Abstract
The Gram-positive bacterium Staphylococcus aureus contains two glyceraldehyde-3-phosphate dehydrogenase (GAPDH) homologues known as GapA and GapB. GapA has been characterized as a functional GAPDH protein, but currently there is no biological evidence for the role of GapB in metabolism in S. aureus. In this study we show through a number of complementary methods that S. aureus GapA is essential for glycolysis while GapB is essential in gluconeogenesis. These proteins are reciprocally regulated in response to glucose concentrations, and both are influenced by the glycolysis regulator protein GapR, which is the first demonstration of the role of this regulator in S. aureus and the first indication that GapR homologues control genes other than those within the glycolytic operon. Furthermore, we show that both GapA and GapB are important in the pathogenesis of S. aureus in a Galleria mellonella model of infection, showing for the first time in any bacteria that both glycolysis and gluconeogenesis have important roles in virulence.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a key glycolytic enzyme, the primary function of which is the oxidative phosphorylation of glyceraldehyde-3-phosphate (G3P) to 1,3-diphosphoglycerate (1,3dPG) during glucose metabolism. However, over the past decade research has shown that GAPDH homologues from both eukaryotes and prokaryotes are in fact multifunctional and can be localized in unexpected cell fractions, such as the cell surface or the nucleus (6, 12, 23, 33). The additional roles identified for GAPDH include protein binding and cell signaling in many organisms, immune evasion in bacteria, and even maintenance of the cell in higher eukaryotes (2, 3, 17, 23, 25, 26, 31). Many of these surface-localized proteins have been shown to be enzymatically active and are usually transcribed from the same open reading frame (ORF) as their cytoplasmic counterpart (8, 12, 23, 24).
While many bacteria contain a single GAPDH gene, a number of species have multiple GAPDH homologues. There are three GAPDH homologues in Escherichia coli: gapA which encodes a GAPDH protein, gapB which encodes an erythrose-4-phosphate dehydrogenase (E4PDH), and gapC, the function of which is unknown (4, 14, 28, 32, 39). The Gram-positive bacterium Bacillus subtilis also has two gap homologues, but in this case they both encode GAPDH proteins, each with opposing roles in glucose metabolism; this differs from the more common case of one GAPDH performing both roles (10). The B. subtilis gapA GAPDH is functional only in glycolysis, converting G3P to 1,3dPG, and has specificity for NAD+ as a cofactor. The gapB GAPDH is gluconeogenic and specifically uses the cofactor NADPH for conversion of 1,3dPG to G3P during gluconeogenesis (10). This is consistent with the normal role of the NADP+/NADPH couple in biosynthetic pathways and NAD+/NADH in catabolism. In gluconeogenesis, B. subtilis gapA expression is repressed by the Deo-like regulator CcgR due to the absence of the glycolytic precursor fructose-1,6-bisphosphate (FBP). In the absence of FBP, CcgR binds to an operator sequence between the ccgR promoter and open reading frame, blocking transcript elongation rather than initiation of transcription and thus repressing expression of the glycolytic operon (9, 21, 40). There is no evidence that CcgR is involved in the regulation of gapB (9, 10).
Staphylococcus aureus is an extremely adaptable and versatile organism and is notably a commensal and opportunistic pathogen capable of causing a diverse range of infections in many human and animal body sites. Therefore, S. aureus is a significant problem for public health care, veterinary medicine, and the food industry (20, 37). Hospital-acquired methicillin-resistant S. aureus (HA-MRSA) still continues to be a concern, and there is an increasing incidence of community-acquired MRSA (CA-MRSA) infections worldwide, which are severe infections in otherwise healthy individuals and so are very unusual for a normally opportunistic pathogen. Furthermore, there is an increasing incidence of MRSA in animals, which is of particular concern as these animals may act as reservoirs for transmission of MRSA to humans and are costly to the food animal production industry. Consequently, further investigation of the fundamental nature of S. aureus physiology is essential.
S. aureus has two GAPDH homologues which share approximately 40% sequence identity with one another; they have been termed gapA (also known as gapC in a bovine mastitis isolate [13]) and gapB. The gapA gene is located within the glycolytic operon alongside several other glycolytic enzymes and has previously been shown to encode a glycolytic GAPDH (35). A homologue of the B. subtilis glycolytic regulator CcgR is also found in the S. aureus glycolytic operon and is known as gapR. The gapB gene is located as a single open reading frame alongside genes involved in DNA replication and repair.
The roles of GapB and GapR in S. aureus have not been determined to date. GapB is assumed to be a gluconeogenic GAPDH in S. aureus due to homology to the Gram-positive model organism B. subtilis; however, no functional evidence has been published to confirm this (7, 30). Although both organisms have gapA and gapB homologues, aspects of carbon metabolism may differ between these species due to the different environments they inhabit, and therefore the assumption that these genes behave in the same way may not be accurate. In fact, there is evidence suggesting that GapB may not be a gluconeogenic GAPDH in S. aureus. GapB protein purified from B. subtilis shows a 50-fold higher catalytic efficiency with NADP+ than with NAD+ (10), whereas purified recombinant GapB protein from an S. aureus bovine mastitis isolate shows very little NAD+-dependent GAPDH activity and no NADP+-dependent GAPDH activity (13). Therefore, the purpose of this work is to investigate the metabolic function of GapB in S. aureus and identify its role in carbon metabolism and virulence.
In this study we used a number of complementary approaches which demonstrate that S. aureus GapB is essential for gluconeogenesis. We also demonstrated that the glycolytic regulator GapR not only regulates gapA expression but also has a novel role in the regulation of gapB. Furthermore, both gapA and gapB showed attenuated virulence in the Galleria mellonella invertebrate model of infection, showing that both glycolysis and gluconeogenesis play important roles in virulence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All of the strains used in this study are listed in Table 1. E. coli strains were grown on Luria agar (LuA) or in broth (LuB), and S. aureus strains were grown in Trypticase soy broth (TSB) (BD Diagnostics) or Tris minimal (TM) medium, a modified minimal Tris succinate medium omitting succinate (29). Where necessary, ampicillin (100 μg/ml), kanamycin (50 μg/ml), tetracycline (10 μg/ml), erythromycin (10 μg/ml), spectinomycin (100 μg/ml), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg/ml) were added to the media. Bacterial cultures were grown at 37°C with aeration unless otherwise stated. Additional carbon sources were added to the medium to a final concentration of 1% (wt/vol) unless otherwise stated.
TABLE 1.
Bacterial strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| E. coli | ||
| TOP10 | E. coli cloning strain | Invitrogen |
| BL21 DE3 | IPTG-inducible T7 expression strain | 34 |
| S. aureus | ||
| RN4220 | Restriction deficient 8325-4 | Laboratory stock |
| 8325-4 | Wild-type 8325 cured of prophages | 15 |
| 8325-4 gapA strain | 8325-4 ΔgapA::tet | This study |
| 8325-4 gapB strain | 8325-4 ΔgapB::spc | This study |
| 8325-4 gapA gapB strain | 8325-4 ΔgapA::tet ΔgapB::spc | This study |
| 8325-4 gapR strain | 8325-4 ΔgapR::spc | This study |
Construction of mutant strains.
The gapA open reading frame and 1,282 bp of flanking sequence were PCR amplified using primers Gap30F and PGKSR, and a 402-bp internal deletion was created by digestion with ClaI. A terminatorless tetracycline cassette was inserted at this position, and the construct was cloned into the E. coli-S. aureus shuttle vector pMAD, which has a temperature-sensitive Gram-positive origin of replication (1). The 5′ and 3′ regions of the gapB open reading frame with flanking sequences were PCR amplified separately using the primer pair GapB5F and GapB5R and the pair GapB3F and GapB3R, to leave a 94-bp deletion within the gapB coding region. A PCR-amplified spectinomycin cassette (primers SpecF and SpecR) was inserted into this deletion site via an NheI site, and the construct was also cloned into pMAD. A similar approach was used for construction of the ΔgapR mutant, using the primer pair GapBF and GapRPR and the pair GapR3′F and GapAPR and a terminatorless spectinomycin cassette (primers SpecF and SpecRNT). All primer sequences are given in Table 2.
TABLE 2.
Primers and probes used in this study
| Description or function | Primer or probe name | Sequence (5′-3′) |
|---|---|---|
| gapA mutagenesis | Gap30f | TTTCTAGATTCGTACCAGCCAGAGGT |
| PGKSR | CAACTGGTTTATGTGGATC | |
| Tetracycline cassette | tetNxF | AAGGCGCCATGCTAACATAGCATTACGG |
| tetNxR | AAGGCGCCCGATTTAGAAATCCCTTTGAG | |
| gapB mutagenesis | GapB5F | GGGGATCCCCTTGAGATTGTTAGAAGA |
| GapB5R | GGGCTAGCCATTAACTATTCCAAACTG | |
| GapB3F | GGGCATATGGCATTATTCCTACTTCTAC | |
| GapB3R | GAAAACTGCTCTCTTGTG | |
| gapR mutagenesis | GapBF | GGGGATCCGCTAATGATAAGTAGTATTTAG |
| gapR probe | GapRPR | GGGGCTAGCCCAGTTACAGCAACTATC |
| GapR3′F | AAAACCCGGGCACAAGGTCAAATTGTCC | |
| gapA probe | GapAPR | CACTCTAGAGCGGAGAAGCGTTTGTGC |
| Spectinomycin cassette | SpecF | GGGGCTAGCGATATAAAATAGGTACTAATC |
| SpecR | GGGGCTAGGGCCATATGCAAGGGTTTATTG | |
| SpecRNT | GGGGCTAGCAAACCCGGGTGTTTCCACCATTTTTTC | |
| Northern probe | GapAPF | CACAGATCTGGAAGGCCATTATAATGGCAG |
| GapBPF | GAATGGTATTACGTATTGC | |
| GapBPR | GTGCTCCAATTTGCTCAG | |
| GapRPF | GTGAAAGACTTATTGCAAG | |
| 16SF | GATCCTGGGTCAGGATG | |
| 16SR | CTAGAGTTGTCAAAGGATG | |
| GapA protein expression | GapA expF | TACTTCCAATCCATGGCAGTAAAAGTAGCAATTAAT |
| GapA expR | TATCCACCTTTACTGTCATTATTTAGAAAGTTCAGCTAAGTA | |
| GapB protein expression | GapB expF | TACTTCCAATCCATGTCAACGAATATTGCAATTAAT |
| GapB expR | TATCCACCTTTACTGTCAGAAGTCAGAGTTAGGCTATAAATTA | |
| GapR protein expression | GapR expF | TACTTCCAATCCGTGAAAGACTTATTGCAAGCACA |
| GapR expR | TATCCACCTTTACTGTCATCTTATTCAAGTATTATCTTTGCT |
The mutagenesis plasmids were introduced into S. aureus strain RN4220, and mutagenesis was carried out as previously described (35), using TM medium for growth and selection of the ΔgapA mutant and Luria medium for growth and selection of the ΔgapB and ΔgapR mutants. Plates also contained 40 μg/ml X-Gal to allow blue/white selection of potential mutants (1). Mutagenesis of ΔgapA and ΔgapR was carried out directly in strain 8325-4, while mutagenesis of ΔgapB was performed in strain RN4220 followed by transduction by phage 11 into strain 8325-4. A ΔgapA ΔgapB double mutant was also produced by phage transduction of the ΔgapB mutation into the ΔgapA mutant background. All mutants were confirmed by PCR analysis (data not shown).
Expression and purification of GAPDH proteins.
The open reading frames of gapA, gapB, and gapR were PCR amplified from 8325-4 chromosomal DNA (Table 2), cloned into the E. coli expression vector pLEICS03 (Protein Expression Laboratory [Protex], University of Leicester, United Kingdom), and transformed into E. coli Top10. The vector pLEIC03 allows inducible expression by isopropyl-β-d-thiogalactopyranoside (IPTG) and introduces an N-terminal His6 tag which can be cleaved by tobacco etch virus (TEV) protease. Clones were sequenced before being transformed into the E. coli expression strain BL21(DE3). Cultures were grown in LuB until an optical density at 600 nm (OD600) of 0.6 was reached, and expression was induced by the addition of IPTG to a final concentration of 100 μM. Cells were grown overnight at 20°C, and the proteins were purified using Ni-nitrilotriacetic acid (NTA) affinity chromatography. Where necessary, the hexahistidine tag was removed with TEV protease. Final protein concentrations were determined using a Bradford standard assay (5).
Growth curves and 5-h growth assays.
Five milliliters of TSB medium (with antibiotics where necessary) was inoculated and grown overnight. The bacteria were then resuspended in fresh medium (TSB or TM medium) to an OD600 of 0.05. Cultures were incubated at 37°C, and the OD600 was taken either every hour for 7 h or after 5 h of growth against a blank reading of medium only. A final reading was then taken after 24 h of growth. Each assay was repeated a minimum of three times on separate days, and the results were averaged and are presented alongside the standard deviation of the data. P values were derived using a Student's t test at 4 h for growth in TSB and at 5 h for growth in TM medium.
GAPDH assay.
For whole-cell GAPDH activity assays (23), cultures were grown in TSB medium overnight, and the OD600 of the suspension was used to equalize the culture for cell growth. Cells were washed in distilled H2O (dH2O) and then resuspended in 1 ml of GAPDH assay buffer (pH 7.5) containing 50 mM Na2HPO4, 5 mM EDTA, 40 mM triethanolamine, 2 mM dl-glyceraldehyde-3-phosphate (G3P), and either 2 mM NAD+ or 2 mM NADP+. Samples were incubated in a 37°C water bath for 30 min; the cells were then pelleted, and the supernatant was extracted. Production of NAD(P)H was detected by measuring the absorption of the suspension at 340 nm. For purified proteins, 200 ng of protein was mixed with GAPDH assay buffer (as above, but 4 mM G3P was used) to a final volume of 200 μl. This was carried out in triplicate for each sample in a 96-well plate, and samples were incubated at 37°C in a FLUOstar Omega plate reader. The absorption was measured at 340 nm after 30 min, and an average reading from the three wells was recorded for each sample. Each GAPDH assay (whole cell and protein) was repeated three times on separate days, and the results were averaged and are presented alongside the standard error of the data. P values were derived using a Student's t test.
Northern blotting.
S. aureus cells were cultured in 10 ml of TM medium for 5 h with aeration, and RNA was extracted and analyzed by Northern blotting as previously described (18). DNA probes for gapA, gapB, gapR, and 16S rRNA were constructed by PCR using the primer pairs presented in Table 2. Each experiment was repeated at least twice, and in each case blots were stripped and reprobed with the 16S probe to demonstrate even loading of the RNA in each well and to quantify expression levels where necessary. Transcripts were evaluated using ImageJ, version 1.41, software (http://rsbweb.nih.gov/ij/).
G. mellonella infection model.
G. mellonella larvae were purchased from Vine House Farms, Ltd., Spalding, United Kingdom, and were stored at 4°C. Prior to use, larvae were maintained at room temperature overnight. Inocula for the larvae were grown overnight with shaking at 37°C in Falcon tubes in 5-ml volumes of CY broth (10 g/liter Casamino Acids, 10 g/liter yeast extract, 5.9 g/liter NaCl) supplemented with appropriate antibiotics. Bacteria were pelleted by centrifugation (13,000 × g for 2 min) and washed and resuspended in phosphate-buffered saline (PBS) to an optical density of 0.125 at 600 nm. Inocula (20 μl; containing approximately 5 × 107 bacteria or PBS alone as a control) were injected into the larval hemolymph adjacent to the fourth proleg using a syringe with a 29-gauge needle and a Tridak Stepper repetitive dispenser (Intertronics, Kidlington, United Kingdom). Groups of 10 infected or control larvae were maintained at 37°C, and viability was determined every 24 h over a 72-h period. The results of three independent experiments were combined, and survival curves were calculated using the Kaplan-Meier method. Significant differences between survival curves were calculated using a log rank test.
RESULTS
Purified recombinant GapB protein does not have GAPDH activity.
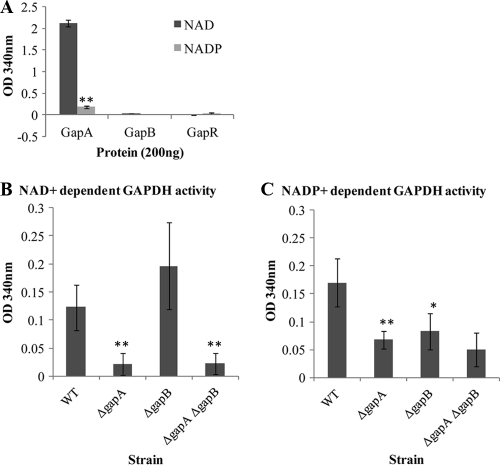
In order to investigate GapB enzymatic function in S. aureus, the gapA and gapB open reading frames from S. aureus strain 8325-4 were cloned into the E. coli protein expression vector pLEICS03 (Protex, University of Leicester, United Kingdom). The glycolytic operon regulator GapR was also cloned into this vector to act as a negative control as it is a regulatory protein and therefore should not have any glycolytic enzyme activity. The recombinant proteins were expressed in E. coli, and 200 ng of purified protein was used to assay for GAPDH activity in the presence of either NAD+ or NADP+ as cofactor.
The GapA protein had very high levels of activity with NAD+ but showed much less activity with NADP+ (P < 0.001) (Fig. 1A), showing that GapA has a reduced affinity for NADP+ and therefore is likely to have a primary role as a glycolytic GAPDH rather than a role in biosynthesis. However, neither GapB nor the negative-control GapR demonstrated any detectable enzyme activity with either NAD+ or NADP+ (Fig. 1A). The possibility that GapB enzyme activity was affected by the presence of the His tag was eliminated by testing for activity after TEV protease cleavage to remove the tag (data not shown). Thus, these results suggest that either GapB is not a GAPDH or the recombinant protein is nonfunctional.
FIG. 1.
Enzyme activity of S. aureus GAPDH proteins in the presence of either NAD+ or NADP+ as a cofactor. (A) GAPDH activity of N-terminal His-tagged proteins His6-GapA, His6-GapB, and His6-GapR. Whole-cell NAD+-dependent GAPDH activity (B) and whole-cell NADP+-dependent GAPDH activity (C) were determined for the 8325-4, 8325-4 ΔgapA, 8325-4 ΔgapB, and 8325-4 ΔgapA ΔgapB strains. Cells were grown overnight in TSB medium and equalized for growth. The data show the average result of three repeats, and the error bars indicate the standard error. Significant differences relative to the wild-type (WT) strain are indicated as follows: *, P < 0.01; **, P < 0.001.
GapB has NADP+- but not NAD+-dependent GAPDH activity in vivo.
Although the purified GapB lacks NADP+-dependent GAPDH activity, the key amino acid residues that determine NADP+ specificity (Ala32 and Asn187) and G3P binding (Thr179 and Thr208) are present, as determined from a number of GAPDH homologues (10), which would suggest a gluconeogenic GAPDH role for this protein. Therefore, to further investigate the function of GapB in S. aureus, deletion/insertion mutations were introduced into the gapA and the gapB loci in strain 8325-4. A terminatorless tetracycline cassette was introduced into the gapA locus to allow expression of the remainder of the glycolytic operon so that only the function of GapA would be impaired. Expression of the downstream genes was confirmed by Northern blotting using a phosphoglycerate kinase (PGK) probe (data not shown). The gapB locus is monocistronic, so a spectinomycin cassette with promoter and terminator was introduced in strain RN4220. The mutation was then transduced by phage into strain 8325-4, removing the possibility of secondary mutations in the genome. A double mutant was also produced by phage transduction of the gapB mutation into the 8325-4 ΔgapA strain.
Whole-cell GAPDH activity was tested for each of the strains in the presence of both NAD+ and NADP+. In both the 8325-4 ΔgapA and 8325-4 ΔgapA ΔgapB strains, the NAD+-dependent GAPDH activity was approximately 5-fold lower than that in the wild-type strain (P < 0.001) (Fig. 1C). There was no significant difference in activity between either the wild type and the ΔgapB mutant or between the ΔgapA mutant and the double mutant, which indicates that in vivo GapB does not show NAD+-dependent GAPDH activity (Fig. 1C). This is consistent with GapA being solely responsible for NAD+-dependent GAPDH activity in S. aureus.
Surprisingly, NADP+-dependent GAPDH activity was found to be significantly lower in both the ΔgapA (P < 0.001) and the ΔgapB mutants (P < 0.01) than in the wild type (Fig. 1D) (by 2.5-fold and 2-fold, respectively). Therefore, both S. aureus GapA and GapB proteins demonstrate NADP+-dependent activity in vivo in contrast to the purified proteins, which show no GapB NADP+-dependent GAPDH activity. However, NADP+-dependent GAPDH activity is not significantly lower in the double mutant than in either of the single mutants, indicating that another enzyme maybe responsible for this residual activity, which may also play a role in this process in vivo.
GapA is not responsible for both glycolytic and gluconeogenic GAPDH activity in S. aureus.
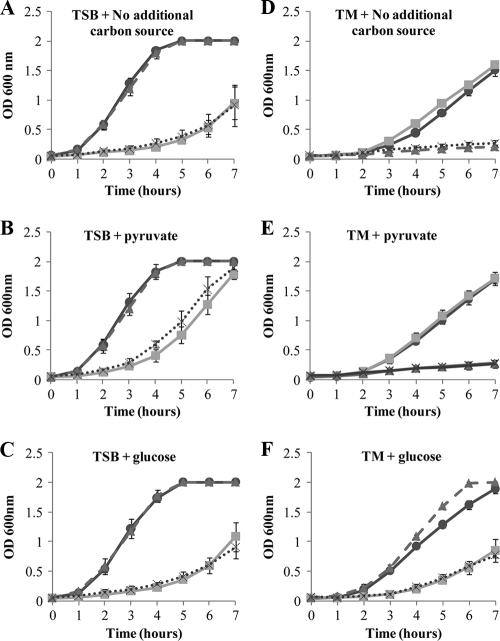
To investigate the role of each GAPDH homologue in the growth of S. aureus, 24-h growth curves were carried out on the 8325-4, 8325-4 ΔgapA, 8325-4 ΔgapB, and 8325-4 ΔgapA ΔgapB strains in the complex medium TSB in the presence of either glucose or pyruvate as alternative carbon sources. TSB contains a mix of Casamino Acids and nitrogenous substances from the digest of casein and soybean meal, with 0.25% glucose (wt/vol) as the primary carbon source. Additional carbon sources were added to the medium to assess the effect of the mutations in response to changes in carbon flow. Glucose and pyruvate were chosen as they both increase carbon flow into the tricarboxylic acid cycle (TCA) cycle and increase the growth rate, but the addition of pyruvate should bypass the need for a functional glycolytic pathway.
Primary carbon metabolism was seen to be unaffected in the ΔgapB mutant as there was no significant difference in growth compared to the wild type under any of the growth conditions tested (Fig. 2). However, both the ΔgapA mutant and the ΔgapA ΔgapB double mutant showed a decrease in growth in both TSB and TSB with glucose compared to growth of the wild-type strain (Fig. 2A and C). This growth defect was significantly alleviated by the addition of pyruvate (P < 0.01) (Fig. 2B). Therefore, glycolysis is disrupted in the ΔgapA strains, but the processes downstream such as the TCA cycle are still functional.
FIG. 2.
Growth of the 8325-4 (black line; •), 8325-4 ΔgapA (gray line; ▪), 8325-4 ΔgapB (dashed line; ▴), and 8325-4 ΔgapA ΔgapB (dotted line; X) strains in TSB without the addition of any additional carbon source (A) or with the addition of 0.056% pyruvate (B) or 0.75% glucose (C), added at time zero. Growth of the same strains, indicated as above, was measured in TM medium without the addition of an additional carbon source (D) and with the addition of 0.056% pyruvate (E) or 1% glucose (F), added at time zero. Cultures were grown at 37°C with aeration, and growth was measured as the optical density of the culture at 600 nm. Experiments were repeated a minimum of three times on different days, and the data presented are the average of three repeats, with error bars indicating the standard deviations.
To further investigate the roles of GapA and GapB in carbon metabolism, growth assays were conducted in a defined medium where the carbon source could be tightly controlled. Tris minimal (TM) medium was chosen as it contains no glucose; in this medium Casamino Acids (1%, vol/vol) act as both the carbon and nitrogen sources. In TM medium in the absence of additional carbon, the ΔgapA mutant grew at a rate comparable to that of the wild type, indicating that this strain is able to utilize secondary carbon sources and that gluconeogenesis must be functional.
In contrast, the growth of both the ΔgapB mutant and the ΔgapA ΔgapB double mutant was severely limited in the absence of glucose (Fig. 2D). Furthermore, the addition of pyruvate enhanced the growth rate of the wild type and the ΔgapA mutant but not that of either ΔgapB mutants, suggesting that a loss of gapB results in a loss of ability to synthesize glucose (Fig. 2E). Addition of glucose significantly increased the growth of the ΔgapB mutant (P < 0.001) to a level comparable to that of the wild-type strain, whereas growth of the ΔgapA mutant was significantly lower in the presence of glucose (P < 0.001) (Fig. 2F). Glucose also enhanced the growth of the ΔgapA ΔgapB double mutant but only to a level similar to that of the ΔgapA mutant under these conditions.
These results show that gluconeogenesis must be functional in the ΔgapA mutant as it is able to grow on Casamino Acids as the sole carbon source. Consequently, the GapA GAPDH is not responsible for both glycolytic and gluconeogenic GAPDH activity in S. aureus. Indeed, our data suggest that GapB is involved in the gluconeogenic pathway as the ΔgapB mutant is unable to grow in the absence of glucose, and pyruvate did not improve the growth, which indicates a loss of gluconeogenesis and an inability to produce de novo glucose for use in other essential processes.
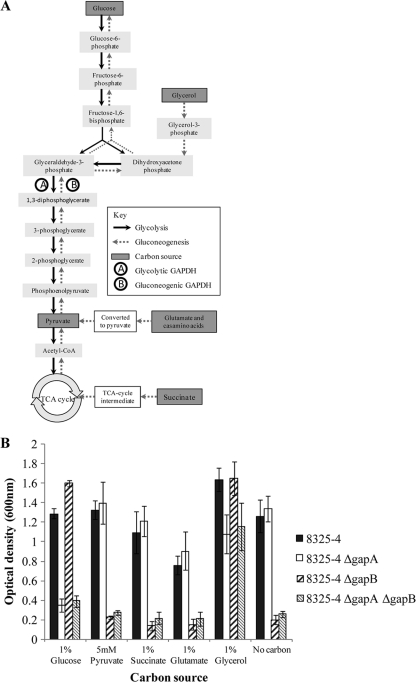
A ΔgapB mutant is unable to utilize secondary carbon sources.
To determine the functional role of GapB in secondary carbon metabolism, the mutant strains were tested for their ability to utilize a wider range of secondary carbon sources. Figure 3A demonstrates where a variety of different carbon sources can enter either glycolysis or gluconeogenesis. The secondary carbon sources tested included succinate, a TCA cycle intermediate, and glutamate, an amino acid (both of which are substrates for gluconeogenesis), and glycerol as it can enter both the glycolytic and gluconeogenic pathways through conversion to G3P. Strains were grown in TM medium with each additional carbon source, and growth was measured after 5 h (Fig. 3B). The ΔgapA mutant was able to grow in the presence of pyruvate, succinate, and glutamate and in the absence of additional carbon at the same rate as the wild type. However, growth was significantly inhibited in the ΔgapA mutant compared to growth of the wild type by the presence of both glucose (P < 0.001) and glycerol (P < 0.01). Conversely, ΔgapB mutant growth was severely inhibited in TM medium unless glucose or glycerol was present. Glycerol enters the gluconeogenic pathway after conversion to G3P (Fig. 3A). Therefore, GapB must function before this point in the pathway for glycerol to recover the growth defect in the ΔgapB mutant. Therefore, the most likely function of GapB in S. aureus is as a gluconeogenic GAPDH.
FIG. 3.
(A) Flow chart indicating where the various carbon sources can enter glycolysis and gluconeogenesis during carbon metabolism. (B) Graph showing the level of growth of the 8325-4, 8325-4 ΔgapA, 8325-4 ΔgapB, and 8325-4 ΔgapA ΔgapB strains after a 5-h incubation in TM medium supplemented with an additional carbon source. Cultures were grown at 37°C with aeration, and growth was measured as the optical density of the culture at 600 nm. Each experiment was repeated three times on different days, and the data presented are averages, with error bars indicating the standard deviations. CoA, coenzyme A.
Transcriptional control of gapA and gapB is also reciprocal with respect to glucose, and both genes are repressed by GapR.
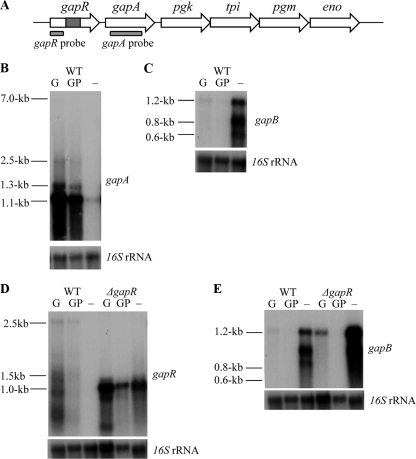
If the GAPDH proteins are active in opposing pathways, then they should be expressed only in response to their individual stimuli and not at the same time. Northern blot analysis was used to determine the expression of gapA and gapB after 5 h of growth in TM medium in the presence and absence of 1% glucose. When glucose is added to the medium at the start of growth, it leads to an increased growth rate, which affects cell density and the pH of the medium, two stimuli that could potentially alter gene expression. To confirm that any transcriptional differences were due to the presence or absence of glucose and not changes in cell density, growth rate, or pH, glucose was added to cultures at either time zero or at 1 h (glucose pulse) before the cells were harvested. The gapA probe hybridizes to multiple transcripts of various sizes due to surrounding genes being cotranscribed with gapA (Fig. 4A). The gapB probe appears to hybridize to three distinct transcripts although the sizes of these suggest that they are all transcribed from within the gapB reading frame and are not due to cotranscription with surrounding genes. Figure 4B and C show that, in the presence of glucose, gapA is induced while gapB is repressed. This occurs under both glucose and glucose pulse conditions, indicating that the effect is due to the presence of glucose and not the changes associated with an increase in growth rate. In contrast, in the absence of glucose and in the presence of Casamino Acids only, gapA transcription is repressed while gapB transcription is induced (Fig. 4B and C and Table 3). Consequently, these data indicate that transcription of gapA and gapB is reciprocally controlled by glucose levels, which is further evidence that they act in opposing pathways.
FIG. 4.
(A) Schematic representation of the glycolytic operon with a ΔgapR mutation, indicating the position of the gapR and gapA probes used for Northern blot analysis. Genes are putative operon regulator (gapR), glyceraldehyde-3-phosphate dehydrogenase (gapA), phosphoglycerate kinase (pgk), triphosphate isomerase (tpi), phosphoglycerate mutase (pgm), and enolase (eno). Northern blot analysis is shown of gapA transcript (B) and gapB transcript (C) expression in response to glucose induction in strain 8325-4 and gapR transcript (D) and gapB transcript (E) expression in response to glucose induction in both wild-type (WT) 8325-4 and 8325-4 ΔgapR strains. Total RNA was extracted from cells grown for 5 h in TM broth with 1% glucose added at time zero (G), 1% glucose added 1 h before cells were harvested (GP), and without glucose (−). Gels presented are representative of experiments that were repeated two times using RNA extracts from cultures grown on different days, with similar results observed each time. Blots were stripped and rehybridized with a control probe (16S) to ensure equal loading of RNA in each case.
TABLE 3.
Densitometry analysis of Northern blots
| Probe (figure no.) | Transcript levels in the WT with:a |
Transcript levels in the ΔgapR strain with:a |
||||
|---|---|---|---|---|---|---|
| Glucose | Glucose pulse | No carbon | Glucose | Glucose pulse | No carbon | |
| gapA (4B) | 1.08 | 0.97 | 0.15 | NAb | NA | NA |
| gapB (4C) | 0.08 | 0.07 | 1.09 | NA | NA | NA |
| gapR (4D) | 0.90 | 0.26 | 0.04 | 0.94 | 0.56 | 0.77 |
| gapB (4E) | 0.08 | 0.07 | 1.09 | 0.33 | 0.15 | 1.42 |
Values represent the ratio of mRNA/16S transcript levels.
NA, not applicable.
In B. subtilis the glycolytic operon is repressed by CcgR under gluconeogenic conditions (8, 35). Sequence analysis shows that there is no evidence of a ccgR-like consensus binding sequence in the putative S. aureus gapR promoter region. However, to determine whether the ccgR homologue gapR plays a similar regulatory role in S. aureus, a mutation was introduced into the gapR locus in strain 8325-4. A deletion/insertion method was used to incorporate a terminatorless spectinomycin resistance cassette into gapR to allow constitutive expression of the downstream genes of the glycolytic operon. To assess transcription of the glycolytic operon by Northern blotting, a gapR probe which hybridized upstream of the inserted spectinomycin cassette was used in place of the gapA probe so that expression of the spectinomycin promoter was not detected. Overall, gapR appears to be expressed at a lower level than gapA in wild-type S. aureus although the response to glucose is consistent (Fig. 4D). This is interesting as it demonstrates differential levels of expression between the operon regulator and the glycolytic genes even though they form a polycistronic transcript. Due to an apparent lack of a promoter upstream of gapA, this variable expression may be a product of posttranscriptional processing of the polycistronic gene transcripts, as seen in B. subtilis (17).
Expression of gapR and the glycolytic operon increased under both glycolytic and gluconeogenic conditions in the ΔgapR mutant compared to the wild type (Fig. 4D). This indicates that GapR represses the glycolytic operon under gluconeogenic conditions and limits the level of expression in the presence of glucose. Interestingly, the expression of gapB compared to that in the wild type was also increased under gluconeogenic and glycolytic conditions in the ΔgapR mutant (Fig. 4E). These data are supported by densitometry analysis which is presented as a ratio of mRNA/16S transcript levels (Table 3).
Therefore, S. aureus GapR also represses the transcription of gapB and limits its expression under gluconeogenic conditions. This indicates that the function of GapR is not limited to control of the glycolytic operon but is also involved in regulating gluconeogenic metabolism in S. aureus. To our knowledge this is the first evidence in any bacteria that a CcgR-like homologue acts to regulate genes other than those of the glycolytic operon.
Glucose inhibits growth in a ΔgapA mutant and promotes growth in a ΔgapB mutant in a reciprocal manner.
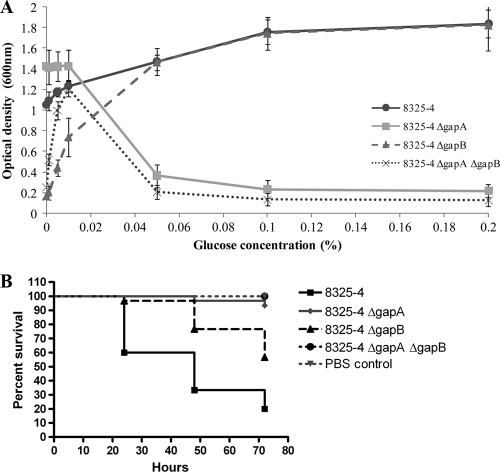
In vivo normal blood glucose levels are within the range of 3.6 to 5.8 mM, and even under trauma conditions the glucose levels increase to only 12 mM (19). Consequently, the 1% glucose concentration used here is much higher than physiological levels. Therefore, to determine the level of glucose required to switch from gapA expression to gapB expression and thus determine whether glucose will have a similar effect on S. aureus growth in vivo, the GAPDH mutant strains were grown in TM medium for 5 h in the presence of varied concentrations of glucose, ranging from 0.001% to 0.2% (Fig. 5A). At 0.001% glucose the ΔgapA mutant and wild-type strains are able to grow, but both the ΔgapB and the ΔgapA ΔgapB mutants have severe growth defects. As glucose concentrations are increased, the growth of the wild type and ΔgapB mutant also increased while the growth of the ΔgapA mutant is decreased. Above 0.05% glucose the ΔgapB mutant grows at a comparable level to that of the wild-type strain while little or no growth can be detected in either the ΔgapA or the ΔgapA ΔgapB double mutant strains. The point at which the switch from gluconeogenic to glycolytic metabolism occurs is approximately 0.01% glucose. The process by which this switch occurs is likely to be carbon catabolite repression (CCR). In the presence of a readily metabolized carbon source such as glucose, the genes responsible for secondary carbon metabolism are repressed, ensuring sequential utilization of available carbon sources based on how favorable they are to the bacteria. When glucose reaches 0.01%, growth of the ΔgapA mutant begins to decrease while growth of the ΔgapB mutant begins to increase, indicating repression of secondary carbon metabolism genes and derepression/induction of glucose metabolism. Therefore, Fig. 5A clearly demonstrates that at 0.01% glucose the bacteria switch from utilizing the highly abundant Casamino Acids as their carbon source to utilizing the favorable but limited carbon source of glucose.
FIG. 5.
(A) Growth of 8325-4 and isogenic ΔgapA, ΔgapB, and ΔgapA ΔgapB mutants after 5 h of incubation in TM medium with varied concentrations of glucose. Cultures were grown at 37°C with aeration, and growth was measured as the optical density of the culture at 600 nm. Each experiment was repeated three times on different days, and results are presented as averages with error bars indicating the standard deviations. (B) Effect of the GAPDH mutations on the pathogenesis of S. aureus infection in G. mellonella. The graph indicates the percent viability of infected G. mellonella larvae at 24, 48, and 72 h postinfection with S. aureus 8325-4 wild-type strain and the isogenic ΔgapA, ΔgapB, and ΔgapA ΔgapB mutants. With the PBS negative control, larvae showed 100% survival at all time points. The results from three independent experiments were combined and used to produce survival curves using the Kaplan-Meier method.
As both pathways are disrupted in the double mutant, it could be assumed that the strain would be unable to grow in any of the concentrations of glucose, but unexpectedly the growth of the double mutant increases with glucose concentration until 0.01%, and then growth levels rapidly decrease as glucose concentrations further rise. This suggests that 0.01% glucose is sufficient for growth without the need for de novo glucose from gluconeogenesis. The reason that we fail to see this level of growth in the ΔgapB single mutant may be because the functional glycolytic pathway will utilize this glucose, limiting its availability for other metabolic functions in S. aureus. Therefore, the regulation of carbon metabolism is tightly controlled in S. aureus to ensure that the bacteria utilize environmental carbon sources in the most efficient way possible.
Both gapA and gapB are required for pathogenesis in an invertebrate model of infection.
To determine the importance of both GapA and GapB in the wider role of host infection, larvae of Galleria mellonella were infected with wild-type 8325-4 and each of the isogenic GAPDH mutant strains. In this infection model, pathogenesis of the bacterial strain is measured by the percent viability of the infected larvae at 24, 48, and 72 h postinfection. The results show that both gapA and gapB are required for a full pathogenic phenotype of S. aureus in this model (Fig. 5B). Survival of the ΔgapA mutant (P < 0.0001), the ΔgapB mutant (P = 0.0003), and the double mutant (P < 0.0001) is significantly reduced compared to that of the wild-type strain. At 72 h postinfection the percent viability of the Galleria was lower in the larvae infected with the wild-type strain (20%) than in larvae infected with either the ΔgapA (93.3%) or the ΔgapB (56.7%) strain, showing that virulence was reduced in each of the mutants. Furthermore, pathogenesis of S. aureus was attenuated entirely in the double mutant strain, with 100% survival of the Galleria seen at all time points. This indicates that the loss of either gapA or gapB results in a reduction in virulence in S. aureus, which shows that at least during infection in this model, S. aureus relies on both primary and secondary carbon metabolism to utilize any available carbon source in the host environment.
DISCUSSION
In this study we have shown that in the pathogenic organism S. aureus, the GAPDH homologue GapB is an essential component in anabolic carbon metabolism. We also showed for the first time that the regulator gapR not only controls expression of gapA and the glycolytic operon but also regulates the level of gapB expression during gluconeogenesis. In addition, we have demonstrated that GapA and GapB are important for virulence in an invertebrate model of infection, showing that both glycolysis and secondary carbon metabolism play important roles during S. aureus infection.
In S. aureus it has been assumed that GapA is a glycolytic GAPDH protein and that GapB is a gluconeogenic GAPDH protein although there has been no published biological evidence (7, 30). In fact, recombinant His6-tagged GapB protein cloned from a bovine mastitis isolate of S. aureus was shown to have little or no GAPDH activity with either NAD+ or NADP+ as a cofactor (13). We confirmed these findings with recombinant GapB from a human S. aureus isolate and were also able to demonstrate that this lack of activity is not due to the presence of the His6 N-terminal tag. In E. coli, the second GAPDH homologue, GapB, functions as an NAD+-dependent E4PDH in the pyridoxine (vitamin B6) biosynthesis pathway (32, 38, 39). We have shown that this is not the case in S. aureus as GapB has no E4PDH activity with either NAD+ or NADP+ (data not shown).
However, our data suggest that although the recombinant GapB protein lacks specific GAPDH activity in vitro, S. aureus GapB is in fact a gluconeogenic enzyme. GapA cannot be solely responsible for gluconeogenic GAPDH activity in S. aureus as the ΔgapA mutant grows as well as the wild-type strain in medium lacking glucose, indicating that glycolysis is disrupted but that secondary carbon metabolism is still functional. In addition the ΔgapB mutant strain shows a reduction in NADP+-dependent GAPDH activity, is unable to grow in medium lacking glucose, and is unable to utilize secondary carbon sources. Thus, these data indicate a loss of anabolic carbon metabolism, most likely due to disruption of the gluconeogenesis pathway. Interestingly, the ΔgapB mutant is able to grow with glycerol as the primary carbon source. Glycerol enters the gluconeogenic pathway as G3P, the product of gluconeogenic GAPDH activity. Therefore, GapB must function before this stage in the gluconeogenic pathway in order for glycerol to recover the growth defect seen in the ΔgapB mutant.
The reciprocal growth response of the ΔgapA and ΔgapB mutants in response to glucose is mirrored in the reciprocal regulation of the transcription of gapA and gapB. Transcriptional analysis confirms that in wild-type S. aureus gapA expression is high in the presence of glucose but repressed under gluconeogenic conditions while gapB is expressed only in the absence of glucose. This is consistent with the GAPDH homologues in B. subtilis although transcription of these genes was determined by using integrated lacZ reporter systems in mutant strains rather than by analysis of wild-type RNA levels (10). By measuring the growth of ΔgapA and ΔgapB strains in variable concentrations of glucose, we have also been able to determine that as little as 0.01% glucose in the medium is sufficient for S. aureus to switch carbon metabolism from gluconeogenesis to glycolysis. Taken together, these data strongly suggest that in S. aureus GapA functions only in glycolysis and GapB functions only in gluconeogenesis.
Based on homology to B. subtilis cggR, the Deo-like regulator gapR has been proposed as the regulator of the glycolytic operon in S. aureus although no data on this gene have been published to date. Our study shows that in S. aureus GapR not only functions to regulate the glycolytic operon but also plays a role in the regulation of gapB transcription. In a ΔgapR knockout strain the expression of the glycolytic operon is derepressed under both glycolytic and gluconeogenic conditions. Interestingly, the expression of gapB is also higher in a gapR mutant under gluconeogenic conditions. This indicates a role of gapR in limiting the expression of gapB when glucose concentrations are low, which could indicate a very complex regulatory relationship between glycolysis and gluconeogenesis in S. aureus. In B. subtilis CggR represses the glycolytic operon in the absence of fructose-1,6-bisphosphate (FBP), an early product of glycolysis (9, 10, 22), by binding to a consensus sequence 32 bp downstream of the transcriptional start site of the operon and blocking elongation of the transcript (9). The transcription of the operon is also indirectly induced by carbon catabolite protein A (CcpA) under glycolytic conditions, providing multiple levels of control (22). Genome-wide sequence homology searches and transcriptome comparison between the B. subtilis wild type and a ΔcggR mutant strongly suggest that the function of CcgR is limited to the glycolytic operon in B. subtilis, whereas CcpA is a key regulator of carbon metabolism and controls many other genes (9, 36). Sequence analysis of the putative promoter regions of S. aureus gapR and gapB does not reveal a similar GapR target sequence in S. aureus, and therefore further work is required to determine the mechanism by which GapR represses transcription of genes in S. aureus. Interestingly, a recent microarray study has shown that in S. aureus CcpA is involved in carbon catabolite repression of gapB in the presence of glucose, but the effect on the induction of gapA expression was less clear (30). Therefore, further study of the regulation of both GAPDH homologues would need to investigate the interplay of GapR and CcpA on controlling gene expression.
The roles of glycolysis and gluconeogenesis in virulence have not been investigated in any bacteria to our knowledge. Therefore, we investigated the impact that each of the GAPDH homologues has on host virulence in a Galleria mellonella model of infection. Larvae of the greater wax moth G. mellonella have been shown to provide a useful insight into the pathogenesis of a wide range of bacterial and eukaryotic microbial infections including S. aureus (27). The Galleria model is particularly useful as these insects share many common aspects with mammalian innate immunity. Furthermore, Galleria infection model results consistently correlate with those of similar mammalian studies as bacterial strains that are attenuated in mammalian models demonstrate lower virulence in Galleria, regardless of the species of bacterium studied (11, 16, 27). Our data showed that both GapA and GapB are required during S. aureus infection. The loss of either of the proteins individually resulted in increased survival of the infected larvae while a loss of both proteins left the bacteria severely attenuated, and all of the infected larvae survived. Primary carbon utilization appears to be more important than secondary carbon utilization during infection as the Galleria showed 93.3% viability at 72 h postinfection with the ΔgapA mutant compared to 56.7% with the ΔgapB mutant, but it is still clear that both pathways are required for S. aureus to make use of all available carbon sources and infect the host to its full virulence potential. The main sugar present in the hemolymph is trehalose, but published literature does suggest that some glucose is also present. S. aureus is able to utilize trehalose which is broken down to glucose-6-phosphate by TreC. This feeds directly into glycolysis before the action of GapA. Therefore, the variation in the abilities of wild-type S. aureus and the Gap mutants to kill Galleria is in part due to the inability of the mutants to use the available carbon sources in the hemolymph, in agreement with the observations made in vitro. It is also possible that each of the proteins may have other “moonlighting” roles in infection, such as seen in Streptococcus pyogenes, where surface-associated GAPDH has been shown to have antiphagocytic properties and is involved in host cell adherence (3), but further study needs to be undertaken to determine any such roles in S. aureus.
In conclusion, the data presented in this study identify GapB as an essential component in secondary carbon use in S. aureus, and the evidence strongly suggests that it is essential for gluconeogenesis. Furthermore, both GapA and GapB are important during S. aureus infection and are required for pathogenesis. Thus, the data presented here may lead to a clearer understanding of the complex control of carbon metabolism in this important pathogen and the role that carbon metabolism plays in virulence during S. aureus infection.
Acknowledgments
This study was partially supported by a Doctoral Training Grant from the BBSRC to Joanne Purves.
We thank Xiaowen Yang at Protex (University of Leicester) for the cloning of pLEIC03 protein expression constructs and the Protein Nucleic Acid Chemistry Laboratory (University of Leicester) for the processing of all sequencing data.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 27 September 2010.
REFERENCES
- 1.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azam, S., N. Jouvet, A. Jilani, R. Vongsamphanh, X. Yang, S. Yang, and D. Ramotar. 2008. Human glyceraldehyde-3-phosphate dehydrogenase plays a direct role in reactivating oxidized forms of the DNA repair enzyme APE1. J. Biol. Chem. 283:30632-30641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boel, G., H. Jin, and V. Pancholi. 2005. Inhibition of cell surface export of group A streptococcal anchorless surface dehydrogenase affects bacterial adherence and antiphagocytic properties. Infect. Immun. 73:6237-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boschi-Muller, S., S. Azza, D. Pollastro, C. Corbier, and G. Branlant. 1997. Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and phosphorylating glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 272:15106-15112. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Campanella, M. E., H. Chu, and P. S. Low. 2005. Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane. Proc. Natl. Acad. Sci. U. S. A. 102:2402-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee, I., S. Schmitt, C. F. Batzilla, S. Engelmann, A. Keller, M. W. Ring, R. Kautenburger, W. Ziebuhr, M. Hecker, K. T. Preissner, M. Bischoff, R. A. Proctor, H. P. Beck, H. P. Lenhof, G. A. Somerville, and M. Herrmann. 2009. Staphylococcus aureus ClpC ATPase is a late growth phase effector of metabolism and persistence. Proteomics 9:1152-1176. [DOI] [PubMed] [Google Scholar]
- 8.Delgado, M. L., J. E. O'Connor, I. Azorin, J. Renau-Piqueras, M. L. Gil, and D. Gozalbo. 2001. The glyceraldehyde-3-phosphate dehydrogenase polypeptides encoded by the Saccharomyces cerevisiae TDH1, TDH2 and TDH3 genes are also cell wall proteins. Microbiology 147:411-417. [DOI] [PubMed] [Google Scholar]
- 9.Doan, T., and S. Aymerich. 2003. Regulation of the central glycolytic genes in Bacillus subtilis: binding of the repressor CggR to its single DNA target sequence is modulated by fructose-1,6-bisphosphate. Mol. Microbiol. 47:1709-1721. [DOI] [PubMed] [Google Scholar]
- 10.Fillinger, S., S. Boschi-Muller, S. Azza, E. Dervyn, G. Branlant, and S. Aymerich. 2000. Two glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J. Biol. Chem. 275:14031-14037. [DOI] [PubMed] [Google Scholar]
- 11.Gao, W., K. Chua, J. K. Davies, H. J. Newton, T. Seemann, P. F. Harrison, N. E. Holmes, H. W. Rhee, J. I. Hong, E. L. Hartland, T. P. Stinear, and B. P. Howden. 2010. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 6:e1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gil-Navarro, I., M. L. Gil, M. Casanova, J. E. O'Connor, J. P. Martinez, and D. Gozalbo. 1997. The glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is a surface antigen. J. Bacteriol. 179:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goji, N., A. A. Potter, and J. Perez-Casal. 2004. Characterization of two proteins of Staphylococcus aureus isolated from bovine clinical mastitis with homology to glyceraldehyde-3-phosphate dehydrogenase. Vet. Microbiol. 99:269-279. [DOI] [PubMed] [Google Scholar]
- 14.Hidalgo, E., A. Limon, and J. Aguilar. 1996. A second Escherichia coli gene with similarity to gapA. Microbiologia 12:99-106. [PubMed] [Google Scholar]
- 15.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jander, G., L. G. Rahme, and F. M. Ausubel. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 182:3843-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin, H., Y. P. Song, G. Boel, J. Kochar, and V. Pancholi. 2005. Group A streptococcal surface GAPDH, SDH, recognizes uPAR/CD87 as its receptor on the human pharyngeal cell and mediates bacterial adherence to host cells. J. Mol. Biol. 350:27-41. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, M., A. Cockayne, and J. A. Morrissey. 2008. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect. Immun. 76:1756-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kummer, U., J. Zobeley, J. C. Brasen, R. Fahmy, A. L. Kindzelskii, A. R. Petty, A. J. Clark, and H. R. Petty. 2007. Elevated glucose concentrations promote receptor-independent activation of adherent human neutrophils: an experimental and computational approach. Biophys. J. 92:2597-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindsay, J. A. 2010. Genomic variation and evolution of Staphylococcus aureus. Int. J. Med. Microbiol. 300:98-103. [DOI] [PubMed] [Google Scholar]
- 21.Ludwig, H., G. Homuth, M. Schmalisch, F. M. Dyka, M. Hecker, and J. Stulke. 2001. Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol. Microbiol. 41:409-422. [DOI] [PubMed] [Google Scholar]
- 22.Ludwig, H., N. Rebhan, H. M. Blencke, M. Merzbacher, and J. Stulke. 2002. Control of the glycolytic gapA operon by the catabolite control protein A in Bacillus subtilis: a novel mechanism of CcpA-mediated regulation. Mol. Microbiol. 45:543-553. [DOI] [PubMed] [Google Scholar]
- 23.Modun, B., and P. Williams. 1999. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect. Immun. 67:1086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pancholi, V., and V. A. Fischetti. 1993. Glyceraldehyde-3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proc. Natl. Acad. Sci. U. S. A. 90:8154-8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pancholi, V., and V. A. Fischetti. 1997. Regulation of the phosphorylation of human pharyngeal cell proteins by group A streptococcal surface dehydrogenase: signal transduction between streptococci and pharyngeal cells. J. Exp. Med. 186:1633-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peleg, A., Y. D. Monga, S. Pillai, E. Mylonakis, R. C. Jr. Moellering, and G. M. Eliopoulos. 2009. Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J. Infect. Dis. 199:532-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schomberg, D., I. Schomberg, and A. Chang (ed.). 2009. Springer handbook of enzymes, supplement, 2nd ed., vol. S1. Springer, Berlin, Germany.
- 29.Sebulsky, M. T., D. Hohnstein, M. D. Hunter, and D. E. Heinrichs. 2000. Identification and characterization of a membrane permease involved in iron-hydroxamate transport in Staphylococcus aureus. J. Bacteriol. 182:4394-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seidl, K., S. Muller, P. Francois, C. Kriebitzsch, J. Schrenzel, S. Engelmann, M. Bischoff, and B. Berger-Bachi. 2009. Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 9:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seifert, K. N., W. P. McArthur, A. S. Bleiweis, and L. J. Brady. 2003. Characterization of group B streptococcal glyceraldehyde-3-phosphate dehydrogenase: surface localization, enzymatic activity, and protein-protein interactions. Can. J. Microbiol. 49:350-356. [DOI] [PubMed] [Google Scholar]
- 32.Seta, F. D., S. Boschi-Muller, M. L. Vignais, and G. Branlant. 1997. Characterization of Escherichia coli strains with gapA and gapB genes deleted. J. Bacteriol. 179:5218-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sirover, M. A. 1997. Role of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in normal cell function and in cell pathology. J. Cell Biochem. 66:133-140. [PubMed] [Google Scholar]
- 34.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, J. M., and D. E. Heinrichs. 2002. Transferrin binding in Staphylococcus aureus: involvement of a cell wall-anchored protein. Mol. Microbiol. 43:1603-1614. [DOI] [PubMed] [Google Scholar]
- 36.Tobisch, S., D. Zuhlke, J. Bernhardt, J. Stulke, and M. Hecker. 1999. Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis. J. Bacteriol. 181:6996-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weese, J. S., and E. van Duijkeren. 2010. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Vet. Microbiol. 140:418-429. [DOI] [PubMed] [Google Scholar]
- 38.Yang, Y., G. Zhao, T. K. Man, and M. E. Winkler. 1998. Involvement of the gapA- and epd (gapB)-encoded dehydrogenases in pyridoxal 5′-phosphate coenzyme biosynthesis in Escherichia coli K-12. J. Bacteriol. 180:4294-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao, G., A. J. Pease, N. Bharani, and M. E. Winkler. 1995. Biochemical characterization of gapB-encoded erythrose 4-phosphate dehydrogenase of Escherichia coli K-12 and its possible role in pyridoxal 5′-phosphate biosynthesis. J. Bacteriol. 177:2804-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zorrilla, S., T. Doan, C. Alfonso, E. Margeat, A. Ortega, G. Rivas, S. Aymerich, C. A. Royer, and N. Declerck. 2007. Inducer-modulated cooperative binding of the tetrameric CggR repressor to operator DNA. Biophys. J. 92:3215-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]