Abstract
Thirty percent of Streptococcus pneumoniae isolates contain pilus islet 1, coding for a pilus composed of the backbone subunit RrgB and two ancillary proteins, RrgA and RrgC. RrgA is the major determinant of in vitro adhesion associated with pilus 1, is protective in vivo in mouse models, and exists in two variants (clades I and II). Mapping of the sequence variability onto the RrgA structure predicted from X-ray data showed that the diversity was restricted to the “head” of the protein, which contains the putative binding domains, whereas the elongated “stalk” was mostly conserved. To investigate whether this variability could influence the adhesive capacity of RrgA and to map the regions important for binding, two full-length protein variants and three recombinant RrgA portions were tested for adhesion to lung epithelial cells and to purified extracellular matrix (ECM) components. The two RrgA variants displayed similar binding abilities, whereas none of the recombinant fragments adhered at levels comparable to those of the full-length protein, suggesting that proper folding and structural arrangement are crucial to retain protein functionality. Furthermore, the two RrgA variants were shown to be cross-reactive in vitro and cross-protective in vivo in a murine model of passive immunization. Taken together, these data indicate that the region implicated in adhesion and the functional epitopes responsible for the protective ability of RrgA may be conserved and that the considerable level of variation found within the “head” domain of RrgA may have been generated by immunologic pressure without impairing the functional integrity of the pilus.
Streptococcus pneumoniae is a main determinant of respiratory tract infections, such as otitis media, sinusitis, and community-acquired pneumonia, and is also responsible for invasive diseases, such as bacteremic pneumonia and meningitis (16, 17, 30, 43, 46, 50, 55). Nonetheless, pneumococci are normal components of the human commensal flora, asymptomatically colonizing the upper respiratory tracts of both children and healthy adults. Colonization is commonly followed by horizontal transmission of S. pneumoniae, leading to its spread within the community (4, 19, 33). Current glycoconjugate vaccines are efficacious against invasive disease caused by serotypes included in the vaccines; however, their potential to prevent carriage and related mucosal diseases, such as otitis media, is not optimal (10, 12, 23, 29, 34, 48). Furthermore, the partial geographic coverage and the phenomenon of serotype replacement associated with the introduction of the 7-valent conjugate vaccine limit to some extent its long-term effectiveness (11, 20, 31, 37). For these reasons, current research is focused on the identification of protein vaccine candidates able to elicit serotype-independent protection against S. pneumoniae infection. In this context, colonization could represent a critical point of intervention, and bacterial components involved in these mechanisms should be studied in order to determine their value as vaccine candidates.
Adhesion of bacteria to the mucosa is considered an essential early step in the colonization process. The ability of S. pneumoniae to adhere to epithelial cells has been ascribed to a number of surface-exposed proteins, including PspC, PsaA, PsrP, PfbB, NanA, PavA, and pili (3, 21, 41, 42, 45, 49, 54).
Pili were recently discovered in many Gram-positive pathogens, and although their biological function has not been fully elucidated, their presence has been mostly related to bacterial adhesion, biofilm formation, and translocation of epithelial barriers (1, 13, 35, 47). These structures are composed of subunits covalently linked by means of intermolecular isopeptide bonds (32, 36, 51-53). Furthermore, intramolecular isopeptide bonds have been found in most pilus subunits characterized to date (9, 26-28). These bonds may play a critical role in maintaining pilus integrity in the face of severe mechanical and chemical stress while bound to host cells and thus may provide a functional mode of stabilization for cell surface proteins involved in host pathogenesis.
In S. pneumoniae, pili (pilus 1 and pilus 2) are encoded by two genomic islets (pilus islet 1 [PI-1] and PI-2) that are not present in all pneumococcal clinical isolates. A number of molecular epidemiological studies have highlighted the presence of PI-1 as a clonal property of S. pneumoniae isolates and have defined, based on sequence analysis, the classification of PI-1 into three major clades (2, 5, 7, 25, 38, 39). Mutants lacking PI-1 are impaired in adhesion to cultured epithelial cells in vitro and are less virulent in murine models of colonization, pneumonia, and bacteremia (6, 41). Interestingly, pilus 1 expression is known to increase host inflammatory responses that might disrupt the mucosal barrier and facilitate subsequent invasion by the bacteria (6).
Pneumococcal pilus 1 is composed of three subunits (RrgA, RrgB, and RrgC); RrgB is the backbone component, and RrgA is the major ancillary protein, localized at the pilus tip and responsible for the adhesion properties of the pilus, whereas RrgC is the minor ancillary protein, likely located at the pilus base (21, 22, 41). In terms of sequence variability, RrgB is classified into three variants and RrgC is conserved, whereas RrgA exists in two major variants (clades I and II) (38). The recombinant form of RrgA clade I adheres in vitro to cultured A549 lung epithelial cells, as well as to purified extracellular matrix (ECM) components (collagen I, fibronectin, and laminin) (21, 41). In addition, RrgA, along with the other two pilus 1 components, is able to elicit protection from lethal challenge with the homologous strain in mouse models of active and passive immunization (18).
In this work, we investigated whether the differences between the two variants had an effect on the biochemical characteristics, biological function, and immunological properties of the molecule. We found that (i) sequence variability was restricted to the “head” domain of RrgA, containing the putative adhesive motifs; (ii) the two RrgA variants were resistant to proteolytic cleavage, and this feature was dependent on the presence of intramolecular isopeptide bonds; (iii) the two variants were able to adhere to epithelial cells and ECM components at comparable levels, whereas a mutant (Asp444Ala) in the RGD tripeptide showed reduced binding; (iv) none of the individual fragments encompassing the N-terminal (NT), central (CP), and C-terminal (CT) portions of RrgA was able to maintain adhesive capacity; (v) antibodies against these fragments revealed the N terminus to be less accessible than the remaining portion of the molecule on the native pilus; and, finally, (vi) antibodies raised against each of the two RrgA variants were cross-reactive and cross-protective in murine passive-immunization studies.
MATERIALS AND METHODS
Protein sequence and structure analysis.
Primary sequences of RrgA proteins representative of the two RrgA variants were aligned using ClustalW. The position-dependent sequence identity was computed between one clade I strain (TIGR4) and one clade II strain (SPEC6B) by averaging on a sliding window of 10 amino acids (aa) shifted by 5 amino acids. Functional domains were predicted using the software SMART (http://smart.embl-heidelberg.de/). The three-dimensional (3D) structure of RrgA was visualized and manipulated using PDBViewer and Chimera software.
Bacterial strains and growth conditions.
S. pneumoniae strains were routinely grown at 37°C in 5% CO2 on tryptic soy agar (TSA) plates (Becton Dickinson) supplemented with 5% defibrinated sheep blood or in Todd-Hewitt broth supplemented with 0.5% (wt/wt) yeast extract (THYE) (Becton Dickinson).
Cloning and site-directed mutagenesis.
Standard recombinant DNA techniques were used to construct expression plasmids (pET21b+; Novagen). Briefly, the coding sequences of the full-length proteins (the N-terminal signal sequence and C-terminal cell wall-sorting signal (CWSS) motif were excluded from the cloning) and protein fragments used in this study were amplified by PCR from chromosomal DNA of S. pneumoniae strain TIGR4 (RrgA clade I) or SPEC6B (RrgA clade II) by using specific primers listed in Table 1. Mutations were introduced in RrgA by PCR-based site-directed mutagenesis: forward and reverse oligonucleotides (Table 1), each containing the desired mutation, were used to create by overlap extension an rrgA fragment containing the mutation. The PCR fragments obtained were then digested with the appropriate restriction enzymes and ligated into the C-terminal 6×His tag expression vector pET21b+ (Novagen); transformants were screened, and recombinant plasmids were identified and confirmed by DNA sequencing.
TABLE 1.
Recombinant proteins used in this studya
| Protein name | Amino acid positionb | Primer sequencec |
|
|---|---|---|---|
| Forward | Reverse | ||
| RrgA clade I | 39-862 | AGTTGCTGCTAGCGAAACGCCTGAAACCAGTCCAGCG | CAGTTCGCTCGAGTTCTCTCTTTGGAGGAATAGGTTC |
| RrgA clade II | 39-859 | GTGCGTGCTAGCGAAACGCCTGAAACCAGTCCAGCG | CAGCGTCTCGAGTTCTCTTTTTGGAGGAATTGGCTC |
| NT | 39-214 | AGTTGCTGCTAGCGAAACGCCTGAAACCAGTCCAGCG | CAGTTCGCTCGAGTGTTTTCCCACTAACCGTCAATTC |
| CP clade I (CP = 1) | 215-637 | AGTTGCTGCTAGCGTGTATGAATAAAAAGATAAGTCT | CATGATCCTCGAGTACAGCTTGTCCATTCTCCAAGCG |
| CP clade II (CP = 2) | 215-634 | AGTTGCTGCTAGCACGGTTGAAACGAAAGAAGCCTCT | CATGATCCTCGAGAGTAGGGACATTATTCACCAACGA |
| CT | 638-862 | AGTTGCTGCTAGCGGTGGTCCACAAAATGATGGTGGT | CAGTTCGCTCGAGTTCTCTCTTTGGAGGAATAGGTTC |
| CT long | 604-862 | AGTTGCTGCTAGCGAGTTAATTGATTTGCAATTGGGC | CATGATCCTCGAGAGTAGGGACATTATTCACCAACGA |
| Asp444 Ala | 39-862 | ATTGTACGCGGAGCTGGGCAAAGTTAC | GTAACTTTGCCCAGCTCCGCGTACAAT |
| Lys191 Ala | 39w-862 | GTACACTTTCAGCGAGAATTTATC | GATAAATTCTCGCTGAAAGTGTAC |
| Asn695 Ala | 39-862 | TTTATGATACCGCTGGTCGAACAA | TTGTTCGACCAGCGGTATCATAAA |
For each recombinant protein used in this study, the fragment name, amino acid position, and forward and reverse primers used to generate the clone are listed.
With respect to the FL sequence.
Nucleotide sequences corresponding to restriction sites are underlined.
Protein expression and purification.
The plasmids containing the rrgA full-length sequence or fragments thereof were transformed into competent Escherichia coli BL21 D3 star (Invitrogen). Protein expression was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) (Sigma; 1 mM final concentration) to a bacterial culture at an optical density at 600 nm (OD600) of 0.4 to 0.5 (LB medium supplemented with 100 μg/ml ampicillin) and growing the bacteria at 25°C for 4 to 5 h to avoid inclusion body formation. The cells were harvested by centrifugation (10 min in a Beckmann JA 81000 rotor at 8,000 × g at 4°C), and the bacterial pellets (5 to 10 g) were lysed with lysozyme (0.25 mg/ml) in 40 ml Bug Buster Reagent (Novagen) supplemented with Benzonase Nuclease (Novagen; 2.5-U/ml final concentration) and Protease Inhibitor Cocktail III (Calbiochem; 2.5 μl/ml lysate). After centrifugation, the soluble fraction was subjected to metal chelate affinity chromatography on His-Trap HP columns (GE Healthcare) according to the manufacturer's instructions. Pooled fractions containing the purified protein were dialyzed overnight (ON) against phosphate-buffered saline (PBS) (1st purification step). An aliquot from the 1st purification step was diluted 3-fold with 20 mM Tris-HCl, pH 7.5, and applied to a 1-ml HiTrap Q HP column. Proteins were eluted with a linear gradient (30 column volumes) from 20 to 500 mM NaCl in 20 mM Tris-HCl, pH 7.5. Pooled fractions containing the purified protein were dialyzed ON against PBS. Protein purity was determined by SDS-PAGE. The protein concentration was determined using a Bradford protein assay, reading the absorbance at 280 nm (NanoDrop). Purified recombinant proteins were subsequently used to immunize CD1 mice (20 μg/mouse) or rabbits (100 μg/rabbit) for antibody generation (Charles River Laboratory).
CD analysis.
Far-UV circular dichroism (CD) spectra from 200 to 260 nm (1-nm steps; 20 nm/min) were obtained at 25°C by averaging 10 scans on a CD spectrometer (Jasco J-810) equipped with a water-cooled Peltier system (PCB1500). Readings were performed at a protein concentration of 0.2 mg/ml in PBS using Suprasil cuvettes (Hellma) with a path length of 0.1 cm. The main compartment of the instrument was flushed with dry nitrogen gas during the measurement. PBS was used as a blank, and its spectrum was subtracted from all recorded CD spectra. The secondary structures of full-length RrgA proteins and RrgA protein fragments were evaluated by deconvolution of the spectra using CDNN v.2.1 (8). Original CD data in millidegrees were converted to Δɛ units.
SDS-PAGE and Western blot analysis.
SDS-PAGE analysis was performed using Nu-Page 4 to 12% Bis-Tris gradient gels (Invitrogen) according to the manufacturer's instructions. A Hi-Mark prestained high-molecular-weight protein standard (Invitrogen) was used. Gels were stained with colloidal Coomassie blue G-250 (Invitrogen) or processed for Western blot analysis (WB) by using standard protocols. Mouse antibodies raised against recombinant His-tagged proteins were used at 1/3,000 dilution. Alkaline phosphatase-conjugated secondary goat anti-mouse IgG antibodies (Promega) were used at 1/5,000, and the signal was developed by using Western Blue Stabilized Substrate for Alkaline Phosphatase (Promega).
Enzymatic digestion of recombinant proteins.
Recombinant proteins were digested with sequencing-grade modified trypsin (Promega), using an enzyme/substrate ratio of 1/100 (wt/wt) in 50 mM ammonium bicarbonate, pH 8, containing 0.1% (wt/vol) Rapigest (Waters) ON at 37°C.
In-gel digestion and MALDI TOF/TOF mass spectrometric analysis.
Spots of colloidal Coomassie blue G-250-stained bands were excised from the SDS-PAGE gel using a Pasteur pipette and destained ON in 200 μl of 50% (vol/vol) acetonitrile, 50 mM ammonium bicarbonate. The spots were then washed with 200 μl of acetonitrile. The acetonitrile was discarded, and the spots were allowed to air dry. Modified trypsin (12 μg/ml) in 5 mM ammonium bicarbonate was added to each spot, and the enzymatic digestion was performed for 3 h at 37°C; 0.8 μl of the digestion was directly spotted on a PAC (Prespotted AnchorChip 96 set for proteomics; Bruker Daltonics) target. The air-dried spots were washed with 0.6 μl of a solution of 70% (vol/vol) ethanol, 0.1% (vol/vol) trifluoroacetic acid (TFA). Peptide mass spectra were recorded with a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF)/TOF mass spectrometer (UltraFlex; Bruker Daltonics, Bremen, Germany). Ions generated by laser desorption at 337 nm (N2 laser) were recorded at an acceleration of 25 kV in the reflector mode. About 200 single spectra were accumulated for improving the signal/noise ratio and analyzed by FlexAnalysis (version 2.4; Bruker Daltonics). External calibration was performed using standard peptides prespotted on the target.
Protein binding to A549 epithelial cells; flow cytometric assay.
A549 lung epithelial cells were nonenzymatically detached from the support by using cell dissociation solution (Sigma), harvested, and resuspended in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1% (wt/vol) bovine serum albumin (BSA), in the absence of serum and antibiotics. The cells were mixed with either medium alone or different concentrations of the purified proteins diluted in DMEM (0.05 to 6 μM) and incubated for 2 h on ice. The A549 cells were then washed twice with 1% BSA in PBS and incubated with antibodies against each protein for 1 h on ice. After two additional washes, the preparations were incubated with Alexa Fluor 488 secondary antibodies (Molecular Probes; 1:200 in PBS-1% BSA), and 10,000 cells were analyzed with a FACS-Canto II flow cytometer (Becton Dickinson). A one-tailed t test was used to compare the adhesion levels of the RrgA clade I wild type (wt) and RrgA Asp444Ala.
Protein binding to ECM components; ELISA.
Ninety-six-well MaxiSorp flat-bottom plates (Nunc) were coated for 1 h at 37°C, followed by an ON incubation at 4°C with 1 μg/well laminin (from human placenta; Sigma) or fibronectin (from human plasma; Sigma) or with 2 μg/well collagen I (from human lung; Sigma) in PBS, pH 7.4. A BSA-coated plate served as a negative control. The plates were washed 3 times with PBS-0.05% Tween 20 and blocked for 2 h at 37°C with 200 μl of PBS-1% BSA, followed by 3 washing steps with PBS-0.05% Tween 20. Recombinant protein samples were initially diluted to 4 μg/ml with PBS and were then transferred into coated, blocked plates in which the samples were serially diluted 2-fold with PBS, obtaining a final volume of 100 μl/well. The plates were incubated for 2 h at 37°C and then ON at 4°C. The plates were washed 3 times and incubated for 2 h at 37°C with the respective primary mouse antibodies (1/10,000 dilutions). After another three washing steps, bound antigen-specific mouse IgGs were revealed with the phosphatase alkaline substrate p-nitrophenyl-phosphate (Sigma) following 2 h of incubation at 37°C with alkaline phosphatase-conjugated goat anti-mouse IgG (Sigma Chemical Co., St. Louis, MO). Readout was performed at 405 nm with an enzyme-linked immunosorbent assay (ELISA) plate reader. A one-tailed t test was used to compare the adhesion levels of the RrgA clade I wt versus RrgA Asp444Ala.
Flow cytometry of entire bacteria.
Bacteria were grown in THYE to exponential phase (OD600 = 0.25), fixed with 2% paraformaldehyde, and then stained with mouse antisera raised against full-length RrgA clades I and II and against RrgA recombinant fragments (final dilution, 1:300). After labeling with a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (Jackson Laboratories), bacterial staining was analyzed by using a FACSCalibur cytometer (Becton Dickinson). Sera from mice immunized with PBS plus adjuvant and with RrgB were used as negative and positive controls, respectively.
Passive-immunization experiments.
Animal studies were done in compliance with the current law, approved by the local animal ethics committee, and authorized by the Italian Ministry of Health. Female 8-week-old BALB/c specific-pathogen-free mice (Charles River) were used. Groups of 8 mice received intraperitoneally 100 μl of rabbit serum raised against either RrgA clade I or clade II. Twenty minutes after antiserum administration, the mice received intraperitoneal challenge with 150 CFU of TIGR4. Blood samples were obtained from each mouse 24 h after challenge, and the bacteremia was quantified by culturing serial dilutions of the blood on agar-blood plates. The plates were incubated overnight at 37°C in a humidified atmosphere (5% CO2), and then the CFU were counted and the bacteremia was calculated. The limit of detection was 125 CFU per ml of blood. After challenge, the animals were monitored for 10 days postchallenge, twice per day for the first 4 days and then daily. Mice were euthanized when they exhibited defined humane endpoints that had been preestablished for the study in agreement with Novartis animal welfare policies, and the day was recorded.
The results of the bacteremia and mortality course (median survival) were analyzed by the Mann-Whitney U test. The survival rates were analyzed by Fisher's exact test. One-tailed tests were used for comparison of the immunized groups to the corresponding control groups. Two-tailed tests were used for the comparison of immunized groups to each other. P values of ≤0.05 were considered and referred to as significant.
RESULTS
Domain organization and sequence variability of the two RrgA variants.
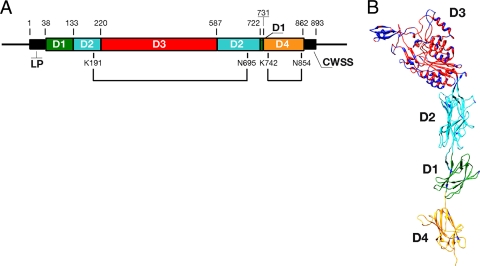
RrgA protein sequence variability studies performed on a collection of 44 pneumococcal isolates highlighted the presence of two genetic variants, clades I and II (893 and 890 amino acids, respectively). The two variants have similar organizations containing an N-terminal leader sequence (aa 1 to 38) and an LPXTG-like (YPRTG) C-terminal CWSS (Fig. 1A). Overall, they share 84% sequence identity, while most of the variability resides in the central portion of the protein (aa 214 to aa 637 of clade I), where the level of sequence identity decreases to 67.7%. The regions of diversity were mapped on the structure of recombinant RrgA clade I obtained from X-ray crystal data (26). We found the variable amino acid cluster in the distal D3 “head” domain, whereas D1, D2, and D4, which form the “stalk” of the molecule, were well conserved (Fig. 1B). Interestingly, the majority of the variable amino acids were located in regions predicted to be surface exposed, thus most likely not affecting the global folding of the domain.
FIG. 1.
Structural organization and sequence variability of RrgA. (A) Protein sequence domain organization of RrgA clade I (aa 1 to 893). The leader peptide (LP), the CWSS, and the two intramolecular isopeptide bonds (K1S1-N6P5, K7u2-N85u) are indicated. (B) The sequence variability between RrgA clade I (TIGR4) and RrgA clade II (SPEC6B) is displayed on the crystal structure of TIGR4 RrgA clade I. Amino acid variability is delineated in dark blue. Domains 1 to 4 are color coded to reflect the schematic organization in panel A.
As described by Izoré and colleagues, RrgA contains a Von Willebrand factor type A (vWA) domain, characteristic of many eukaryotic ECM binding proteins, like the α2β1 integrin, an important human receptor for collagen (15, 16, 26). The vWA factor-like domain, present in both RrgA clades I and II, resides within the D3 distal region, encompassing amino acids 224 to 450 of RrgA clade I. Interestingly, the MIDAS motif, composed of Asp232-X-Ser234-X-Ser236 and Asp387, known to be crucial for collagen recognition and binding through coordination of metal ions, is conserved in both RrgA variants, although its position is internal to the predicted structure and therefore unlikely to be directly engaged in a superficial interaction with the collagen helix.
As RrgA has been shown to be important for bacterial recognition of laminin and fibronectin (21), Izoré and colleagues hypothesized that this property could be associated with the two elongated arms, also located within the D3 domain. The two arms form a U-shaped cradle rich in basic residues, which could be engaged in the recognition of negatively charged molecules, such as glycosaminoglycans (GAGs), that are directly associated with laminin and fibronectin (26). Interestingly, the corresponding cradle-shaped region present in variant II lacked the majority of positively charged amino acids, which in most instances were replaced by hydrophobic residues (Lys294→Ala; Arg298→Ile; Lys476→Val; Lys511→Trp).
Finally, the RrgA sequence contains an Arg-Gly-Asp (RGD) tripeptide, which is implicated in the interactions of a number of bacterial adhesins with integrin receptors present on the basolateral surfaces of eukaryotic cells (24). The RGD motif is conserved between the two RrgA variants and is contained within the D3 domain (aa 442 to 444 of RrgA clade I). However, in contrast to its expected surface exposure, in recombinant RrgA clade I this domain is buried within the structure. Therefore, assuming that the proposed structural model corresponds to the native protein folding, the implication of the RGD motif in binding could be possible only if a conformational change occurs during the interaction of RrgA with its ligands.
RrgA resistance to proteolytic cleavage is due to the presence of two intramolecular isopeptide bonds.
The backbone subunits of pili in Gram-positive bacteria have been shown to resist proteolytic cleavage, a feature associated with the presence of intramolecular isopeptide bonds (9, 14, 27, 28). As RrgA contains two intramolecular isopeptide bonds (26), we investigated whether the two variants were also protease resistant and if this feature was associated with the presence of isopeptide bonds.
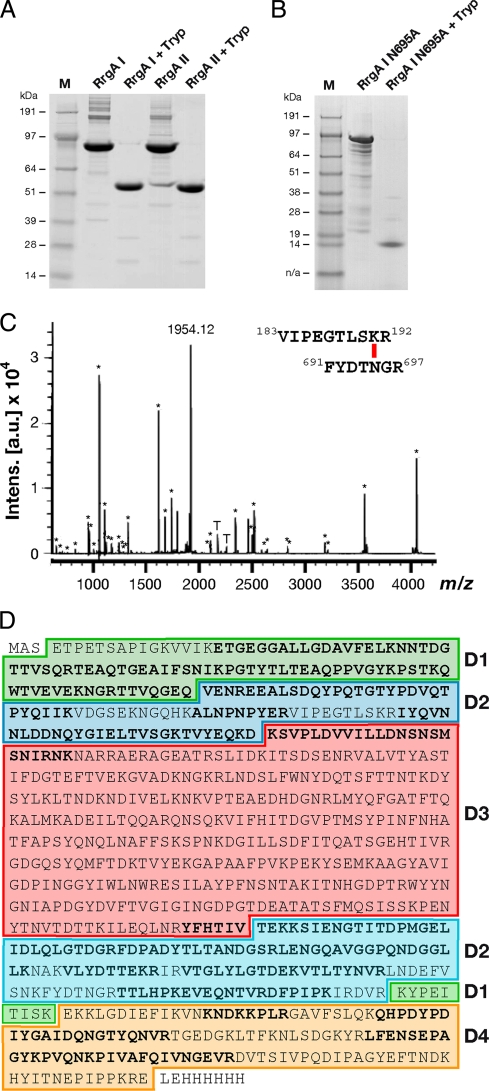
Trypsin proteolytic-cleavage reactions were performed on the purified recombinant full-length (FL) RrgA clades I and II. Both RrgA variants were significantly resistant to enzymatic digestion compared to other recombinant S. pneumoniae proteins, which, under the same reaction conditions, were completely digested (data not shown). Despite significant sequence diversity, the cleavage patterns of RrgA clade I and RrgA clade II were similar. In fact, for both digestions, the SDS-PAGE pattern revealed one major polypeptidic fragment with an apparent molecular mass of 55 kDa (Fig. 2A). Analysis by peptide mass fingerprinting (PMF) showed that the peptides derived from the 55-kDa fragment covered two distinct sequence regions located at the N- and C termini of RrgA (Fig. 2C and D), while the “head” D3 domain was completely digested. Interestingly, one of the major peaks (m/z, 1,954.12) could not be assigned to any theoretical tryptic peptide (Fig. 2C) but was consistent with the molecular mass of the peptide 183VIPEGTLSKR192, linked by an isopeptide bond to the peptide 691FYDTNGR697 (expected m/z, 1,953.992) (underlining indicates amino acids involved in forming an intramolecular isopeptide bond). To confirm the sequences of the two fragments involved in the isopeptide bond, the ion of m/z 1,954.12 was fragmented by MALDI TOF/TOF mass spectrometry (MS), and the primary sequence deduced from the tandem MS (MS/MS) spectrum was in agreement with the sequences of the cross-linked peptides (data not shown). The same cross-linked peptide was identified in RrgA clade II by PMF (data not shown).
FIG. 2.
RrgA intramolecular isopeptide bonds confer resistance to proteolysis. (A and B) SDS-PAGE analysis of overnight trypsin digestion of RrgA clade I and RrgA clade II (A) and RrgA clade I mutant Asn695Ala (B). (C) Peptide mass fingerprint of the RrgA clade I 55-kDa trypsin-resistant fragment. Each m/z signal labeled with an asterisk corresponds to an RrgA clade I tryptic peptide (underlined sequences in panel D). The signal of m/z 1,954.12 is consistent with the molecular mass of the peptide 183VIPEGTLSKR192, linked by an isopeptide bond to the peptide 691FYDTNGR697 (expected m/z, 1,953.99). Signals labeled with a T are trypsin autolysis peptides. (D) Protein sequence of FL RrgA clade I as cloned in the pET21b+ expression vector. Sequences in boldface letters correspond to the peptides identified by PMF. The colored boxes represent the RrgA clade I domains identified within the crystal structure. The color codes are as follows: D1, green; D2, turquoise; D3, light red; D4, dark yellow.
In order to define the role of the intramolecular isopeptide bond in conferring proteolytic resistance on the full-length RrgA molecule, two site-directed mutants were generated in RrgA clade I in amino acids involved in the formation of the isopeptide bond linking the N- and C-terminal portions of RrgA (Lys191Ala and Asn695Ala). Analysis of the digested products indicated that the structured trypsin-resistant core of 55 kDa was absent in both RrgA mutants, thus confirming the critical role of the intramolecular isopeptide bond in conferring resistance to proteolysis (Fig. 2B). In the recombinant Asn695Ala form, one major tryptic-resistant fragment of 12 kDa (apparent molecular mass) was generated. As assessed by PMF, this fragment corresponds to the C-terminal D4 domain. This result could be explained by the presence of a second isopeptide bond within D4 (Lys742-Asn854), as recently reported by Izoré and colleagues (26). Overall, these studies indicate that both RrgA variants are resistant to tryptic digestion and that this resistance is due to the presence of intramolecular isopeptide bonds, which are necessary to maintain the integrity of the stalk. On the other hand, the central domain, D3, comprising the “head” of the molecule, was more susceptible to proteolysis and underwent complete digestion.
Clade I and II RrgA display similar binding activities.
As previously shown, RrgA clade I is able to adhere to epithelial cells, as well as to purified ECM components, such as collagen, laminin, and fibronectin (21, 41). However, thus far, no functional data on RrgA clade II have been reported. In order to investigate whether the two variants maintain the same binding properties, and the molecular mechanisms underlying this function, we tested the in vitro adhesion of the two recombinant full-length RrgA variants, along with the two mutant forms of RrgA clade I, Lys191Ala and Asn695Ala (which are unable to form the isopeptide bond in D2), and a mutant obtained from the replacement of the aspartic residue of the RGD tripeptide motif with alanine (Asp444Ala).
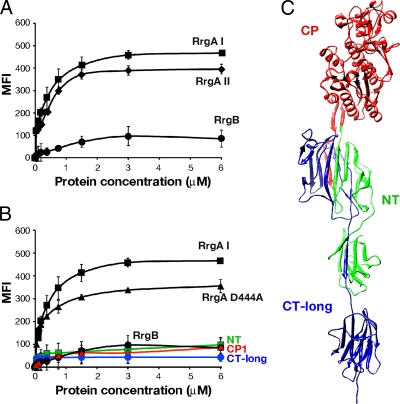
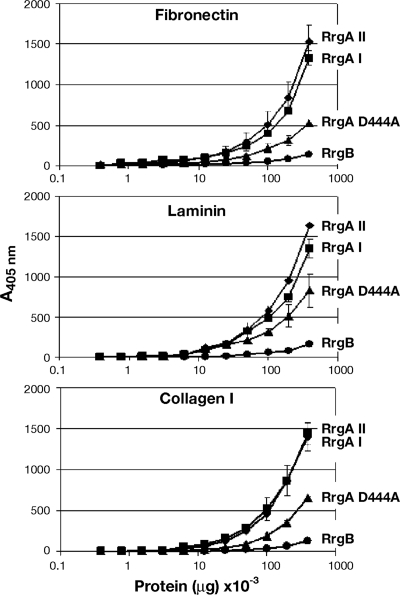
The binding of the recombinant proteins to A549 cells in suspension was evaluated by antibody detection (polyclonal mouse antisera raised against RrgA clades I and II) and fluorescence-activated cell sorter (FACS) analysis at a range of protein concentrations (from 6 to 0.05 μM). The assay revealed that the binding was dose dependent for both RrgA clades I and II, with both proteins reaching the plateau at a concentration of 3 μM with similar mean fluorescence intensities (MFI), significantly higher than the RrgB negative control (Fig. 3A). Additionally, the FL RrgA mutants (Lys191Ala and Asn695Ala) were as adhesive as the wild-type form (data not shown). Moreover, FL RrgA proteins of both variants revealed similar adhesion levels to endothelial human brain microvascular epithelial cells (HBMEC) and Detroit 562 epithelial cells (data not shown). As shown in Fig. 3B, adhesion to A549 cells was also retained in the RrgA Asp444Ala mutant, although the MFI was significantly reduced by 24% with respect to the wt (P = 0.007). To confirm the presence of similar secondary structures, the wt RrgA clade I and the RrgA Asp444Ala mutant were analyzed, along with RrgA clade II, by CD. Analysis of the spectra indicated that the three recombinant proteins were folded and displayed highly similar contents of secondary structural elements, indicating that the Asp→Ala mutation also caused no major changes in the global folding of the molecule (Table 2; see Fig. S1 in the supplemental material). The two RrgA variants also revealed comparable dose-dependent binding properties to laminin, fibronectin, and collagen ECM components (range of concentrations tested, 0.4 to 0.004 μg/ml), with the adhesion capacity of the RrgA Asp444Ala mutant significantly reduced with respect to wt RrgA clade I (P = 0.001, P = 0.03, and P = 0.002 for the three ECM components) (Fig. 4).
FIG. 3.
Full-length RrgA is required for adhesion to epithelial cells in vitro. (A) Binding (4°C) of recombinant full-length RrgA clade I and clade II (dose range, 0.05 to 6 μM) to A549 epithelial cells in suspension as measured by FACS analysis compared to the pilus backbone, RrgB (used as a negative control). (B) Binding at 4°C of RrgA recombinant fragments and RrgA Asp444Ala to A549 cells in suspension (dose range, 0.05 to 6 μM). In panels A and B, for each protein concentration, the net mean fluorescence intensity was calculated from three independent experiments (the error bars represent the standard deviations). (C) Mapping of the RrgA fragments used in the adhesion experiments onto the RrgA clade I crystal structure. NT, green; CP, red; CT long, blue.
TABLE 2.
Secondary-structure analysis of RrgA recombinant proteins by CD spectroscopya
| Structure | Content (%) |
||||||
|---|---|---|---|---|---|---|---|
| RrgA clade I | RrgA D444A | RrgA clade II | NT | CP-1 | CP-2 | CT-long | |
| Helix | 23.60 | 25.70 | 21.30 | 19.80 | 26.80 | 29.90 | 10.90 |
| Antiparallel | 19.00 | 17.60 | 21.00 | 10.90 | 14.60 | 13.60 | 27.40 |
| Parallel | 6.10 | 6.20 | 6.20 | 4.10 | 5.70 | 5.90 | 5.20 |
| Beta turn | 15.30 | 15.00 | 15.50 | 28.60 | 16.70 | 15.50 | 22.20 |
| Random coil | 31.40 | 30.80 | 31.90 | 35.40 | 31.20 | 30.20 | 36.50 |
| Total | 95.40 | 95.20 | 95.80 | 98.90 | 95.20 | 95.00 | 102.10 |
Analysis of the CD spectra (see Fig. S1 in the supplemental material) was performed using CDNN v.2.1, including unit conversion to Δɛ units.
FIG. 4.
Full-length RrgA clades I and II bind extracellular matrix components in vitro. Different doses of recombinant RrgA clades I and II, along with RrgA Asp444Ala, were incubated with purified fibronectin (top), laminin (middle), and collagen I (bottom); the pilus backbone, RrgB, was used as a negative control. Binding was quantified by ELISA at an absorbance of 405 nm. The points represent the means (the error bars indicate the standard deviations of the means) of measurements made in triplicate.
RrgA fragments are not able to bind to epithelial cells and to ECM components.
To obtain further insight into the regions of RrgA implicated in the adhesive function, full-length proteins were divided into three nonoverlapping portions on the basis of (i) sequence similarity between the two clades, (ii) prediction of functional domains, and (iii) analysis of the available crystallographic data (Fig. 3C). The two highly conserved NT (aa 39 to 214) and CT (aa 638 to 862) domains were cloned from TIGR4, whereas the variable central portion, mainly encompassing the D3 domain, was cloned from strain TIGR4 (CP-1; aa 215 to 637) and from strain SPEC-6B (CP-2; aa 215 to 634). Three out of the four selected fragments were successfully expressed and purified as described in Materials and Methods. In contrast, the CT portion was insoluble and therefore was replaced by a CT long protein fragment (aa 604 to 862) that was obtained in soluble form. The four recombinant fragments were tested in in vitro adhesion experiments with A549 epithelial cells and ECM components, as described above for the full-length proteins. To verify whether they maintained their secondary structures, the fragments were analyzed by CD. The results show that each of the fragments was folded, with CP-1 and CP-2 displaying similar secondary-structure compositions (Table 2; see Fig. S1 in the supplemental material). None of the recombinant fragments was able to bind to A549 cells (Fig. 3B) or to ECM components (data not shown) at a level similar to that of FL RrgA, thus suggesting that adhesive properties of RrgA might be due to interplay between different protein domains.
The NT portion is not accessible to antibodies in the native RrgA.
The four structural domains of RrgA make few contacts with each other and are mostly associated through short linkers, potentially conferring a significant level of flexibility on the protein (26). In order to investigate to what extent the observed conformation of crystallized recombinant RrgA reflects the dynamic properties of the native protein once associated with the RrgB backbone fiber, the reactivity of antibodies raised against the FL, as well as single recombinant fragments, was analyzed by FACS on S. pneumoniae.
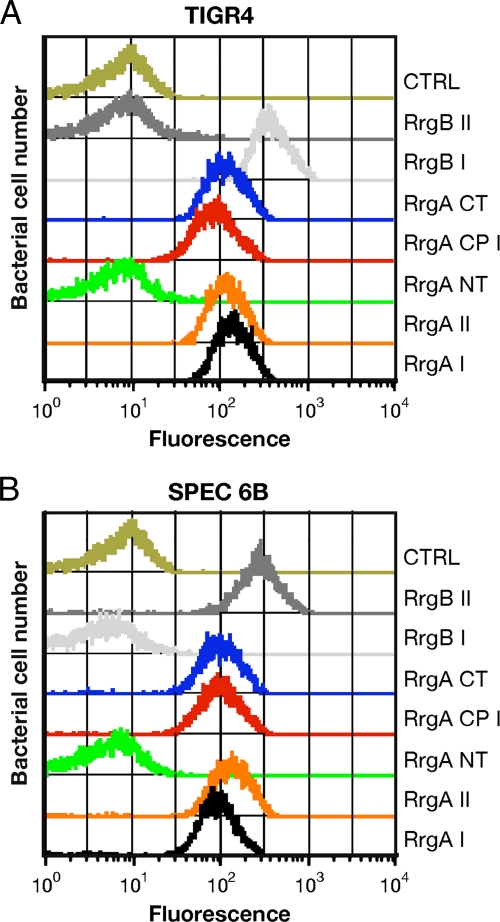
Antisera raised against FL RrgA clades I and II were able to recognize at comparable levels whole bacteria of two S. pneumoniae strains expressing RrgA clade I and clade II, respectively, by FACS (Fig. 5), thus indicating that the two RrgA variants are cross-reactive in vitro. Similarly, antisera against each single recombinant NT, CT, and CP protein fragment were able to recognize FL recombinant proteins and fragments thereof, as well as the typical pilus ladder of both RrgA variants, by WB (see Fig. S2 and S3 in the supplemental material). In contrast, FACS analysis performed on the TIGR4 and SPEC-6B strains revealed that while antisera against the CP-1, CP-2, and CT portions recognized the native pili of both strains, sera raised against the NT fragment were always FACS negative (Fig. 5), indicating that this region was not accessible once RrgA was incorporated into the native pilus fiber. Interestingly, a similar result was obtained when the antisera were tested against an RrgB-defective TIGR4 mutant, which is no longer able to assemble a pilus fiber but still exposes a functional RrgA molecule on its surface (reference 41 and data not shown).
FIG. 5.
The RrgA NT fragment is not exposed on the S. pneumoniae pilus. Two S. pneumoniae clinical isolates, TIGR4 (A) and SPEC6B (B), expressing pilus 1 were stained with anti-RrgA antibodies and then analyzed by flow cytometry (FACSCalibur). Bacteria were labeled with mouse or rabbit polyclonal antibodies (1:400) raised against FL RrgA (clades I and II); FL RrgB (clades I and II);and NT, CP (clades I and II), and CT fragments. CTRL, sera serving as negative controls. Anti-mouse and -rabbit IgG antibodies were FITC labeled (1:100 dilution).
Polyclonal antibodies against FL RrgA are cross-protective in vivo.
RrgA was previously shown to elicit protection in active-immunization studies against homologous TIGR4 challenge. Similarly, polyclonal antisera raised against the full-length RrgA clade I were protective in passive-immunization studies, showing that antibodies were functional in vivo (18).
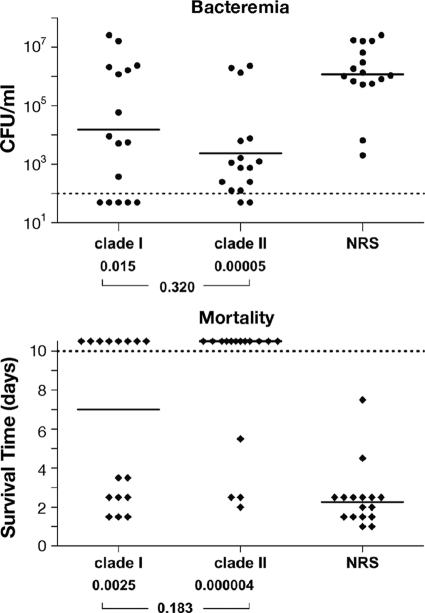
To investigate whether protection was clade specific, we tested whether rabbit polyclonal antisera raised against full-length RrgA clades I and II were cross-protective in passive-immunization experiments (Fig. 6). Mice that received RrgA clade I rabbit antiserum were significantly protected against subsequent challenge with TIGR4, both in terms of bacteremia, with a significant reduction of CFU/ml compared with controls that received normal rabbit serum (NRS) (P = 0.015), and in terms of the mortality course (median survival was 5.5 days longer than that of the control group; P = 0.0025). At the end of the observation period, the survival rate for this group was 50% (P = 0.001). Protection was also achieved in mice upon administration of RrgA clade II rabbit antiserum prior to TIGR4 challenge, with significant reduction of bacteremia (P < 0.0001), increase of median survival by 8 days compared to that of the control group (P < 0.0001), and a survival rate of 75% (P < 0.0001), indicating that immunization with RrgA clade II elicits antibodies that are cross-protective against RrgA clade I. No significant differences were observed between the two immunized groups for all the parameters examined: bacteremia (P = 0.320), median survival (P = 0.183), and survival rate (P = 0.273). This result suggests that protective epitopes are composed of regions that are conserved in the two protein variants.
FIG. 6.
RrgA adhesin is cross-protective in passive-immunization experiments in mice. Passive transfer of rabbit antisera (100 μl) raised against polyclonal anti-RrgA (clade I or II, as indicated) was performed. Mice were then challenged intraperitoneally with TIGR4 (clade I). NRS was used as a negative control. (A) The solid circles represent values (CFU/ml) for single animals, the horizontal bars represent the geometric mean for each group, and the dashed line indicates the detection limit (i.e., no CFU were detected in samples positioned below the dashed line). (B) The solid diamonds represent survival days for single animals, the horizontal bars represent the median survival time for each group, and the dashed line indicates the endpoint of observation (i.e., animals whose survival time was above the dashed line were alive at the 10th day postchallenge). P values are indicated at the bottom of each panel, both for each immunized group compared with the control (upper values) and for immunized groups compared to each other (lower value). The results shown come from the combination of two independent experiments performed under the same conditions.
DISCUSSION
In recent years, many Gram-positive bacteria were discovered to have pili on their surfaces. In S. pneumoniae, only a number of strains were shown to express pilus 1, known to be involved in adherence to lung epithelial cells in vitro and in colonization in vivo (6, 41). Additionally, the subunits that constitute the pilus were demonstrated to elicit protection in a murine model of infection (18). RrgA, the major pilus 1 adhesin, has a direct role in binding to epithelial cells and to purified ECM components (21, 41). In PI-1-positive pneumococcal strains, RrgA exists as two distinct variants, named clade I and clade II.
Recently, the high-resolution crystal structure of the recombinant form of RrgA clade I was published (23), describing a flexible molecule organized into four nested structural domains (D1 to D4). In this report, we investigated the structural variability among the two genetic variants of RrgA and how this variability impacts the global folding of the two RrgA variants, their functional roles in binding epithelial cells and ECM components in vitro, and their protective role in vivo.
Analysis of the primary sequence diversity within the available 3D structure revealed that most of the variability found within RrgA is restricted to the distal D3 domain, which contains the motifs predicted to be implicated in adhesion. Notably, despite the significant level of diversity found within D3, the predicted binding domains are well conserved between the two RrgA variants. In agreement with this observation, both full-length RrgA proteins were able to adhere to epithelial and endothelial cells, as well as to purified ECM components. On the other hand, regardless of the conditions employed for testing, none of the recombinant portions of RrgA (NT, CT, and CP) were able to adhere, indicating that the global folding of the FL molecule, as well as the interplay of the D1 to D4 domains, is crucial to retain its functionality. Furthermore, a site-specific mutation in the aspartic acid residue of the RGD tripeptide in the D3 domain significantly reduced protein binding in the in vitro assays. Interestingly, the RGD tripeptide of the published RrgA structural model obtained from X-ray data is internal and not exposed on the surface, as would be expected for a functionally active domain. Since this point mutation does not change the secondary-structure content, as verified by CD analysis, this finding could be explained in two ways. First, the point mutation could induce local destabilization, altering the exposure of residues involved in adhesion; second, the RGD motif is directly involved in RrgA binding and becomes accessible only during the interaction between RrgA and its ligands upon a conformational change, similar to that reported for thrombin (44).
Regardless of local variability, the two protein variants could share similar overall folding, as suggested by comparable secondary-structure contents (CD analysis) and by the presence of conserved intramolecular isopeptide bonds. Moreover, the two variants behaved similarly when exposed to proteolytic cleavage; in fact, while the variable distal D3 domain was protease sensitive in both RrgA clades I and II, the stalk of the molecule was highly resistant to digestion and to thermal denaturation, probably due to the presence of the two intramolecular isopeptide bonds (25).
On the basis of the results presented here, the two RrgA variants are likely to display similar folding and adhesive properties; however, the conformation of the molecule once incorporated into the native pilus can only be speculative. Evidence for a conformational change is provided by the observation that antibodies directed against the NT part of the protein (Fig. 3C), are not able to bind the NT fragment of the native RrgA (in both clade I and clade II strains) in S. pneumoniae. Notably, the same antibodies recognize the recombinant proteins and the typical pilus ladder in bacterial cell extracts. Under the same experimental conditions, the CP and CT portions (the latter containing the LPXTG-like sequence anchoring RrgA to the backbone) are surface exposed and accessible to antibodies. A similar result was observed both in wild-type isolates and in RrgB-defective mutants expressing a functional RrgA but unable to polymerize a pilus fiber. This suggests that the conformation of native RrgA, independent of its association with the pilus, may differ from that of the recombinant crystallized form.
Interestingly, a report published by Muzzi et al. suggested that RrgA variability could be an example of positive selection driven by host immune pressure (40). However, the cross-protection observed between the two clades in passive-immunization experiments indicates that the functional antibodies directed against conserved epitopes are sufficient to warrant protection; hence, the model proposed is not supported by these data.
Finally, we hypothesize that the mechanism of protection afforded by RrgA could be 2-fold: on one hand, anti-RrgA antibodies could have an opsonizing effect on S. pneumoniae and therefore prevent disease; on the other hand, they could inhibit initial binding of RrgA to epithelial cells and thus play a role in protection from pneumococcal colonization.
In conclusion, the two RrgA variants display comparable folding and similar adhesive properties, despite the sequence variability in the surface-exposed D3 “head” domain. Additionally, in animal models of infection, antibodies to both variants provided cross-protection, thus supporting the potential of RrgA as a component of an effective protein-based vaccine against S. pneumoniae.
Supplementary Material
Editor: A. Camilli
Footnotes
Published ahead of print on 7 September 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Abbot, E. L., W. D. Smith, G. P. Siou, C. Chiriboga, R. J. Smith, J. A. Wilson, B. H. Hirst, and M. A. Kehoe. 2007. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell Microbiol. 9:1822-1833. [DOI] [PubMed] [Google Scholar]
- 2.Aguiar, S. I., I. Serrano, F. R. Pinto, J. Melo-Cristino, and M. Ramirez. 2008. The presence of the pilus locus is a clonal property among pneumococcal invasive isolates. BMC Microbiol. 8:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderton, J. M., G. Rajam, S. Romero-Steiner, S. Summer, A. P. Kowalczyk, G. M. Carlone, J. S. Sampson, and E. W. Ades. 2007. E-cadherin is a receptor for the common protein pneumococcal surface adhesin A (PsaA) of Streptococcus pneumoniae. Microb. Pathog. 42:225-236. [DOI] [PubMed] [Google Scholar]
- 4.Auranen, K., J. Mehtala, A. Tanskanen, and S. Kaltoft. 2010. Between-strain competition in acquisition and clearance of pneumococcal carriage: epidemiologic evidence from a longitudinal study of day-care children. Am. J. Epidemiol. 171:169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagnoli, F., M. Moschioni, C. Donati, V. Dimitrovska, I. Ferlenghi, C. Facciotti, A. Muzzi, F. Giusti, C. Emolo, A. Sinisi, M. Hilleringmann, W. Pansegrau, S. Censini, R. Rappuoli, A. Covacci, V. Masignani, and M. A. Barocchi. 2008. A second pilus type in Streptococcus pneumoniae is prevalent in emerging serotypes and mediates adhesion to host cells. J. Bacteriol. 190:5480-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barocchi, M. A., J. Ries, X. Zogaj, C. Hemsley, B. Albiger, A. Kanth, S. Dahlberg, J. Fernebro, M. Moschioni, V. Masignani, K. Hultenby, A. R. Taddei, K. Beiter, F. Wartha, A. von Euler, A. Covacci, D. W. Holden, S. Normark, R. Rappuoli, and B. Henriques-Normark. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 103:2857-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basset, A., K. Trzcinski, C. Hermos, K. L. O'Brien, R. Reid, M. Santosham, A. J. McAdam, M. Lipsitch, and R. Malley. 2007. Association of the pneumococcal pilus with certain capsular serotypes but not with increased virulence. J. Clin. Microbiol. 45:1684-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Böhm, G., R. Muhr, and R. Jaenicke. 1992. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 5:191-195. [DOI] [PubMed] [Google Scholar]
- 9.Budzik, J. M., C. B. Poor, K. F. Faull, J. P. Whitelegge, C. He, and O. Schneewind. 2009. Intramolecular amide bonds stabilize pili on the surface of bacilli. Proc. Natl. Acad. Sci. U. S. A. 106:19992-19997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dagan, R. 2004. The potential effect of widespread use of pneumococcal conjugate vaccines on the practice of pediatric otolaryngology: the case of acute otitis media. Curr. Opin. Otolaryngol. Head Neck Surg. 12:488-494. [DOI] [PubMed] [Google Scholar]
- 11.Dagan, R. 2009. Serotype replacement in perspective. Vaccine 27(Suppl. 3):C22-C24. [DOI] [PubMed] [Google Scholar]
- 12.Dagan, R., N. Givon-Lavi, O. Zamir, M. Sikuler-Cohen, L. Guy, J. Janco, P. Yagupsky, and D. Fraser. 2002. Reduction of nasopharyngeal carriage of Streptococcus pneumoniae after administration of a 9-valent pneumococcal conjugate vaccine to toddlers attending day care centers. J. Infect. Dis. 185:927-936. [DOI] [PubMed] [Google Scholar]
- 13.Dramsi, S., E. Caliot, I. Bonne, S. Guadagnini, M. C. Prevost, M. Kojadinovic, L. Lalioui, C. Poyart, and P. Trieu-Cuot. 2006. Assembly and role of pili in group B streptococci. Mol. Microbiol. 60:1401-1413. [DOI] [PubMed] [Google Scholar]
- 14.ElMortaji, L., R. Terrasse, A. Dessen, T. Vernet, and A. M. Di Guilmi. 2010. Stability and assembly of pilus subunits of Streptococcus pneumoniae. J. Biol. Chem. 285:12405-12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emsley, J., M. Cruz, R. Handin, and R. Liddington. 1998. Crystal structure of the von Willebrand factor A1 domain and implications for the binding of platelet glycoprotein Ib. J. Biol. Chem. 273:10396-10401. [DOI] [PubMed] [Google Scholar]
- 16.Emsley, J., S. L. King, J. M. Bergelson, and R. C. Liddington. 1997. Crystal structure of the I domain from integrin alpha2beta1. J. Biol. Chem. 272:28512-28517. [DOI] [PubMed] [Google Scholar]
- 17.Fletcher, M. A., and B. Fritzell. 2007. Brief review of the clinical effectiveness of PREVENAR against otitis media. Vaccine 25:2507-2512. [DOI] [PubMed] [Google Scholar]
- 18.Gianfaldoni, C., S. Censini, M. Hilleringmann, M. Moschioni, C. Facciotti, W. Pansegrau, V. Masignani, A. Covacci, R. Rappuoli, M. A. Barocchi, and P. Ruggiero. 2007. Streptococcus pneumoniae pilus subunits protect mice against lethal challenge. Infect. Immun. 75:1059-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Givon-Lavi, N., D. Fraser, N. Porat, and R. Dagan. 2002. Spread of Streptococcus pneumoniae and antibiotic-resistant S. pneumoniae from day-care center attendees to their younger siblings. J. Infect. Dis. 186:1608-1614. [DOI] [PubMed] [Google Scholar]
- 20.Hanage, W. P. 2008. Serotype-specific problems associated with pneumococcal conjugate vaccination. Future Microbiol. 3:23-30. [DOI] [PubMed] [Google Scholar]
- 21.Hilleringmann, M., F. Giusti, B. C. Baudner, V. Masignani, A. Covacci, R. Rappuoli, M. A. Barocchi, and I. Ferlenghi. 2008. Pneumococcal pili are composed of protofilaments exposing adhesive clusters of Rrg A. PLoS Pathog. 4:e1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hilleringmann, M., P. Ringler, S. A. Muller, G. De Angelis, R. Rappuoli, I. Ferlenghi, and A. Engel. 2009. Molecular architecture of Streptococcus pneumoniae TIGR4 pili. EMBO J. 28:3921-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, S. S., V. L. Hinrichsen, A. E. Stevenson, S. L. Rifas-Shiman, K. Kleinman, S. I. Pelton, M. Lipsitch, W. P. Hanage, G. M. Lee, and J. A. Finkelstein. 2009. Continued impact of pneumococcal conjugate vaccine on carriage in young children. Pediatrics 124:e1-e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hynes, R. O., E. E. Marcantonio, M. A. Stepp, L. A. Urry, and G. H. Yee. 1989. Integrin heterodimer and receptor complexity in avian and mammalian cells. J. Cell Biol. 109:409-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imai, S., Y. Ito, T. Ishida, T. Hirai, I. Ito, K. Maekawa, S. Takakura, Y. Iinuma, S. Ichiyama, and M. Mishima. 2009. High prevalence of multidrug-resistant pneumococcal molecular epidemiology network clones among Streptococcus pneumoniae isolates from adult patients with community-acquired pneumonia in Japan. Clin. Microbiol. Infect. 15:1039-1045. [DOI] [PubMed] [Google Scholar]
- 26.Izoré, T., C. Contreras-Martel, L. ElMortaji, C. Manzano, R. Terrasse, T. Vernet, A. M. Di Guilmi, and A. Dessen. 2010. Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure 18:106-115. [DOI] [PubMed] [Google Scholar]
- 27.Kang, H. J., and E. N. Baker. 2009. Intramolecular isopeptide bonds give thermodynamic and proteolytic stability to the major pilin protein of Streptococcus pyogenes. J. Biol. Chem. 284:20729-20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang, H. J., N. G. Paterson, A. H. Gaspar, H. Ton-That, and E. N. Baker. 2009. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc. Natl. Acad. Sci. U. S. A. 106:16967-16971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Käyhty, H., K. Auranen, H. Nohynek, R. Dagan, and H. Makela. 2006. Nasopharyngeal colonization: a target for pneumococcal vaccination. Expert Rev. Vaccines 5:651-667. [DOI] [PubMed] [Google Scholar]
- 30.Kim, K. S. 2010. Acute bacterial meningitis in infants and children. Lancet Infect. Dis. 10:32-42. [DOI] [PubMed] [Google Scholar]
- 31.Klugman, K. P. 2009. The significance of serotype replacement for pneumococcal disease and antibiotic resistance. Adv. Exp. Med. Biol. 634:121-128. [DOI] [PubMed] [Google Scholar]
- 32.Lauer, P., C. D. Rinaudo, M. Soriani, I. Margarit, D. Maione, R. Rosini, A. R. Taddei, M. Mora, R. Rappuoli, G. Grandi, and J. L. Telford. 2005. Genome analysis reveals pili in Group B Streptococcus. Science 309:105. [DOI] [PubMed] [Google Scholar]
- 33.Lipsitch, M., K. O'Neill, D. Cordy, B. Bugalter, K. Trzcinski, C. M. Thompson, R. Goldstein, S. Pelton, H. Huot, V. Bouchet, R. Reid, M. Santosham, and K. L. O'Brien. 2007. Strain characteristics of Streptococcus pneumoniae carriage and invasive disease isolates during a cluster-randomized clinical trial of the 7-valent pneumococcal conjugate vaccine. J. Infect. Dis. 196:1221-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucero, M. G., V. E. Dulalia, L. T. Nillos, G. Williams, R. A. Parreno, H. Nohynek, I. D. Riley, and H. Makela. 2009. Pneumococcal conjugate vaccines for preventing vaccine-type invasive pneumococcal disease and X-ray defined pneumonia in children less than two years of age. Cochrane Database Syst. Rev. 7:CD004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manetti, A. G., C. Zingaretti, F. Falugi, S. Capo, M. Bombaci, F. Bagnoli, G. Gambellini, G. Bensi, M. Mora, A. M. Edwards, J. M. Musser, E. A. Graviss, J. L. Telford, G. Grandi, and I. Margarit. 2007. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol. Microbiol. 64:968-983. [DOI] [PubMed] [Google Scholar]
- 36.Manzano, C., T. Izore, V. Job, A. M. Di Guilmi, and A. Dessen. 2009. Sortase activity is controlled by a flexible lid in the pilus biogenesis mechanism of gram-positive pathogens. Biochemistry 48:10549-10557. [DOI] [PubMed] [Google Scholar]
- 37.Melegaro, A., Y. H. Choi, R. George, W. J. Edmunds, E. Miller, and N. J. Gay. 2010. Dynamic models of pneumococcal carriage and the impact of the Heptavalent Pneumococcal Conjugate Vaccine on invasive pneumococcal disease. BMC Infect. Dis. 10:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moschioni, M., G. De Angelis, S. Melchiorre, V. Masignani, E. Leibovitz, M. A. Barocchi, and R. Dagan. 2009. Prevalence of pilus encoding islets among acute otitis media Streptococcus pneumoniae isolates from Israel. Clin. Microbiol. Infect. doi: 10.1111/j.1469-0691.2009.03105. [DOI] [PubMed]
- 39.Moschioni, M., C. Donati, A. Muzzi, V. Masignani, S. Censini, W. P. Hanage, C. J. Bishop, J. N. Reis, S. Normark, B. Henriques-Normark, A. Covacci, R. Rappuoli, and M. A. Barocchi. 2008. Streptococcus pneumoniae contains 3 rlrA pilus variants that are clonally related. J. Infect. Dis. 197:888-896. [DOI] [PubMed] [Google Scholar]
- 40.Muzzi, A., M. Moschioni, A. Covacci, R. Rappuoli, and C. Donati. 2008. Pilus operon evolution in Streptococcus pneumoniae is driven by positive selection and recombination. PLoS One 3:e3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson, A. L., J. Ries, F. Bagnoli, S. Dahlberg, S. Falker, S. Rounioja, J. Tschop, E. Morfeldt, I. Ferlenghi, M. Hilleringmann, D. W. Holden, R. Rappuoli, S. Normark, M. A. Barocchi, and B. Henriques-Normark. 2007. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol. Microbiol. 66:329-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noske, N., U. Kammerer, M. Rohde, and S. Hammerschmidt. 2009. Pneumococcal interaction with human dendritic cells: phagocytosis, survival, and induced adaptive immune response are manipulated by PavA. J. Immunol. 183:1952-1963. [DOI] [PubMed] [Google Scholar]
- 43.O'Brien, K. L., L. J. Wolfson, J. P. Watt, E. Henkle, M. Oria-Knoll, N. McCall, E. Lee, K. Mulholland, O. S. Levine, and T. Cherian. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893-902. [DOI] [PubMed] [Google Scholar]
- 44.Papaconstantinou, M. E., C. J. Carrell, A. O. Pineda, K. M. Bobofchak, F. S. Mathews, C. S. Flordellis, M. E. Maragoudakis, N. E. Tsopanoglou, and E. Di Cera. 2005. Thrombin functions through its RGD sequence in a non-canonical conformation. J. Biol. Chem. 280:29393-29396. [DOI] [PubMed] [Google Scholar]
- 45.Papasergi, S., M. Garibaldi, G. Tuscano, G. Signorino, S. Ricci, S. Peppoloni, I. Pernice, P. C. Lo, G. Teti, F. Felici, and C. Beninati. 2010. Plasminogen- and fibronectin-binding protein B is involved in the adherence of Streptococcus pneumoniae to human epithelial cells. J. Biol. Chem. 285:7517-7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pelton, S. I., and E. Leibovitz. 2009. Recent advances in otitis media. Pediatr. Infect. Dis. J. 28:S133-S137. [DOI] [PubMed] [Google Scholar]
- 47.Pezzicoli, A., I. Santi, P. Lauer, R. Rosini, D. Rinaudo, G. Grandi, J. L. Telford, and M. Soriani. 2008. Pilus backbone contributes to group B Streptococcus paracellular translocation through epithelial cells. J. Infect. Dis. 198:890-898. [DOI] [PubMed] [Google Scholar]
- 48.Rinta-Kokko, H., R. Dagan, N. Givon-Lavi, and K. Auranen. 2009. Estimation of vaccine efficacy against acquisition of pneumococcal carriage. Vaccine 27:3831-3837. [DOI] [PubMed] [Google Scholar]
- 49.Rose, L., P. Shivshankar, E. Hinojosa, A. Rodriguez, C. J. Sanchez, and C. J. Orihuela. 2008. Antibodies against PsrP, a novel Streptococcus pneumoniae adhesin, block adhesion and protect mice against pneumococcal challenge. J. Infect. Dis. 198:375-383. [DOI] [PubMed] [Google Scholar]
- 50.Ryan, M. W., and P. J. Antonelli. 2000. Pneumococcal antibiotic resistance and rates of meningitis in children. Laryngoscope 110:961-964. [DOI] [PubMed] [Google Scholar]
- 51.Telford, J. L., M. A. Barocchi, I. Margarit, R. Rappuoli, and G. Grandi. 2006. Pili in gram-positive pathogens. Nat. Rev. Microbiol. 4:509-519. [DOI] [PubMed] [Google Scholar]
- 52.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50:1429-1438. [DOI] [PubMed] [Google Scholar]
- 53.Ton-That, H., and O. Schneewind. 2004. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 12:228-234. [DOI] [PubMed] [Google Scholar]
- 54.Uchiyama, S., A. F. Carlin, A. Khosravi, S. Weiman, A. Banerjee, D. Quach, G. Hightower, T. J. Mitchell, K. S. Doran, and V. Nizet. 2009. The surface-anchored NanA protein promotes pneumococcal brain endothelial cell invasion. J. Exp. Med. 206:1845-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Poll, T., and S. M. Opal. 2009. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet 374:1543-1556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.