparse antioxidant defense system of Treponema pallidum. Proc. Natl. Acad. Sci. U. S. A. 107:6240-6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Penn, C. W. 1981. Avoidance of host defences by Treponema pallidum in situ and on extraction from infected rabbit testes. J. Gen. Microbiol. 126:69-75. [DOI] [PubMed] [Google Scholar]
- 96.Penn, C. W., and J. G. Rhodes. 1982. Surface-associated antigens of Treponema pallidum concealed by an inert outer layer. Immunology 46:9-16. [PMC free article] [PubMed] [Google Scholar]
- 97.Perine, P. L., R. S. Weiser, and S. J. Klebanoff. 1973. Immunity to syphilis. I. Passive transfer in rabbits with hyperimmune serum. Infect. Immun. 8:787-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pinne, M., and D. A. Haake. 2009. A comprehensive approach to identification of surface-exposed, outer membrane-spanning proteins of Leptospira interrogans. PLoS One 4:e6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Postle, K., and H. Vakharia. 2000. TolC, a macromolecular periplasmic ‘chunnel.’ Nat. Struct. Biol. 7:527-530. [DOI] [PubMed] [Google Scholar]
- 100.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qu, J., C. Mayer, S. Behrens, O. Holst, and J. H. Kleinschmidt. 2007. The trimeric periplasmic chaperone Skp of Escherichia coli forms 1:1 complexes with outer membrane proteins via hydrophobic and electrostatic interactions. J. Mol. Biol. 374:91-105. [DOI] [PubMed] [Google Scholar]
- 102.Radolf, J. D. 1995. Treponema pallidum and the quest for outer membrane proteins. Mol. Microbiol. 16:1067-1073. [DOI] [PubMed] [Google Scholar]
- 103.Radolf, J. D., T. E. Fehniger, F. J. Silverblatt, J. N. Miller, and M. A. Lovett. 1986. The surface of virulent Treponema pallidum: resistance to antibody binding in the absence of complement and surface association of recombinant antigen 4D. Infect. Immun. 52:579-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Radolf, J. D., K. R. O. Hazlett, and S. A. Lukehart. 2006. Pathogenesis of syphilis, p. 197-236. In J. D. Radolf and S. A. Lukehart (ed.), Pathogenic treponemes: cellular and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 105.Radolf, J. D., and S. A. Lukehart. 2006. Immunology of syphilis, p. 285-322. In J. D. Radolf and S. A. Lukehart (ed.), Pathogenic treponemes: cellular and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 106.Radolf, J. D., E. J. Robinson, K. W. Bourell, D. R. Akins, S. F. Porcella, L. M. Weigel, J. D. Jones, and M. V. Norgard. 1995. Characterization of outer membranes isolated from Treponema pallidum, the syphilis spirochete. Infect. Immun. 63:4244-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Radolf, J. D., B. Steiner, and D. Shevchenko. 1999. Treponema pallidum: doing a remarkable job with what it's got. Trends Microbiol. 7:7-9. [DOI] [PubMed] [Google Scholar]
- 108.Randall, A., J. Cheng, M. Sweredoski, and P. Baldi. 2008. TMBpro: secondary structure, beta-contact and tertiary structure prediction of transmembrane beta-barrel proteins. Bioinformatics 24:513-520. [DOI] [PubMed] [Google Scholar]
- 109.Remmert, M., D. Linke, A. N. Lupas, and J. Soding. 2009. HHomp—prediction and classification of outer membrane proteins. Nucleic Acids Res. 37:W446-W451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rice, M., and T. J. Fitzgerald. 1985. Immune immobilization of Treponema pallidum: antibody and complement interactions revisited. Can. J. Microbiol. 31:1147-1151. [DOI] [PubMed] [Google Scholar]
- 111.Ruiz, N., D. Kahne, and T. J. Silhavy. 2006. Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 4:57-66. [DOI] [PubMed] [Google Scholar]
- 112.Schulz, G. E. 2002. The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta 1565:308-317. [DOI] [PubMed] [Google Scholar]
- 113.Sell, S., and S. J. Norris. 1983. The biology, pathology, and immunology of syphilis. Int. Rev. Exp. Pathol. 24:203-276. [PubMed] [Google Scholar]
- 114.Setubal, J. C., M. Reis, J. Matsunaga, and D. A. Haake. 2006. Lipoprotein computational prediction in spirochaetal genomes. Microbiology 152:113-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shevchenko, D. V., D. R. Akins, E. J. Robinson, M. Li, O. V. Shevchenko, and J. D. Radolf. 1997. Identification of homologs for thioredoxin, peptidyl prolyl cis-trans isomerase, and glycerophosphodiester phosphodiesterase in outer membrane fractions from Treponema pallidum, the syphilis spirochete. Infect. Immun. 65:4179-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shevchenko, D. V., T. J. Sellati, D. L. Cox, O. V. Shevchenko, E. J. Robinson, and J. D. Radolf. 1999. Membrane topology and cellular location of the Treponema pallidum glycerophosphodiester phosphodiesterase (GlpQ) ortholog. Infect. Immun. 67:2266-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Silhavy, T. J., D. Kahne, and S. Walker. 2010. The bacterial cell envelope. Cold Spring Harbor Perspect. Biol. 2:a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stamm, L. V., R. L. Hodinka, P. B. Wyrick, and P. J. Bassford, Jr. 1987. Changes in the cell surface properties of Treponema pallidum that occur during in vitro incubation of freshly extracted organisms. Infect. Immun. 55:2255-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stegmeier, J. F., and C. Andersen. 2006. Characterization of pores formed by YaeT (Omp85) from Escherichia coli. J. Biochem. 140:275-283. [DOI] [PubMed] [Google Scholar]
- 120.Subbarao, G. V., and B. van den Berg. 2006. Crystal structure of the monomeric porin OmpG. J. Mol. Biol. 360:750-759. [DOI] [PubMed] [Google Scholar]
- 121.Swancutt, M. A., B. S. Riley, J. D. Radolf, and M. V. Norgard. 1989. Molecular characterization of the pathogen-specific, 34-kilodalton membrane immunogen of Treponema pallidum. Infect. Immun. 57:3314-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sykes, J. A., J. N. Miller, and A. J. Kalan. 1974. Treponema pallidum within cells of a primary chancre from a human female. Br. J. Vener. Dis. 50:40-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Turner, T. B., and D. H. Hollander. 1957. Biology of the treponematoses. World Health Organization, Geneva, Switzerland.
- 124.Vogt, J., and G. E. Schulz. 1999. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure 7:1301-1309. [DOI] [PubMed] [Google Scholar]
- 125.Weiser, R. S., D. Erickson, P. L. Perine, and N. N. Pearsall. 1976. Immunity to syphilis: passive transfer in rabbits using serial doses of immune serum. Infect. Immun. 13:1402-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Werner, J., A. M. Augustus, and R. Misra. 2003. Assembly of TolC, a structurally unique and multifunctional outer membrane protein of Escherichia coli K-12. J. Bacteriol. 185:6540-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wimley, W. C. 2003. The versatile beta-barrel membrane protein. Curr. Opin. Struct. Biol. 13:404-411. [DOI] [PubMed] [Google Scholar]
- 128.Yu, C. S., C. J. Lin, and J. K. Hwang. 2004. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13:1402-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu, N. Y., J. R. Wagner, M. R. Laird, G. Melli, S. Rey, R. Lo, P. Dao, S. C. Sahinalp, M. Ester, L. J. Foster, and F. S. Brinkman. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zeth, K. 2010. Structure and evolution of mitochondrial outer membrane proteins of beta-barrel topology. Biochim. Biophys. Acta 1797:1292-1299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.