Abstract
Cryptococcus neoformans is an opportunistic fungal pathogen that causes disease in individuals with suppressed cell-mediated immunity. Recent studies in our laboratory have shown that increases in pulmonary Th1-type and interleukin-17A (IL-17A) cytokine production, classical macrophage activation, and sterilizing immunity are elicited in response to infection with a gamma interferon (IFN-γ)-producing C. neoformans strain, H99γ. IL-17A-treated macrophages, compared to IL-4-treated macrophages, have been demonstrated to exhibit increased microbicidal activity in vitro, a characteristic consistent with classical macrophage activation. The purpose of these studies is to determine the role of IL-17A in the induction of classically activated macrophages following infection with C. neoformans. Immunohistochemistry and real-time PCR were used to characterize the macrophage activation phenotype in lung tissues of mice treated with isotype control or anti-IL-17A antibodies and given an experimental pulmonary infection with C. neoformans strain H99γ. The pulmonary fungal burden was resolved, albeit more slowly, in mice depleted of IL-17A compared to the fungal burden in isotype control-treated mice. Nonetheless, no difference in classical macrophage activation was observed in IL-17A-depleted mice. Similarly, classical macrophage activation was evident in mice deficient in IL-17A or the IL-17 receptor A, which mediates IL-17A signaling, following pulmonary infection with wild-type C. neoformans strain H99 or H99γ. These studies suggest that IL-17A may play a role in the early immune response to C. neoformans but is not required for classical macrophage activation in mice experimentally infected with C. neoformans.
Cryptococcus neoformans is an opportunistic fungal pathogen and a frequent cause of life-threatening infection in individuals with suppressed cell-mediated immunity (CMI) (25). C. neoformans is the most common mycological agent of morbidity and mortality in AIDS patients (31). Infection is initiated following the inhalation of desiccated basidiospores or yeast into lung alveoli, resulting in asymptomatic disease or mild bronchopneumonia in immune-competent individuals (25). However, bronchial infection is severe in immunocompromised patients and often leads to dissemination, resulting in meningoencephalitis. As inhalation is the principal route of entry for C. neoformans, clearance from the lungs is largely dependent upon the ability of resident alveolar macrophages to degrade the yeast cells, thereby preventing dissemination.
The resolution of pulmonary C. neoformans infection in experimental murine models is associated with the induction of Th1-type cytokine responses characterized by the production of interleukin-2 (IL-2), IL-12, tumor necrosis factor alpha (TNF-α), and gamma interferon (IFN-γ) (3, 4, 6, 11, 16, 22, 23, 26, 28). These cytokines, in turn, induce lymphocyte and phagocyte recruitment and are associated with the induction of classically activated macrophages (caMac) (13). Classically activated macrophages are induced in environments with high levels of IFN-γ cytokine and are microbicidal in nature, whereas alternatively activated macrophages (aaMac) are induced by Th2-type cytokines (specifically, IL-4 and IL-13) and mediate wound healing (4, 27-29, 41). Previous studies demonstrated that experimental pulmonary infection with a C. neoformans strain engineered to express IFN-γ, strain H99γ, results in the generation of a polarized Th1-type cytokine response in mice (36). Mice given a pulmonary infection with C. neoformans strain H99γ resolve the acute infection and are completely protected from a secondary infection with wild-type (WT) cryptococci (36, 38, 40). Recently, we demonstrated that clearance of infection with C. neoformans strain H99γ is associated with the induction of inducible nitric oxide synthase (iNOS) expression, caMac, and pulmonary Th1-type cytokine production (15). Conversely, we observed increases in the expression of hallmark markers of aaMac (arginase, CD206, FIZZ1, and Ym1), uncontrolled microbial growth, and Th2-type cytokine secretion in the lungs of mice during infection with wild-type cryptococci (15). In addition to a robust Th1-type cytokine response, infection with C. neoformans strain H99γ is also associated with a significant increase in IL-17A cytokine expression in the lungs (15, 38).
IL-17A is a proinflammatory cytokine recently demonstrated to play a role in protective immune responses to several fungal pathogens (7, 20, 28, 41). caMac are known to produce cytokines that are important for Th17 differentiation, such as IL-6 and IL-23 (24, 27), and it has been suggested that IL-17A induces macrophage production of proinflammatory cytokines (18, 27). The role for IL-17A in the induction of the classical macrophage phenotype has not been fully elucidated, but one report has demonstrated that intracellular proliferation rates of C. neoformans within the alveolar macrophage-like cell line J774 and human primary monocyte-derived macrophages were lower following pretreatment with IL-17A than in macrophages stimulated with IL-4 or IL-13 (35). The present studies seek to investigate the necessity for IL-17A in the induction of classical macrophage activation in mice given an experimental pulmonary C. neoformans infection.
MATERIALS AND METHODS
Mice.
Female BALB/c (H-2d) and C57BL/6 (H-2b) mice (National Cancer Institute/Charles River Laboratories), IFN-γ−/− mice (The Jackson Laboratory, Bar Harbor, ME), IL-17RA−/− mice on a BALB/c background (a kind gift of Jay K. Kolls, Louisiana State University Health Sciences Center, New Orleans, LA), IL-17A−/− mice on a C57BL/6 background (a kind gift of Jay K. Kolls), and IL-17RA−/− mice on a C57BL/6 background (Amgen Inc., Thousand Oaks, CA), all with an average weight of 20 to 25 g, were used throughout these studies. Mice were housed at the University of Texas at San Antonio Small Animal Laboratory vivarium and handled according to guidelines approved by the Institutional Animal Care and Use Committee.
Strains and media.
C. neoformans strains H99 (serotype A, Mat α) and H99γ (an interferon-γ-producing C. neoformans strain derived from H99 [36]) were recovered from 15% glycerol stocks stored at −80°C prior to use in the experiments described herein. C. neoformans strains were maintained on yeast extract-peptone-dextrose (YPD) medium (Becton, Dickinson and Company). Yeast cells were grown for 14 to 16 h at 30°C with shaking in YPD broth (Becton, Dickinson and Company), collected by centrifugation, and washed three times with sterile phosphate-buffered saline (PBS), and viable yeast was quantified using trypan blue dye exclusion in a hemacytometer.
Murine model.
Pulmonary C. neoformans infections were initiated by nasal inhalation as previously described (8, 9). Briefly, mice were anesthetized with 2% isoflurane by using a rodent anesthesia device (Eagle Eye Anesthesia, Jacksonville, FL) and then given a yeast inoculum of 1 × 104 CFU of C. neoformans strain H99 or H99γ in 50 μl of sterile PBS pipetted directly into the nares. The inocula used for nasal inhalation were verified by quantitative culture on YPD agar. Mice were treated 5 h postinoculation with 100 μg of rat anti-mouse neutralizing antibodies against IL-17A (R&D Systems, Minneapolis, MN) or isotype-matched rat IgG2a (eBioscience, San Diego, CA) by intranasal inhalation as described above. Animals were treated with either isotype control or IL-17A antibody every 4 days thereafter. The mice were fed ad libitum and monitored by inspection twice daily. Mice were euthanized on predetermined days following inoculation, and lung tissues were excised using an aseptic technique, homogenized in 1 ml of sterile PBS, and cultured by 1:10 dilutions on YPD agar supplemented with chloramphenicol (Mediatech, Inc., Herndon, VA). CFU were enumerated following incubation at 30°C for 48 h.
Pulmonary leukocyte isolation.
Lungs were excised on days 7 and 14 postinoculation and digested enzymatically at 37°C for 30 min in 10 ml of digestion buffer (RPMI 1640 and 1 mg/ml of collagenase type IV [Sigma Chemical Co., St. Louis, MO]) with intermittent (every 10 min) stomacher homogenizations. The enzymatically digested tissues were then successively filtered through sterile nylon filters of various pore sizes (70 and 40 μm) (BD Biosciences, San Diego, CA) and washed with sterile Hanks’ balanced salt solution (HBSS) to enrich for leukocytes. Erythrocytes were lysed by incubation in NH4Cl buffer (0.859% NH4Cl, 0.1% KHCO3, 0.0372% Na2EDTA [pH 7.4]; Sigma Chemical Co.) for 3 min on ice followed by a 10-fold excess of PBS. The resulting leukocyte population was then collected by centrifugation (800 × g) for 5 min, washed twice with sterile PBS, resuspended in sterile PBS containing 2% heat-inactivated fetal bovine serum (fluorescence-activated cell sorter [FACS] buffer), and enumerated in a hemacytometer using trypan blue dye exclusion. For gene expression analysis and macrophage microbicidal activity assays, the leukocyte population was enriched for macrophages by positive selection using magnetic beads labeled with CD11b antibody according to the manufacturer's recommendations (Miltenyi Biotec, Auburn, CA).
Antibodies.
For flow cytometry experiments, rat anti-mouse CD16/CD32 (Fc Block, BD Biosciences), rat anti-mouse CD45 (BD Biosciences) conjugated to phycoerythrin-Cy7 (PE-Cy7), and rat anti-mouse F4/80 conjugated to allophycocyanin (APC) (Caltag Laboratories, Burlingame, CA) were used. For immunohistochemistry experiments, rabbit anti-mouse Arg1 (Santa Cruz Biotechnology Inc., Santa Cruz, CA), rat anti-mouse CD206 (AbD Serotec, Raleigh, NC), rat anti-mouse Ym1 (R&D Systems, Minneapolis, MN), rat anti-mouse F4/80 (AbD Serotec), and rabbit anti-mouse iNOS (Axxora, LLC, San Diego, CA) were used. Primary antibodies were detected using appropriate Alexa Fluor 488-conjugated goat anti-rat IgG (Invitrogen, Carlsbad, CA) or goat anti-rabbit IgG (Invitrogen) secondary antibodies.
Flow cytometry.
Standard methodology was employed for the direct and indirect immunofluorescence of pulmonary leukocytes. Briefly, in 96-well U-bottom plates, 1 × 106 leukocyte-enriched lung cells were incubated with Fc Block in 50 μl of PBS for 5 min to block nonspecific binding of antibodies to cellular Fc receptors. Subsequently, an optimal concentration of CD45 and F4/80 antibodies was added to allow for dual staining in 50 μl of FACS buffer. Following 30 min of incubation on ice, the cells were washed three times with FACS buffer and then fixed in 200 μl of 2% ultrapure formaldehyde (Polysciences, Inc., Warrington, PA). Cells were incubated with either FACS buffer alone or single fluorochrome-conjugated antibodies to determine positive staining and spillover/compensation calculations, and background fluorescence was determined by the flow cytometer. Samples were analyzed using the software provided with a BD FACSArray flow cytometer (BD Biosciences). Dead cells were excluded on the basis of forward-angle and 90° light scatter. For data analyses, 30,000 events (cells) were evaluated from a predominantly leukocytic population identified by backgating from CD45+-stained cells. The absolute number of leukocytes was quantified by multiplying the total number of cells observed through hemacytometer counting by the percentage of CD45+ cells determined by flow cytometry. The absolute number of macrophage cells was determined by multiplying the percentage of F4/80 and CD45 double-positive cells by the total number of CD45+ cells.
Macrophage killing assay.
Lungs were excised from mice on day 7 postinoculation with wild-type C. neoformans strain H99 and used to prepare a single-cell suspension as described above. Macrophages were positively selected using CD11b antibody and the Miltenyi system according to the manufacturer's instructions (Miltenyi Biotec). Cell viability and quantity were determined using trypan blue exclusion in a hemacytometer. Macrophages were then cultured at a density of 1 × 105 cells per well in RPMI supplemented with 2 mM l-glutamine and 100 μg/ml penicillin-streptomycin in a 96-well tissue culture plate and incubated at 37°C and 5% CO2 for 24 h. Cell culture supernatants were then collected, and an aliquot (50 μl) was used to determine nitric oxide production using Griess reagent (Sigma Chemical Co.) according to the manufacturer's instructions. Light absorbance values were measured at 540 nm using a BioTek Elx 808 absorbance microplate reader with Gen5 version 1.04.5 software (BioTek Instruments, Winooski, VT). Cryptococci were subsequently liberated from within adherent macrophages using 0.1% Triton X-100 in PBS. CFU were enumerated following incubation of 1:10 dilutions at 30°C for 48 h on YPD agar supplemented with chloramphenicol (Mediatech, Inc.).
Real-time PCR.
Total RNA was isolated from purified CD11b+ cells using TRIzol reagent (Invitrogen) and then DNase treated (Invitrogen) to remove possible traces of contaminating DNA according to the manufacturer's instructions. First-strand cDNA was synthesized from 1 μg of total RNA by using the oligo(dT) primer and reagents supplied in the SuperScript III reverse transcriptase (RT) kit (Invitrogen) according to the manufacturer's instructions. The cDNA was used as a template for analysis by real-time PCR by using the TaqMan gene expression assay (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. All real-time PCRs were performed using the 7300 real-time PCR system (Applied Biosystems). For each real-time PCR, a master mix was prepared on ice with TaqMan gene expression assays specific for iNOS, IFN-γ, IL-17A, Arg1, CD206, FIZZ1, Ym1, IL-4, and IL-13 (Applied Biosystems). TaqMan rodent glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Applied Biosystems) was used as an internal control. The thermal cycling parameters contained an initial denaturing cycle of 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Results of the real-time PCR data were derived using the comparative CT method as previously described (1, 5, 34) to detect relative gene expression. The parameter CT is defined as the cycle number at which the amplification plot passes a fixed threshold above the baseline. Each reaction was run in triplicate in separate wells and normalized to a control endogenous gene product, GAPDH. The following formula was used to quantify the fold differential expression of each specific gene in macrophage-enriched populations following pulmonary inoculation with C. neoformans strain H99γ compared to gene expression in wild-type-infected mice: 2−ΔΔCT, where ΔΔCT = (CT of IL-17A-depleted sample − CT GAPDH of IL-17A-depleted sample) − (CT of isotype control sample − CT GAPDH of isotype control sample). CT represents the mean CT value of each sample in triplicate. Therefore, the result represents the fold change in the expression of the gene in question following IL-17A depletion during infection with C. neoformans strain H99γ compared to the gene expression in isotype control-treated mice.
Immunohistochemistry.
Mice were continuously sedated at predetermined time points by using 2% isoflurane as previously stated. Lungs were perfused with sterile PBS by transcardial perfusion through the right ventricle. The pericardium and trachea were exposed by dissection, and an incision was made in the trachea for the insertion of a sterile, flexible cannula attached to a 3-ml syringe. The lungs were then slowly inflated with 0.5 to 1.0 ml of a Tissue-Tek optimal cutting temperature (OCT) compound (Sakura Finetek, Torrance, CA)-2 M sucrose (1:1 [vol/vol]) solution. The lungs were then excised and immediately preserved in cryomolds containing OCT medium on dry ice and stored at −80°C until use. Serial frozen tissue sections were cut at a thickness of 10 μm and processed for immunohistochemical analysis to visualize the leukocyte infiltration. Tissue sections were fixed at −20°C in acetone for 10 min and then were placed in cold (−20°C) 70% ethanol for 5 min and washed in PBS for 3 min. Nonspecific binding was inhibited by blocking for 30 min at room temperature with serum (10% in PBS) from the same species from which the fluorochrome-conjugated antibodies were derived. Tissue sections were incubated overnight at 4°C with primary antibodies diluted in species-specific serum (3% in PBS) at preoptimized concentrations. Subsequently, the sections were washed seven times in Tris-NaCl-Tween 20 (TNT) buffer solution for 3 min each time. Sections were then incubated with secondary antibodies for 30 min at room temperature. Slides were washed seven times in TNT buffer for 3 min each time, one time in PBS containing 1% Triton X to minimize background fluorescence (3 min), and a final time in TNT buffer (3 min). Sections were then mounted with FluorSave reagent (Calbiochem, La Jolla, CA) containing 0.3 μM 4′,6′- diamidino-2-phenylindole (DAPI) dilactate (Molecular Probes, Eugene, OR). Fluorescence was visualized with a Leica digital monitor R (DMR) epifluorescence microscope (Leica Microsystems, Wetzlar, Germany). Images were acquired using a cooled SPOT real-time charge-coupled device camera (Diagnostic Instruments Inc., Sterling Heights, MI).
Cytokine analysis.
Cytokine production in lung tissues was analyzed using the Bio-Plex protein array system (Luminex-based technology) (Bio-Rad Laboratories, Hercules, CA). Briefly, lung tissue was excised and homogenized in ice-cold, sterile PBS (1 ml) and kept on ice. An aliquot (50 μl) was taken to quantify the pulmonary fungal burden, and an antiprotease buffer solution (1 ml) containing PBS, protease inhibitors (inhibiting cysteine, serine, and other metalloproteinases), and 0.05% Triton X-100 was added to the homogenate that was then clarified by centrifugation (800 × g) for 5 min. Previous studies in our laboratory have confirmed no decrease in cytokine levels in the interval prior to the addition of protease inhibitors to homogenates. Pulmonary homogenates were assayed undiluted for cytokine production using the Bio-Plex protein array system (Bio-Rad).
Statistical analysis.
The unpaired Student's t test (two-tailed) using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA) was used to detect statistically significant differences. Statistically significant differences were defined as P values <0.05.
RESULTS
Effect of IL-17A depletion on fungal burden and macrophage recruitment during infection with C. neoformans strain H99γ.
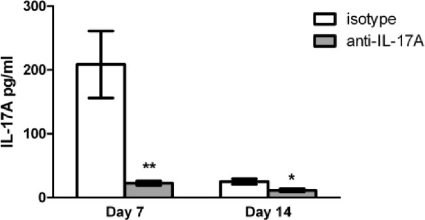
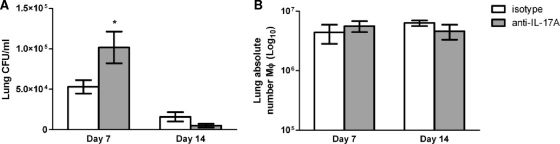
The IL-17 family is comprised of several isoforms, the best-characterized being IL-17A and IL-17F. Previous studies have demonstrated a significant increase of IL-17A levels in lung tissues derived from mice given a pulmonary infection with C. neoformans strain H99γ compared to levels in mice infected with wild-type C. neoformans (15, 38). IL-17A is 10- to 30-fold more potent than IL-17F in stimulating downstream gene transcription (12). To assess the contribution of IL-17 to the control of fungal burden and macrophage recruitment during C. neoformans strain H99γ infection, BALB/c mice were given an intranasal inoculation with C. neoformans strain H99γ followed by treatment with rat anti-mouse IL-17A antibody or rat anti-mouse IgG2a isotype control antibody, and pulmonary fungal burden and macrophage infiltration were quantified on days 7 and 14 postinoculation. We observed average reductions of approximately 89.1% and 53.7% in IL-17A levels in mice that received the anti-IL-17A depletion antibody at days 7 and 14 postinfection, respectively, compared to the levels in isotype control-treated mice (Fig. 1). The lack of a robust percent decrease in IL-17A levels observed on day 14 in mice treated with the anti-IL-17A antibody compared to levels in isotype control-treated mice is likely due to an overall reduction in IL-17A production in the lungs of infected mice at that time point, perhaps due to reduced fungal burden in isotype control-treated mice on day 14 (Fig. 2A). IL-17A production was similar on days 7 and 14 in mice that received the anti-IL-17A neutralization antibody (Fig. 1). Figure 2A demonstrates that the pulmonary fungal burden was significantly increased in IL-17A-depleted mice on day 7 postinoculation compared to the fungal burden in isotype-treated mice. However, no significant difference in fungal burden was observed in IL-17A-depleted mice compared to isotype control-treated mice on day 14 postinfection (Fig. 2A).
FIG. 1.
IL-17A depletion in mice infected with C. neoformans strain H99γ. BALB/c mice received an intranasal inoculation with C. neoformans strain H99γ and were treated with IL-17A antibody or isotype control IgG2a antibody. The lungs from each group of mice were excised at days 7 and 14 postinoculation and assayed for IL-17A cytokine expression. Data represent cumulative results from four experiments utilizing five mice per group. Asterisks indicate where significant decreases in IL-17A levels (**, P < 0.001; *, P < 0.05) were observed in IL-17A-depleted mice compared to the levels in isotype control-treated mice following infection with C. neoformans strain H99γ.
FIG. 2.
Depletion of IL-17A during C. neoformans strain H99γ infection does not impair the resolution of infection or macrophage recruitment. BALB/c mice received an intranasal inoculation with C. neoformans strain H99γ and were treated with IL-17A antibody or isotype control IgG2a antibody. The lungs from each group of mice were excised at days 7 and 14 postinoculation, and fungal burden was quantified (A). Alternatively, pulmonary leukocytes were enzymatically dispersed, and the absolute number of F4/80+/CD45+ macrophages was quantified by flow cytometry (B). Pulmonary fungal burden data represent cumulative results from three experiments using five mice per group per time point. Results are expressed as mean numbers of CFU per milliliter of lung homogenate ± SEM. Flow cytometry data represent cumulative results from three independent experiments using pooled leukocytes from five mice per group per experiment. Results are expressed as the absolute number of F4/80+/CD45+ dual-positive cells. Asterisks indicate where significant increases in the number of F4/80+/CD45+ cells (*, P < 0.05) were observed in IL-17A-depleted mice compared to the number of cells in isotype control-treated mice.
To evaluate macrophage infiltration in response to IL-17A depletion, pulmonary leukocytes were isolated from the lungs of IL-17A-depleted or isotype control-treated mice on days 7 and 14 postinoculation with C. neoformans strain H99γ and analyzed by flow cytometry. Depletion of IL-17A appeared to have no impact on the absolute number of F4/80+/CD45+ cells (macrophages) in mice infected with C. neoformans strain H99γ compared to mice that received isotype control IgG antibody (Fig. 2B). Thus, IL-17A depletion during pulmonary infection with C. neoformans strain H99γ resulted in an initial, but not sustained, delay in C. neoformans clearance that was not associated with a shift in macrophage infiltration into infected lungs.
Effect of IL-17A depletion on macrophage activation phenotype.
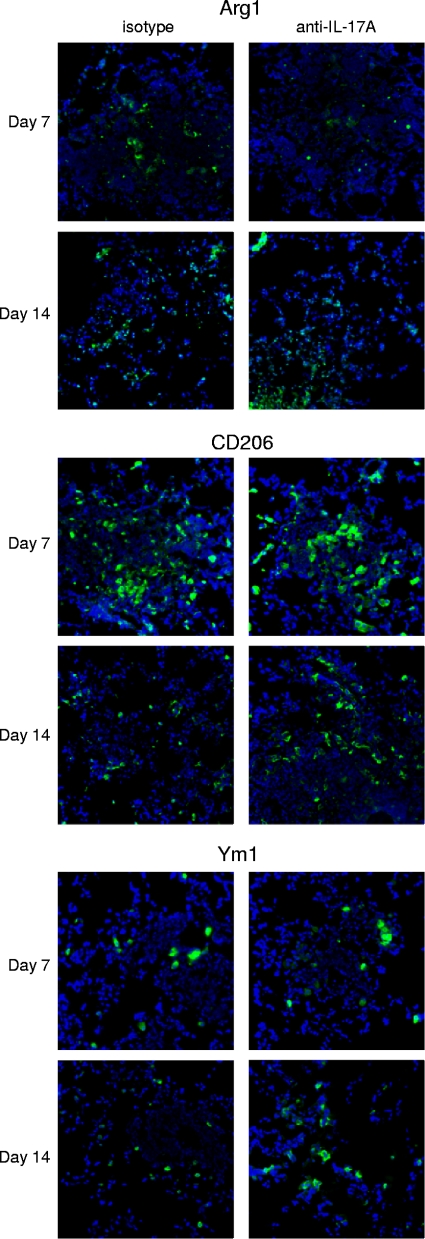
We investigated whether potential differences in macrophage activation phenotype accounted for the delayed response in IL-17A-depleted mice on day 7 postinoculation compared to isotype-treated mice. Lungs were excised on days 7 and 14 postinoculation from mice treated with IL-17A antibody or isotype control IgG2a during infection with C. neoformans strain H99γ and evaluated for hallmark markers of caMac and aaMac by immunohistological examination. Tissue sections were labeled for F4/80+ cells (macrophages) or markers of classical (iNOS) and alternative (Arg1, CD206, and Ym1) macrophage activation. No difference in F4/80 staining was observed in the lungs of mice depleted of IL-17A during infection with C. neoformans strain H99γ compared to that in mice treated with the isotype control antibody at any time point evaluated (Fig. 3), a finding that corroborates previous results using flow cytometry (Fig. 2A). Staining for iNOS, a biomarker for caMac, was no different on day 7 postinoculation in mice depleted of IL-17A during infection with C. neoformans strain H99γ compared to that in isotype control-treated mice. However, increased iNOS expression was observed in IL-17A-depleted mice on day 14 postinfection compared to that in isotype control-treated mice (Fig. 3). Indeed, iNOS expression was undetectable on day 14 postinfection in isotype control-treated mice, a finding consistent with previous results (15). The perpetuation of iNOS expression in IL-17A-depleted mice on day 14 of infection (Fig. 3) may reflect a prolonged macrophage response in compensation for the significant increase in pulmonary fungal burden observed on day 7 postinfection (Fig. 2A). Alternatively, the aaMac hallmark proteins Arg1, CD206, and Ym1 were expressed at similarly low levels in lung tissues derived from IL-17-depleted mice or isotype control-treated mice on days 7 and 14 postinfection (Fig. 4). The modest expression of aaMac markers in conjunction with robust iNOS expression suggests that macrophages responding to C. neoformans strain H99γ infection in both IL-17A-depleted and isotype control-treated mice are classically activated.
FIG. 3.
IL-17A depletion during experimental pulmonary infection with C. neoformans strain H99γ does not change macrophage recruitment and prolongs iNOS expression in the lungs of C. neoformans strain H99γ-infected mice. BALB/c mice received an intranasal inoculation with C. neoformans strain H99γ and were treated with IL-17A antibody or isotype control IgG2a antibody. Lungs were excised on days 7 and 14 postinoculation and immediately frozen in OCT medium. Lungs were cryosectioned, and macrophage infiltration and activation were evaluated using immunofluorescence staining with anti-F4/80 or anti-iNOS antibodies. Nuclei were counterstained with DAPI. Data shown are representative lung sections from two independent experiments (three mice per group). Digital photographs show representative areas of lungs (20× objective).
FIG. 4.
IL-17A depletion during experimental pulmonary infection with C. neoformans strain H99γ does not induce aaMac. BALB/c mice received an intranasal inoculation with C. neoformans strain H99γ and were treated with IL-17A antibody or isotype control IgG2a antibody. Lungs were excised on days 7 and 14 postinoculation and immediately frozen in OCT medium. Lungs were cryosectioned, and alternative macrophage activation was evaluated using immunofluorescence staining with anti-Arg1, anti-CD206, or anti-Ym1 antibodies. Nuclei were counterstained with DAPI. Data shown are representative lung sections from two independent experiments (three mice per group). Digital photographs show representative areas of lungs (20× objective).
Effect of IL-17A depletion on macrophage gene expression in pulmonary macrophage-enriched cell populations.
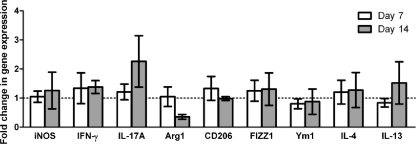
To confirm the macrophage activation phenotype, we quantified relative mRNA expression levels of various genes associated with classical or alternative macrophage activation in pulmonary macrophage-enriched cell populations obtained from C. neoformans strain H99γ-infected lungs of IL-17A-depleted or isotype control-treated mice. Pulmonary leukocytes were isolated from enzymatically dispersed lungs at days 7 and 14 postinoculation with C. neoformans strain H99γ. We subsequently extracted total RNA from CD11b+ cell populations and evaluated macrophage activation using real-time PCR (Fig. 5). We observed no significant difference in transcript levels of iNOS, IFN-γ, and IL-17 (caMac markers) or Arg1, CD206, FIZZ1, Ym1, IL-4, and IL-13 (aaMac markers) on day 7 or 14 postinoculation with C. neoformans strain H99γ in mice depleted of IL-17 compared to the levels in isotype-control treated mice. These observations confirm our immunohistochemistry results, indicating that depletion of IL-17A does not result in the inhibition of classical macrophage activation during pulmonary infection with C. neoformans strain H99γ.
FIG. 5.
Real-time PCR analysis of macrophage-enriched populations show no difference in expression of caMac and aaMac transcripts in mice depleted of IL-17A during infection with C. neoformans strain H99γ compared to expression in isotype control-treated mice. BALB/c mice received an intranasal inoculation with C. neoformans strain H99γ and were treated with IL-17A antibody or isotype control IgG2a antibody. Pulmonary leukocytes were isolated from lung tissues by enzymatic digestion on days 7 and 14 postinoculation, and macrophages were enriched for by positive selection of CD11b+ cells. Real-time PCR analyses of total mRNA from macrophage-enriched populations were conducted for iNOS, IFN-γ, IL-17, Arg1, CD206, FIZZ1, Ym1, IL-4, IL-13, and GAPDH. Bars represent the fold changes in gene expression in macrophages from mice depleted of IL-17A during infection with C. neoformans strain H99γ compared to gene expression in isotype control-treated mice. Data represent cumulative results from three independent experiments utilizing pooled leukocytes of five mice per group per experiment.
Effect of IL-17A depletion on pulmonary cytokine production.
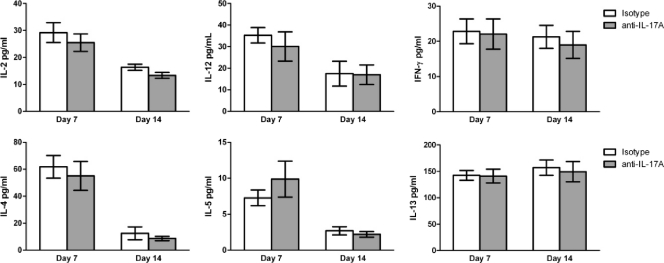
The local cytokine milieu appears critical in determining macrophage activation (13). Previous studies have shown that pulmonary infection with C. neoformans strain H99γ results in the induction of local Th1-type and IL-17A cytokine production in mice (15, 36, 38). Our goal herein was to determine whether depletion of IL-17A during infection with C. neoformans strain H99γ would result in changes in the overall pulmonary cytokine milieu. We therefore evaluated Th1-type (IL-2, IL-12, and IFN-γ) and Th2-type (IL-4, IL-5, and IL-13) cytokine production in total lung homogenates derived from IL-17A-depleted or isotype control-treated mice given an experimental pulmonary infection with C. neoformans strain H99γ on days 7 and 14 postinoculation. We observed no significant differences in lung Th1- or Th2-type cytokine expression in IL-17A-depleted mice compared to that in isotype-control treated mice on day 7 or 14 postinoculation (Fig. 6). These results suggest that depletion of IL-17A does not affect the overall Th1/Th2 balance during infection with C. neoformans strain H99γ.
FIG. 6.
The depletion of IL-17A during pulmonary infection with C. neoformans strain H99γ does not alter the pulmonary cytokine environment. BALB/c mice received an intranasal inoculation with C. neoformans strain H99γ and were treated with IL-17A antibody or isotype control IgG2a antibody. Lung homogenates were prepared from lungs excised on days 7 and 14 postinoculation and assayed for IL-2, IL-4, IL-5, IL-12, IL-13, and IFN-γ cytokine production. Data represent cumulative results from three experiments utilizing five mice per group.
Macrophage activation phenotype in IL-17RA−/− mice.
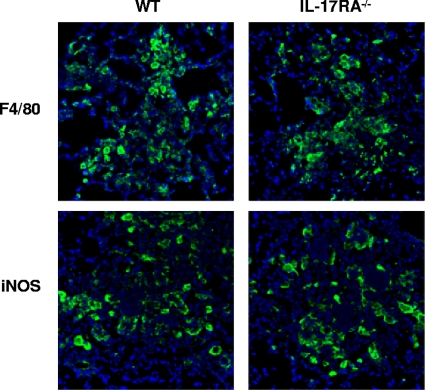
IL-17A and IL-17F signaling occur through IL-17RA. In order to confirm that a lack of IL-17A signaling has a minimal, if any, impact on classical macrophage activation in our model, we evaluated macrophage infiltration and iNOS expression in IL-17RA−/− mice following infection with C. neoformans strain H99γ by using immunohistochemistry. Lungs were excised from IL-17RA−/− and wild-type BALB/c mice on day 7 postinoculation with C. neoformans strain H99γ and evaluated for pulmonary fungal burden, F4/80+ cells (macrophages), or iNOS expression, a hallmark marker for caMac, using immunohistochemistry. The pulmonary fungal burden was significantly increased in IL-17RA−/− mice (2.52 × 105 ± 7.30 × 104, mean ± standard error of the mean [SEM]) compared to that in wild-type mice (6.78 × 104 ± 1.14 × 104) on day 7 postinoculation (P < 0.05 in IL-17RA−/− mice compared to wild-type mice). However, we observed no difference in macrophage recruitment or iNOS expression in IL-17RA−/− mice compared to that in wild-type mice (Fig. 7). These findings corroborate previous fungal burden (Fig. 2A), flow cytometry (Fig. 2B), and immunohistochemistry (Fig. 3 and 4) results demonstrating that IL-17A has a minimal effect on macrophage recruitment and classical macrophage activation during infection with C. neoformans strain H99γ.
FIG. 7.
IL-17RA−/− mice have similar pulmonary F4/80 and iNOS expression during experimental pulmonary infection with C. neoformans strain H99γ compared to expression in wild-type mice. IL-17RA−/− and wild-type BALB/c mice received an intranasal inoculation with C. neoformans strain H99γ. Lungs were excised on day 7 postinoculation and immediately frozen in OCT medium. Lungs were cryosectioned and expression of F4/80 and iNOS was evaluated using immunofluorescence staining with anti-F4/80 or anti-iNOS antibodies. Nuclei were counterstained with DAPI. Data shown are representative lung sections from two independent experiments (three mice per group). Digital photographs show representative areas of lungs (20× objective).
Macrophage activation phenotype in IL-17A−/−, IL-17RA−/−, and wild-type C57BL/6 mice during experimental pulmonary cryptococcosis.
Our results presented herein have thus far utilized a model system in which we have demonstrated classical macrophage activation in response to pulmonary cryptococcosis (15). Although we were careful to use an inoculum of C. neoformans strain H99γ shown not to elicit significant IFN-γ production compared to production in wild-type cryptococcus-infected BALB/c mice (15), the effect of IL-17A on macrophage activation could theoretically be overshadowed by the IFN-γ produced by strain H99γ. Previous studies in our laboratory have demonstrated that IFN-γ is necessary for the resolution of an experimental pulmonary infection with C. neoformans strain H99γ (38). We therefore sought to determine whether IFN-γ was necessary for IL-17A production in response to pulmonary infection with C. neoformans strain H99γ. Our studies showed no significant difference in the production of IL-17A in the lungs of IFN-γ−/− mice (226 pg/ml ± 35.58 pg/ml) compared to that in wild-type mice (235 pg/ml ± 86.76 pg/ml) on day 7 after pulmonary inoculation with C. neoformans strain H99γ. Thus, IL-17A production in the lungs of mice during infection with C. neoformans strain H99γ is not dependent on IFN-γ. Nonetheless, we evaluated the effect of IL-17A using C57BL/6 mice, in which there is a natural induction of classically activated macrophages in response to pulmonary C. neoformans infection (4).
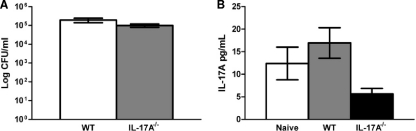
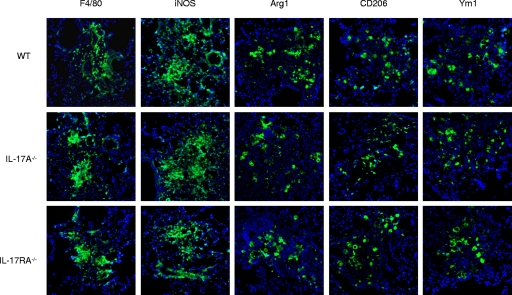
To evaluate pulmonary fungal burden and local IL-17A cytokine production, homogenates were prepared from pulmonary tissues excised from naïve, uninfected C57BL/6 mice and from C57BL/6 and IL-17A−/− mice (C57BL/6 background) infected with the wild-type, non-IFN-γ-producing C. neoformans strain H99. As shown in Fig. 8A, there was no significant difference in pulmonary fungal burden in IL-17A−/− mice compared to that in wild-type mice on day 7 postinoculation. Also, IL-17A production was not significantly different in naïve, uninfected mice compared to that in wild-type and IL-17A−/− mice (Fig. 8B), demonstrating that IL-17A expression is not induced by infection with C. neoformans strain H99. Subsequently, we evaluated classical and alternative macrophage activation in IL-17A−/− and IL-17RA−/− mice (C57BL/6 background) compared to that in wild-type C57BL/6 mice following infection with C. neoformans strain H99 on day 7 postinoculation. The numbers of infiltrating macrophages were similar for wild-type and IL-17A-deficient mice, and similar levels of classical (iNOS) and alternative (Arg1, CD206, and Ym1) macrophage activation markers were observed (Fig. 9). These results suggest that macrophages are classically activated in C57BL/6 mice during infection with C. neoformans strain H99 and that IL-17A does not contribute to classical activation. Furthermore, deletion of IL-17A does not result in a change in markers for alternative macrophage activation. To confirm the macrophage activation phenotype, we quantified the relative mRNA expression levels of genes associated with classical or alternative macrophage activation in pulmonary macrophage-enriched cell populations obtained from C57BL/6 wild-type and IL-17A−/− mice infected with C. neoformans strain H99. Pulmonary leukocytes were isolated from enzymatically dispersed lungs at day 7 postinoculation with C. neoformans strain H99γ. We subsequently extracted total RNA from CD11b+ cell populations and evaluated macrophage activation using real-time PCR (Fig. 10). We observed no significant difference in transcript levels of IFN-γ or Arg1, CD206, FIZZ1, Ym1, IL-4, and IL-13 (aaMac markers) on day 7 postinoculation with C. neoformans strain H99 in IL-17A−/− mice compared to the levels in wild-type animals. Transcript levels of the caMac marker iNOS, however, were significantly decreased in pulmonary macrophages from IL-17A−/− mice compared to the levels in wild-type C57BL/6 mice (Fig. 10). It is important to note, however, that iNOS protein expression was unchanged (Fig. 9) even though gene expression was decreased.
FIG. 8.
C57BL/6 mice do not produce IL-17A in response to infection with C. neoformans strain H99. C57BL/6 and IL-17A−/− mice received an intranasal inoculation with C. neoformans strain H99. The lungs from each group of mice were excised at day 7 postinoculation, and fungal burden was quantified (A). Alternatively, pulmonary homogenates were prepared from infected lungs excised on day 7 postinoculation or from naive mice and assayed for IL-17A cytokine production (B). Data represent cumulative results from two experiments utilizing four mice per group.
FIG. 9.
C57BL/6, IL-17A−/−, and IL-17RA−/− mice have similar pulmonary macrophage infiltration and express similar levels of classical and alternative macrophage activation markers during experimental pulmonary infection with C. neoformans strain H99. IL-17A−/−, IL-17RA−/−, and WT C57BL/6 mice received an intranasal inoculation with C. neoformans strain H99. Lungs were excised on day 7 postinoculation and immediately frozen in OCT medium. Tissues were cryosectioned, and expression of F4/80, iNOS, Arg1, CD206, and Ym1 was evaluated using immunofluorescence staining with antibodies directed against each marker. Data shown are representative lung sections from two independent experiments (three mice per group). Digital photographs show representative areas of lungs (20× objective).
FIG. 10.
Real-time PCR analysis of macrophage-enriched populations shows decreased iNOS expression and no difference in expression of aaMac transcripts in IL-17A−/− mice compared to expression in WT mice during infection with C. neoformans strain H99. C57BL/6 WT and IL-17A−/− mice received an intranasal inoculation with C. neoformans strain H99. Pulmonary leukocytes were isolated from lung tissues by enzymatic digestion on day 7 postinoculation, and macrophages were enriched for by positive selection of CD11b+ cells. Real-time PCR analyses of total mRNA from macrophage-enriched populations were conducted for iNOS, IFN-γ, Arg1, CD206, FIZZ1, Ym1, IL-4, IL-13, and GAPDH. Bars represent the fold change in gene expression in macrophages from IL-17A−/− mice during infection with C. neoformans strain H99 compared to that in WT mice. Data shown represent cumulative results from three independent experiments utilizing pooled leukocytes of five mice per group per experiment (*, P < 0.05).
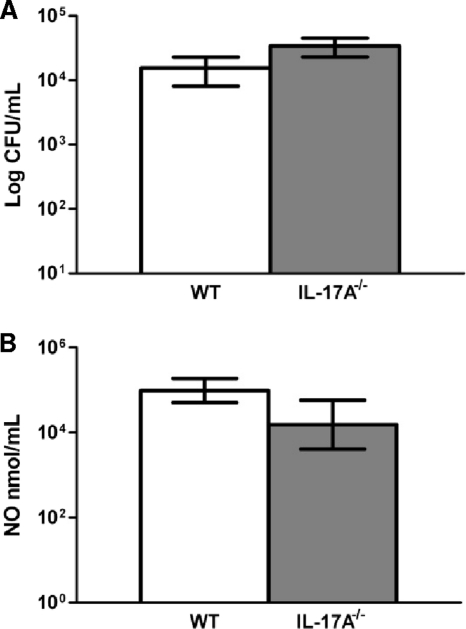
Next, we sought to determine the functional activity of pulmonary macrophages from IL-17A−/− and wild-type mice. We subsequently cultured primary macrophages isolated from the lungs of wild-type C57BL/6 and IL-17A−/− mice on day 7 postinoculation with C. neoformans strain H99 for 24 h and evaluated macrophage killing and iNOS production. There was no need to add additional cryptococci to cultures since the macrophages were derived from actively infected lungs; the macrophage preparation already contained a significant amount of live cryptococci, as previously noted (4). No significant difference in macrophage killing activity (Fig. 11A) or iNOS production (Fig. 11B) was observed in IL-17A−/− mice compared to that in wild-type mice. These findings support our results using IL-17RA−/− and wild-type BALB/c mice given a pulmonary infection with C. neoformans strain H99γ (Fig. 7), further suggesting that IL-17A has a minimal effect on macrophage recruitment and classical macrophage activation during experimental pulmonary cryptococcosis.
FIG. 11.
Macrophages derived from IL-17A−/− mice, compared to those derived from wild-type mice, do not have a deficiency in killing C. neoformans strain H99 yeasts or in iNOS activity. Pulmonary leukocytes were isolated from lung tissues on day 7 postinoculation, and macrophages were enriched for by positive selection of CD11b+ cells. (A) Macrophages were grown in complete medium, cultured for 24 h, lysed, and subsequently plated on YPD agar for CFU determination. (B) Culture supernatants were also utilized to determine iNOS activity. Data represent cumulative results from two experiments utilizing four mice per group.
DISCUSSION
Previous studies in our laboratory have demonstrated that mice given an experimental pulmonary infection with C. neoformans strain H99γ have levels of pulmonary macrophage infiltration comparable to those in mice infected with wild-type H99 yeast yet proceed to resolve the infection (15, 36, 38). We subsequently determined the phenotype of infiltrating macrophages and demonstrated that macrophages responding to infection with C. neoformans strain H99γ are classically activated, whereas aaMac respond to infection with the parental C. neoformans strain H99 (15). Indeed, others have reported increased resistance to infection and decreased pulmonary pathology in response to abrogation of aaMac development during C. neoformans infection in the absence of IL-4, IL-13, or both cytokines (28, 33, 41). Zhang et al. were able to demonstrate robust induction of iNOS, a hallmark marker of caMac, in enriched macrophage populations from C. neoformans-resistant IL-4−/−/-13−/− mice following C. neoformans infection. These results coincided with a decrease in pulmonary eosinophilia and marked induction of Th1-type and IL-17A cytokine expression and neutrophil recruitment (41). Conversely, IFN-γ−/− mice demonstrated progressive susceptibility to C. neoformans infection characterized by pulmonary fibrosis, decreased fungistasis, eosinophilia, and the induction of aaMac (4).
The induction of Th1-type CMI responses is associated with resistance to C. neoformans infections (17). In addition to the induction of Th1-type cytokines in the protective response to C. neoformans strain H99γ, we observed robust IL-17A cytokine expression (15, 38). Recent evidence suggests a role for IL-17 in resistance to C. neoformans infections. Administration of IL-23, a cytokine that plays a role in Th17 maintenance and proliferation (21), induces IL-17 production during C. neoformans infection and results in significantly increased survival rates and decreased fungal burden in mice. This is achieved without an alteration in the expression of the Th1-type cytokines IFN-γ and TNF-α or a change in IgE class switching, a marker for Th2-type immune activation (20). Furthermore, IL-23 administration is sufficient to significantly reduce fungal burden during C. neoformans infection in mice deficient in both IL-12 and IL-23 and to induce the proinflammatory cytokines TNF-α, IL-1β, and IL-6 (19, 20). These studies indicate a role for IL-17 in protective immunity to C. neoformans that is distinct from Th1- or Th2-type polarized responses, consistent with our observation that a lack of IL-17 does not affect pulmonary cytokine polarization (Fig. 6). Conversely, in the absence of IL-13 or both IL-4 and IL-13, robust IL-17 expression is observed in mice in response to C. neoformans infection (28, 41), implicating Th2-type cytokines in the negative regulation of IL-17 (32).
caMac are induced in environments with high IFN-γ cytokine levels (13, 15, 27, 41), but recent evidence points to a distinct role for IL-17 in the proinflammatory, microbicidal responses of macrophages (18, 20, 27, 35). IL-17 has been demonstrated to stimulate production of the proinflammatory cytokines IL-1β, IL-6, and TNF-α in peripheral blood mononuclear cell (PBMC)-derived human macrophages (18). During C. neoformans infection in IL-12- and IL-23-deficient p40−/− mice, the administration of IL-23 stimulates IL-17 production and results in the production of IL-1β, IL-6, and TNF-α that is persistent in B-cell- and T-cell-deficient Rag−/− mice (20). Treatment of J774 alveolar macrophages or PBMC-derived macrophages with IL-17 results in decreased intracellular proliferation of C. neoformans compared to that in macrophages stimulated with IL-4 or IL-13 (35). Furthermore, infection with the less virulent C. neoformans serotype D strain RC-2 results in increased inflammation and elevated IL-17 levels that are abrogated upon macrophage depletion (14). We find in the present study that, while IL-17A and caMac are induced in the protective response to C. neoformans strain H99γ, IL-17 deficiency does not change the phenotype of activated macrophages. These data suggest that IL-17 may augment macrophage effector function rather than induce classical macrophage differentiation. Macrophages indeed secrete IL-17 in response to proinflammatory stimuli. Da Silva et al. found that chitin, a fungal cell wall component, will induce IL-17 secretion from bone marrow-derived macrophages in a TLR2- and MyD88-dependent manner (10). The majority of IL-17 produced in response to ovalbumin (OVA)-induced pulmonary inflammation is produced by macrophages (32). Also, Guerrero et al. found that depletion of macrophages will reduce IL-17 production in a proinflammatory response to C. neoformans infection (14). However, using a model system in which infection with C. neoformans strain H99 results in the induction of classically activated macrophages (4), we were able to demonstrate functional activity of IL-17-deficient macrophages that is similar to that of macrophages derived from wild-type mice (Fig. 11). Indeed, intracellular yeast proliferation and cryptococcal expulsion rates were similar in the J774 alveolar macrophage-like cell line and primary human macrophages cultured in the presence or absence of recombinant IL-17 (35). The observation that we do not see any significant difference between pulmonary fungal burden in IL-17A−/− mice and that in C57BL/6 mice following infection with wild-type C. neoformans strain H99 suggests that IL-17A does not mediate significant protection during H99 infection. In addition, the significant increase in pulmonary fungal burden in IL-17A-depleted and IL-17RA−/− BALB/c mice compared to that in wild-type BALB/c mice following infection with the transgenic C. neoformans strain H99γ in the presence of caMac suggests that the mechanism for increased fungal burden in IL-17-deficient mice is unrelated to classical macrophage activation. Studies are under way to determine the mechanism for the increased fungal burden observed in IL-17-deficient mice following infection with C. neoformans strain H99γ.
IL-17 is also known to recruit neutrophils to sites of infection and inflammation and to enhance neutrophil phagocytosis and antimicrobial respiratory burst (30, 39). Macrophages and neutrophils work in concert to generate densely packed granulomata, which function in the containment of inflammatory pathogens (2). Neutrophil infiltration is crucial to the protective responses to C. neoformans infections (36, 37, 41). Though we do not see a significant difference in neutrophil influx following depletion of IL-17A during infection with C. neoformans strain H99γ (unpublished observations), we do see a prolonged iNOS response (Fig. 3). This suggests compensation for a lack of efficacy of the innate immune response, which may be due to improperly stimulated neutrophils in the absence of IL-17.
In conclusion, while there is an early delay in the control of fungal burden in IL-17A-depleted mice during infection with C. neoformans strain H99γ compared to that in isotype control-treated mice, we observed no difference in resolution of infection, macrophage infiltration, activation phenotype, or polarized cytokine response. Classical macrophage activation in response to pulmonary infection with C. neoformans strain H99γ was retained in IL-17RA−/− mice. In addition, although pulmonary infection of C57BL/6 mice with wild-type cryptococci did not result in the production of IL-17A, classical macrophage activation was still evident. All together, the evidence suggests that IL-17A does not contribute to classical macrophage activation during experimental pulmonary cryptococcosis.
Acknowledgments
This work was supported by research grant RO1 AI071752-03 (F.L.W.) and RO1 HL079142 (J.K.K.) from the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH).
This content is solely the responsibility of the authors and does not reflect the official views of the NIAID or the NIH.
Editor: G. S. Deepe, Jr.
Footnotes
Published ahead of print on 4 October 2010.
REFERENCES
- 1.Aarskog, N. K., and C. A. Vedeler. 2000. Real-time quantitative polymerase chain reaction. A new method that detects both the peripheral myelin protein 22 duplication in Charcot-Marie-Tooth type 1A disease and the peripheral myelin protein 22 deletion in hereditary neuropathy with liability to pressure palsies. Hum. Genet. 107:494-498. [DOI] [PubMed] [Google Scholar]
- 2.Adams, D. O. 1976. The granulomatous inflammatory response. A review. Am. J. Pathol. 84:164-192. [PMC free article] [PubMed] [Google Scholar]
- 3.Aguirre, K., E. A. Havell, G. W. Gibson, and L. L. Johnson. 1995. Role of tumor necrosis factor and gamma interferon in acquired resistance to Cryptococcus neoformans in the central nervous system of mice. Infect. Immun. 63:1725-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arora, S., Y. Hernandez, J. R. Erb-Downward, R. A. McDonald, G. B. Toews, and G. B. Huffnagle. 2005. Role of IFN-gamma in regulating T2 immunity and the development of alternatively activated macrophages during allergic bronchopulmonary mycosis. J. Immunol. 174:6346-6356. [DOI] [PubMed] [Google Scholar]
- 5.Chang, J. T., I. H. Chen, C. T. Liao, H. M. Wang, Y. M. Hsu, K. F. Hung, C. J. Lin, L. L. Hsieh, and A. J. Cheng. 2002. A reverse transcription comparative real-time PCR method for quantitative detection of angiogenic growth factors in head and neck cancer patients. Clin. Biochem. 35:591-596. [DOI] [PubMed] [Google Scholar]
- 6.Collins, H. L., and G. J. Bancroft. 1992. Cytokine enhancement of complement-dependent phagocytosis by macrophages: synergy of tumor necrosis factor-alpha and granulocyte-macrophage colony-stimulating factor for phagocytosis of Cryptococcus neoformans. Eur. J. Immunol. 22:1447-1454. [DOI] [PubMed] [Google Scholar]
- 7.Conti, H. R., F. Shen, N. Nayyar, E. Stocum, J. N. Sun, M. J. Lindemann, A. W. Ho, J. H. Hai, J. J. Yu, J. W. Jung, S. G. Filler, P. Masso-Welch, M. Edgerton, and S. L. Gaffen. 2009. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J. Exp. Med. 206:299-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox, G. M., H. C. McDade, S. C. Chen, S. C. Tucker, M. Gottfredsson, L. C. Wright, T. C. Sorrell, S. D. Leidich, A. Casadevall, M. A. Ghannoum, and J. R. Perfect. 2001. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 39:166-175. [DOI] [PubMed] [Google Scholar]
- 9.Cox, G. M., J. Mukherjee, G. T. Cole, A. Casadevall, and J. R. Perfect. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Da Silva, C. A., D. Hartl, W. Liu, C. G. Lee, and J. A. Elias. 2008. TLR-2 and IL-17A in chitin-induced macrophage activation and acute inflammation. J. Immunol. 181:4279-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flesch, I. E., G. Schwamberger, and S. H. Kaufmann. 1989. Fungicidal activity of IFN-gamma-activated macrophages. Extracellular killing of Cryptococcus neoformans. J. Immunol. 142:3219-3224. [PubMed] [Google Scholar]
- 12.Gaffen, S. L. 2009. Structure and signaling in the IL-17 receptor family. Nat. Rev. Immunol. 9:556-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon, S. 2003. Alternative activation of macrophages. Nat. Rev. Immunol. 3:23-35. [DOI] [PubMed] [Google Scholar]
- 14.Guerrero, A., N. Jain, X. Wang, and B. C. Fries. 2010. Cryptococcus neoformans variants generated by phenotypic switching differ in virulence through effects on macrophage activation. Infect. Immun. 78:1049-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardison, S. E., S. Ravi, K. L. Wozniak, M. L. Young, M. A. Olszewski, and F. L. Wormley, Jr. 2010. Pulmonary infection with an interferon-gamma-producing Cryptococcus neoformans strain results in classical macrophage activation and protection. Am. J. Pathol. 176:774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huffnagle, G. B., and M. F. Lipscomb. 1998. Cells and cytokines in pulmonary cryptococcosis. Res. Immunol. 149:387-396. [DOI] [PubMed] [Google Scholar]
- 17.Jain, A. V., Y. Zhang, W. B. Fields, D. A. McNamara, M. Y. Choe, G. H. Chen, J. Erb-Downward, J. J. Osterholzer, G. B. Toews, G. B. Huffnagle, and M. A. Olszewski. 2009. Th2 but not Th1 immune bias results in altered lung functions in a murine model of pulmonary Cryptococcus neoformans infection. Infect. Immun. 77:5389-5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jovanovic, D. V., J. A. Di Battista, J. Martel-Pelletier, F. C. Jolicoeur, Y. He, M. Zhang, F. Mineau, and J. P. Pelletier. 1998. IL-17 stimulates the production and expression of proinflammatory cytokines, IL-beta and TNF-alpha, by human macrophages. J. Immunol. 160:3513-3521. [PubMed] [Google Scholar]
- 19.Kleinschek, M. A., U. Muller, S. J. Brodie, W. Stenzel, G. Kohler, W. M. Blumenschein, R. K. Straubinger, T. McClanahan, R. A. Kastelein, and G. Alber. 2006. IL-23 enhances the inflammatory cell response in Cryptococcus neoformans infection and induces a cytokine pattern distinct from IL-12. J. Immunol. 176:1098-1106. [DOI] [PubMed] [Google Scholar]
- 20.Kleinschek, M. A., U. Muller, N. Schutze, R. Sabat, R. K. Straubinger, W. M. Blumenschein, T. McClanahan, R. A. Kastelein, and G. Alber. 2010. Administration of IL-23 engages innate and adaptive immune mechanisms during fungal infection. Int. Immunol. 22:81-90. [DOI] [PubMed] [Google Scholar]
- 21.Korn, T., E. Bettelli, M. Oukka, and V. K. Kuchroo. 2009. IL-17 and Th17 cells. Annu. Rev. Immunol. 27:485-517. [DOI] [PubMed] [Google Scholar]
- 22.Levitz, S. M., and D. J. DiBenedetto. 1988. Differential stimulation of murine resident peritoneal cells by selectively opsonized encapsulated and acapsular Cryptococcus neoformans. Infect. Immun. 56:2544-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loke, P., M. G. Nair, J. Parkinson, D. Guiliano, M. Blaxter, and J. E. Allen. 2002. IL-4 dependent alternatively-activated macrophages have a distinctive in vivo gene expression phenotype. BMC Immunol. 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez, F. O., L. Helming, and S. Gordon. 2009. Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 27:451-483. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell, T. G., and J. R. Perfect. 1995. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8:515-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mody, C. H., C. L. Tyler, R. G. Sitrin, C. Jackson, and G. B. Toews. 1991. Interferon-gamma activates rat alveolar macrophages for anticryptococcal activity. Am. J. Respir. Cell Mol. Biol. 5:19-26. [DOI] [PubMed] [Google Scholar]
- 27.Mosser, D. M., and J. P. Edwards. 2008. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8:958-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller, U., W. Stenzel, G. Kohler, C. Werner, T. Polte, G. Hansen, N. Schutze, R. K. Straubinger, M. Blessing, A. N. McKenzie, F. Brombacher, and G. Alber. 2007. IL-13 induces disease-promoting type 2 cytokines, alternatively activated macrophages and allergic inflammation during pulmonary infection of mice with Cryptococcus neoformans. J. Immunol. 179:5367-5377. [DOI] [PubMed] [Google Scholar]
- 29.Osterholzer, J. J., R. Surana, J. E. Milam, G. T. Montano, G. H. Chen, J. Sonstein, J. L. Curtis, G. B. Huffnagle, G. B. Toews, and M. A. Olszewski. 2009. Cryptococcal urease promotes the accumulation of immature dendritic cells and a non-protective T2 immune response within the lung. Am. J. Pathol. 174:932-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peck, A., and E. D. Mellins. 2010. Precarious balance: Th17 cells in host defense. Infect. Immun. 78:32-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powderly, W. G. 1993. Cryptococcal meningitis and AIDS. Clin. Infect. Dis. 17:837-842. [DOI] [PubMed] [Google Scholar]
- 32.Song, C., L. Luo, Z. Lei, B. Li, Z. Liang, G. Liu, D. Li, G. Zhang, B. Huang, and Z. H. Feng. 2008. IL-17-producing alveolar macrophages mediate allergic lung inflammation related to asthma. J. Immunol. 181:6117-6124. [DOI] [PubMed] [Google Scholar]
- 33.Stenzel, W., U. Muller, G. Kohler, F. L. Heppner, M. Blessing, A. N. McKenzie, F. Brombacher, and G. Alber. 2009. IL-4/IL-13-dependent alternative activation of macrophages but not microglial cells is associated with uncontrolled cerebral cryptococcosis. Am. J. Pathol. 174:486-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voelz, K., D. A. Lammas, and R. C. May. 2009. Cytokine signaling regulates the outcome of intracellular macrophage parasitism by Cryptococcus neoformans. Infect. Immun. 77:3450-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wormley, F. L., Jr., J. R. Perfect, C. Steele, and G. M. Cox. 2007. Protection against cryptococcosis by using a murine gamma interferon-producing Cryptococcus neoformans strain. Infect. Immun. 75:1453-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wozniak, K. L., and S. M. Levitz. 2008. Cryptococcus neoformans enters the endolysosomal pathway of dendritic cells and is killed by lysosomal components. Infect. Immun. 76:4764-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wozniak, K. L., S. Ravi, S. Macias, M. L. Young, M. A. Olszewski, C. Steele, and F. L. Wormley. 2009. Insights into the mechanisms of protective immunity against Cryptococcus neoformans infection using a mouse model of pulmonary cryptococcosis. PLoS One 4:e6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye, P., F. H. Rodriguez, S. Kanaly, K. L. Stocking, J. Schurr, P. Schwarzenberger, P. Oliver, W. Huang, P. Zhang, J. Zhang, J. E. Shellito, G. J. Bagby, S. Nelson, K. Charrier, J. J. Peschon, and J. K. Kolls. 2001. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J. Exp. Med. 194:519-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young, M., S. Macias, D. Thomas, and F. L. Wormley, Jr. 2009. A proteomic-based approach for the identification of immunodominant Cryptococcus neoformans proteins. Proteomics 9:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, Y., F. Wang, K. C. Tompkins, A. McNamara, A. V. Jain, B. B. Moore, G. B. Toews, G. B. Huffnagle, and M. A. Olszewski. 2009. Robust Th1 and Th17 immunity supports pulmonary clearance but cannot prevent systemic dissemination of highly virulent Cryptococcus neoformans H99. Am. J. Pathol. 175:2489-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]