Abstract
One of the most crucial events during infection with the dimorphic fungus Paracoccidioides brasiliensis is adhesion to pulmonary epithelial cells, a pivotal step in the establishment of disease. In this study, we have evaluated the relevance of a 32-kDa protein, a putative adhesion member of the haloacid dehalogenase (HAD) superfamily of hydrolases, in the virulence of this fungus. Protein sequence analyses have supported the inclusion of PbHad32p as a hydrolase and have revealed a conserved protein only among fungal dimorphic and filamentous pathogens that are closely phylogenetically related. To evaluate its role during the host-pathogen interaction, we have generated mitotically stable P. brasiliensis HAD32 (PbHAD32) antisense RNA (aRNA) strains with consistently reduced gene expression. Knockdown of PbHAD32 did not alter cell vitality or viability but induced morphological alterations in yeast cells. Moreover, yeast cells with reduced PbHAD32 expression were significantly affected in their capacity to adhere to human epithelial cells and presented decreased virulence in a mouse model of infection. These data support the hypothesis that PbHad32p binds to extracellular matrix (ECM) proteins and modulates the initial immune response for evasion of host defenses. Our findings point to PbHAD32 as a novel virulence factor active during the initial interaction with host cells in P. brasiliensis.
The adherence of pathogenic microorganisms to host tissues is considered indispensable for their initial colonization and successful infection and dissemination (38). The internalization process has been shown to depend greatly upon the adherence to the host cell surface that generates a cytoplasmic uptake signal (24). In the case of dimorphic fungal pathogens, of which the site of primary infection is generally the lung, the ability to adhere to epithelial cells represents a mechanism by which the infecting agent avoids the entrapment by respiratory tract mucus and, later on, elimination by the action of mucigen ciliary cells (24). Paracoccidioides brasiliensis is the etiological agent of paracoccidioidomycosis (PCM), one of the most common endemic systemic mycoses in Latin America (30, 32). As in other thermodimorphic fungi, P. brasiliensis mycelial fragments and microconidia act as the infectious propagules, reaching the lung alveoli, where, at the temperature of the host's tissues (37°C), it shifts to the parasitic yeast form (6). During this process, adherence of P. brasiliensis to pulmonary epithelial cells is considered an essential event.
Extracellular matrix (ECM) proteins have been shown to play an important role during the initial interaction and adherence between host cells and clinically relevant dimorphic fungi, such as P. brasiliensis, Penicillium marneffei, and Histoplasma capsulatum (17, 18, 22, 25). In P. brasiliensis, the major immunogenic antigen, Gp43 (a 43-kDa glycoprotein), detected in the cell wall and as an exocellular compound of both the yeast and mycelial phases, was proven to bind to laminin, a major ECM protein (27, 37, 39). More recently, Gonzalez and coworkers identified a 32-kDa protein in cell wall protein extracts of both forms of P. brasiliensis that was capable of binding to various ECM proteins, including laminin, fibronectin, and fibrinogen (14). Additionally, they demonstrated that this 32-kDa protein is involved in the initial conidial adherence to pulmonary epithelial cells that express ECM proteins on the surface, acting as a bridge between the two cell types (13).
The main goal of this work was to further characterize this 32-kDa protein and its true role during the P. brasiliensis infectious process. Protein sequence analysis revealed a putative adhesion member of the haloacid dehalogenase (HAD) superfamily of hydrolases, P. brasiliensis Had32p (PbHad32p). Using antisense RNA (aRNA) technology and Agrobacterium tumefaciens-mediated transformation (ATMT), we constructed a mitotically stable P. brasiliensis PbHAD32 aRNA strain with consistently reduced gene expression (1, 2). Yeast cells with reduced PbHAD32 expression were significantly affected in their capacity to adhere to epithelial cells. Moreover, the knockdown strain presented decreased virulence in a mouse model of infection, pointing to PbHAD32 as a novel virulence factor during the initial interaction with host cells.
MATERIALS AND METHODS
Microorganisms and culture media.
P. brasiliensis yeast cells (strain ATCC 60855) were maintained at 36°C by subculturing them in brain heart infusion (BHI) medium supplemented with 1% glucose (Becton Dickinson and Company, Sparks, MD). Unless indicated otherwise, yeast cells were grown in BHI liquid medium at 36°C with aeration on a mechanical shaker and were routinely collected during their exponential phase of growth (72 to 96 h). Conidial production was carried out as described previously (31).
A. tumefaciens strain LBA1100 (5) was used as the recipient for the binary vectors constructed in this study. Bacterial cells were maintained at 28°C in Luria Bertani (LB) medium containing kanamycin (100 mg/ml). Escherichia coli XL1-Blue was grown at 37°C in LB medium supplemented with appropriate antibiotics and was used as the host for plasmid amplification and cloning (34). Morphological transition from yeast to mycelia was performed in BHI liquid medium at 20°C (29). The conidium-to-yeast transition process was carried out by incubating conidia at 36°C in BHI liquid medium. Samples were collected during the transition process for RNA extraction and quantification of gene expression (11).
To evaluate cell morphology, the strains were exponentially grown, collected, and fixed in a slide and visualized with an AxiosterPlus microscope (Zeiss). Images were acquired with a PowerShot G5 camera (Canon).
Protein sequence analysis.
BLAST analysis of the amino acid sequence of the 32-kDa hydrolase protein reported by Gonzalez et al. (14) was performed at the Broad Institute (http://www.broadinstitute.org/science/data). The P. brasiliensis genome database was used to obtain the putative complete gene and protein sequence. Multiple-sequence analysis of homologous HAD hydrolases of the following organisms was also performed using the CLUSTAL program (version 2.0.12; http://www.ebi.ac.uk/): P. brasiliensis (GenBank accession number EEH46031.1), Blastomyces dermatitidis (NCBI reference sequence XP_002625690), Histoplasma capsulatum (GenBank accession number EEH06199.1), Coccidioides posadasii (NCBI reference sequence XP_003069355.1), Penicillium marneffei (NCBI reference sequence XP_002147878.1), Aspergillus fumigatus (NCBI reference sequence XP_753809.1), and Fusarium graminearum (NCBI reference sequence XP_385308.1).
Generating P. brasiliensis PbHAD32 aRNA strains.
P. brasiliensis wild-type (PbWt) strain ATCC 60855 DNA was extracted from yeast cultures during exponential growth using the TRIzol reagent (Invitrogen, Carlsbad, CA). A Platinum high-fidelity Taq DNA polymerase (Invitrogen) was employed to amplify aRNA oligonucleotides for PbHAD32 (AS1, AS2, and AS3).
Plasmid construction for aRNA and ATMT of P. brasiliensis were performed as described previously (1, 2). Briefly, the amplified PbHAD32 aRNA oligonucleotides were inserted into the pCR35 plasmid under the control of the calcium-binding protein 1 (CBP-1) promoter region from H. capsulatum (28). The pUR5750 plasmid was used as a parental binary vector to harbor this aRNA cassette within the transfer DNA (T-DNA). The constructed binary vectors were introduced into A. tumefaciens LBA1100 ultracompetent cells by electroporation as described previously (9) and isolated by kanamycin selection (100 mg/ml).
ATMT of P. brasiliensis yeast cells was performed using A. tumefaciens cells harboring the desired binary vector, as described by Almeida et al. (1). A 1:10 yeast/bacterium ratio was employed during the 3-day period of cocultivation at 28°C. Selection of P. brasiliensis transformants was performed in BHI solid medium with hygromycin B (Hyg; 50 mg/ml) over a 15-day incubation period at 36°C. Randomly selected Hyg-resistant transformants were tested for mitotic stability. P. brasiliensis yeast cells were also transformed with the empty parental vector, pUR5750, as a control during assays carried out in this study.
Gene and protein expression analysis.
Total RNA was obtained from PbWt and P. brasiliensis PbHAD32 aRNA yeast cells using the TRIzol reagent (Invitrogen). Total RNA was treated with DNase I (Invitrogen) and tested using a conventional PCR with β-tubulin primers to confirm the absence of chromosomal DNA contamination (12); cDNA was synthesized using 2 μg of total RNA with SuperScript III reverse transcriptase, according to the manufacturer's instructions (Invitrogen).
Real-time PCR was done using a SuperScript III Platinum two-step quantitative reverse transcription-PCR (qRT-PCR) kit with SYBR green, according to the manufacturer's instructions (Invitrogen). The CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA) was used to measure gene expression levels. PbHAD32 expression was evaluated in both PbWt and PbHAD32 aRNA yeast cells at different time points. Melting curve analysis was performed after the amplification phase to eliminate the possibility of nonspecific amplification or primer-dimer formation. Fold changes in mRNA expression were calculated using the 2ΔΔCT formula, where ΔΔCT is the difference in the threshold cycle (CT) between the target gene and the β-tubulin gene (a housekeeping gene) (21). Each experiment was done in triplicate, and the expression level was measured in triplicate.
Protein expression analysis was performed by Western blotting, as described by Gonzalez and coworkers, using a specific monoclonal antibody against PbHad32p (14).
Viability and vitality of P. brasiliensis yeast cells.
PbWt and PbHAD32 aRNA yeast cells were grown in BHI liquid medium at 36°C. After various subcultures, we evaluated their viability using ethidium bromide-fluorescence staining (7) and determining the numbers of CFU. For this purpose, several dilutions of the cultures were plated in BHI medium supplemented with 0.5% glucose, 4% horse serum, and EDTA (300 mM) (19), and the numbers of CFU were counted after 7 days of culture.
Vitality was evaluated as the ability to absorb glucose with later activation of a cell membrane proton pump (35) and subsequent acidification of the medium due to released H+. PbWt and PbHAD32 aRNA yeast cells were grown in liquid medium, and measurement of vitality was made at different time points. Cell samples were collected, washed twice with sterile water (pH 7.0), and suspended in a final volume of 8 ml of water (pH 7.0). Two milliliters of this suspension was add to a beaker with 38 ml of water, and when the pH became stable (pH between 5.5 and 6), 10 ml of 20% glucose was added. The pH of the experimental medium was evaluated each 3 min up to 60 min to evaluate the increase in the level of H+ in the medium.
Adherence of P. brasiliensis to A549 cells.
The human lung epithelial cell line A549, corresponding to type II epithelial cells from an adenocarcinoma cell line, was obtained from the European Collection of Cell Cultures (ECACC). Cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS). For the assays, we used confluent monolayers obtained by adding 4 × 105 cells per well to 24-well tissue culture plates (Nunc, Kamstrup, Denmark), and then the plates were incubated for 24 h at 36°C with 5% CO2 prior to evaluation of the interaction with PbWt and PbHAD32 aRNA yeast cells. A549 cell monolayers were washed once with culture medium (DMEM), cocultured with P. brasiliensis yeast cells at a concentration of 8 × 104 yeast cells per well (corresponding to a ratio of 1:5 for P. brasiliensis/A549 cells), and incubated for 1 and 3 h at 36°C with 5% CO2. The supernatants of the cultures were then removed, the monolayers were lysed, yeasts adherent to epithelial cells were collected, and dilutions of these suspensions were plated on BHI medium plates supplemented with 0.5% glucose, 4% horse serum, and EDTA (300 mM) (19). These results were compared with the number of yeast cells added to each well. The percent adherence was expressed as the number of CFU obtained from each experimental well (P. brasiliensis yeast cells and A549 cells) divided by the number of CFU obtained from the control wells (P. brasiliensis yeast cells alone). The viability of P. brasiliensis yeasts was also evaluated after 24 h of infection by determining the number of CFU and ethidium bromide-fluorescence staining procedures, as described above.
To determine cytokine expression, we used confluent monolayers and the RT-PCR procedures described above. Total RNA was extracted using the TRIzol reagent, while ubiquitin was used as the housekeeping gene. We evaluated the expression of interleukin-6 (IL-6), IL-10, IL-12p40, and tumor necrosis factor alpha (TNF-α) by the A549 cell line infected with PbWt and PbHAD32 aRNA strains at different time points (15 and 30 min and 1, 3, 6, 12, 24, and 48 h). Each experiment was done thrice, and the expression level was measured in triplicate.
In vivo infection.
Isogenic 8-week-old BALB/c male mice, obtained from the breeding colony of the Corporación para Investigaciones Biológicas (CIB), Medellín, Colombia, were used in all experiments and were kept with food and water ad libitum (33). Regulations given by the Colombian government (law 84 of 1983, resolution no. 8430 of 1993) and the regulations of the European Communities and the Canadian Council of Animal Care (1998) were fully complied with.
Animals were infected intratracheally (i.t.) with 2 × 106 P. brasiliensis yeast cells harvested at the exponential phase of growth in BHI liquid medium (yeast cell viability, above 95%). Prior to infection, cells were washed thrice with PBS, passed through a syringe to eliminate cell clumps, and submitted to counting procedures in a Neubauer chamber (each mother cell was considered a single cell).
Statistics.
Data are reported as averages ± standard errors of the means (SEMs), and all assays were done at least three times. All statistical analyses, including analysis of variance (ANOVA), were performed using the SPSS (version 17.0) statistics program. A P value of <0.05 was considered statistically significant. The survival rate, determined from two independent infections in a mouse model (n = 8 mice for each P. brasiliensis strain), was analyzed using the Kaplan-Meyer and log-rank tests.
RESULTS
The 32-kDa protein is conserved among pathogenic dimorphic fungi.
The previously reported amino acid sequence (14) of the 32-kDa protein was compared with the sequenced genome of P. brasiliensis by analysis with the BLAST program at the Broad Institute. A match was obtained for a 1,716-bp genome sequence with 2 exons and 1 intron (PbHAD32) encoding a protein with 244 amino acids and a conserved HAD superfamily hydrolase domain. Sequence analysis revealed between 58% and 84% identity with the sequences of other dimorphic and filamentous fungal hypothetical HAD superfamily hydrolases (Fig. 1). No homologues were identified in more evolutionarily distant fungal species (e.g., Cryptococcus neoformans, Candida spp., and Saccharomyces cerevisiae), Drosophila melanogaster, or mammalian cells.
FIG. 1.
Multiple-sequence alignment of sequences of HAD superfamily hydrolase proteins from diverse human fungal pathogens: Blastomyces dermatitidis (Bd), Histoplasma capsulatum (Hc), Coccidioides posadasii (Cp), Penicillium marneffei (Pm), Aspergillus fumigatus (Af), and Fusarium graminearum (Fg) revealed 84%, 82%, 77%, 67%, 66%, and 58% identities, respectively, compared with the sequence of P. brasiliensis (Pb).
Knockdown of PbHAD32 expression.
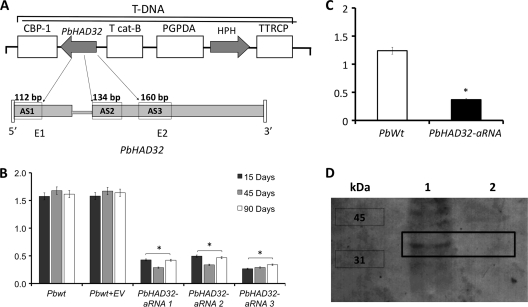
Using aRNA technology and ATMT, we generated P. brasiliensis PbHAD32 aRNA strains to further study the function of this protein. Three different aRNA oligonucleotides were designed from the 1st (AS1) and 2nd (AS2 and AS3) exons of PbHAD32 and inserted individually into PbWt yeast cells (Fig. 2A). For control experiments, PbWt yeast cells were transformed with the empty vector. Decreases in the level of expression of PbHAD32 ranging from 72 to 80% were obtained when the results were compared to those for both the PbWt strain and the strain harboring the empty vector (Fig. 2B). No major differences in PbHAD32 expression were detected among the generated PbHAD32 aRNA strains. PbHAD32 expression in PbHAD32 aRNA strains was also determined after 15, 45, and 90 days of subculturing of yeast cells, confirming knockdown of gene expression and stable genomic integration of the T-DNA (Fig. 2B). As a control, we also produced conidia from a PbHAD32 aRNA strain (AS2) to perform a conidium-to-yeast (C-Y) transition and to confirm a decrease in the level of PbHAD32 expression after the morphological transition (Fig. 2C). Furthermore, protein expression analysis by Western blotting confirmed the decrease in PbHAD32 protein levels in PbHAD32 aRNA strains (Fig. 2D). Yeast cells from a PbHAD32 aRNA strain generated with aRNA oligonucleotide AS2 were selected for analysis during the remaining assays.
FIG. 2.
Generation of P. brasiliensis PbHAD32 aRNA strains. (A) T-DNA construct for aRNA silencing of PbHAD32 in P. brasiliensis via ATMT. PbHAD32 aRNA oligonucleotides AS1 (base pairs 1 to 112 of PbHAD32; exon 1), AS2 (base pairs 175 to 309 of PbHAD32; exon 2), and AS3 (base pairs 376 to 536 of PbHAD32; exon 2) were amplified, individually placed under the control of the calcium-binding protein (CBP1) promoter, and later on inserted into the T-DNA of the binary vector pUR5750 for ATMT of P. brasiliensis. (B) Gene expression levels of PbHAD32 in PbWt, PbWt transformed with the empty vector (PbWt + EV), and PbHAD32 aRNA yeast cells after subculture for 15, 45, and 90 days (gene expression levels obtained by RT-PCR were normalized to the level of expression of the internal reference, TUB2; *, P < 0.05 compared with PbWt and PbWt transformed with the empty vector). (C) Gene expression levels of PbHAD32 in PbWt and PbHAD32 aRNA yeast cells after the yeast-to-mycelium transition (Y-M), production of conidia (M-C), and transition into yeast cells (C-Y) (the complete process was Y-M-C-Y) (gene expression levels obtained by RT-PCR were normalized to the level of expression of the internal reference, TUB2; *, P < 0.05 compared with PbWt). (D) Western blot analysis of total protein extracts from PbHAD32 in PbWt and PbHAD32 aRNA yeast cells. Lane 1, PbWt; lane 2, PbHAD32 aRNA.
PbHAD32 silencing alters yeast cell morphology without affecting cell viability and vitality.
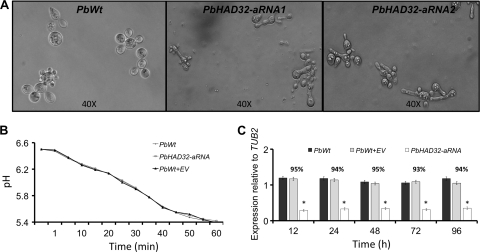
Microscopic observations indicated that PbHAD32 aRNA yeast cells were more elongated than the wild-type cells and cells harboring the empty vector (Fig. 3A); however, no morphological alterations were observed in conidia or the mycelial form (data not shown). The decrease in expression of PbHAD32 did not alter yeast cell vitality or viability (Fig. 3B). Moreover, no significant differences between PbHAD32 aRNA strains and the controls were detected during batch culture growth of yeast cells (data not shown). We also studied the viabilities of PbHAD32 aRNA and PbWt yeast cells at different time points during batch culture. No major differences in either viability or PbHAD32 expression were observed throughout the assay (Fig. 3C).
FIG. 3.
Silencing of PbHAD32 leads to distinct P. brasiliensis yeast cell morphology without affecting cell vitality. (A) Microscopic evaluation of PbWt yeast cells and yeast cells from two different PbHAD32 aRNA strains generated with different aRNA oligonucleotides (PbHAD32 aRNA1, AS1; PbHAD32 aRNA2, AS2). Magnifications, ×40. (B) Vitality of PbWt, PbWt transformed with the empty vector (PbWt + EV), and PbHAD32 aRNA yeast cells. (C) Cell viability (represented on top of the bars) and gene expression levels of PbHAD32 in PbWt, PbWt transformed with the empty vector, and PbHAD32 aRNA yeast cells during batch culture growth (gene expression levels obtained by RT-PCR were normalized to the level of expression of the internal reference, TUB2; *, P < 0.05 compared with PbWt and PbWt transformed with the empty vector).
PbHAD32 plays a role in adherence to human epithelial cells without altering cytokine expression.
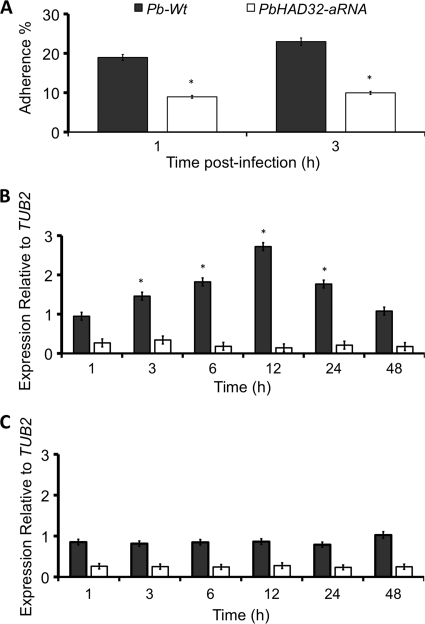
To elucidate the role of PbHAD32 in adherence of P. brasiliensis to host cells, we infected A549 epithelial human lung cells with yeast cells of PbWt and PbHAD32 aRNA strains. At 1 h of interaction, the adherence of PbHAD32 aRNA yeast cells was significantly decreased to half, numbers that were approximately maintained after 3 h (Fig. 4A) and 24 h (data not shown). PbHAD32 expression was also evaluated during infection of epithelial cells. Yeast cells of either the PbWt or PbHAD32 aRNA strain placed alone in culture medium showed no alterations in gene expression (Fig. 4B). However, contrary to what was observed in PbHAD32 aRNA yeast cells during infection, a continuous increase in PbHAD32 expression was detected in PbWt cells up until 12 h, decreasing later on until 48 h after the initial infection (Fig. 4C).
FIG. 4.
PbHad32p plays an essential role during adherence to epithelial cells. (A) Adherence of PbWt and PbHAD32 aRNA yeast cells to A549 epithelial human lung cells at different times postinfection (1 and 3 h). The percent adherence was expressed as the number of CFU adhered to epithelial cells divided by the number of CFU from wells without epithelial cells (*, P < 0.05 compared with PbWt). (B) Gene expression levels of PbHAD32 during infection of epithelial cells with PbWt yeasts cells (*, P < 0.05 compared with PbHAD32 aRNA). (C) Gene expression levels of PbHAD32 in PbWt and PbHAD32 aRNA yeast cells growing in the absence of epithelial cells.
We also evaluated cytokine gene expression during infection of A549 epithelial cells. IL-6, IL-10, IL12p40, and TNF-α levels were measured at different time points after infection. P. brasiliensis wild-type and PbHAD32 aRNA strains did not induce cytokine gene expression throughout the assay (at least at measurable mRNA levels).
PbHAD32 is an important virulence factor for P. brasiliensis infection.
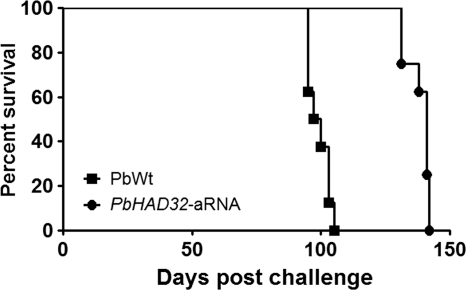
The relevance of PbHAD32 during the host-pathogen interaction was further evaluated using a mouse model of infection. Animals infected with the P. brasiliensis control strain (PbWt) started to die at day 95, with the average survival being 99 days. Contrarily, mice infected with the PbHAD32 aRNA strain started to die at day 131, with the average survival being 141 days (Fig. 5).
FIG. 5.
Silencing of PbHAD32 decreases the virulence of P. brasiliensis in a murine model of infection. Representative survival curve of an experimental infection carried out in BALB/c mice via i.t. infection with 2 × 106 PbWt or PbHAD32 aRNA yeast cells (P < 0.0001).
DISCUSSION
One of the pivotal events during infection with P. brasiliensis is the interaction and adhesion between fungal and host cells followed by adhesion to epithelial pulmonary cells (24). Although some P. brasiliensis molecules that may participate in adhesion to host tissues have been identified (e.g., malate synthase, enolase, thriose phosphate isomerase, an adaptin-like protein, and glyceraldehyde-3-phosphate dehydrogenase, among others), the exact role of these proteins remains uncharacterized due to a lack of gene-based evidence (3, 4, 8, 10, 26).
In the present study we have focused on a 32-kDa protein, PbHad32p, previously shown to play an important role in the adherence of P. brasiliensis to host cells and in the subsequent immune response in experimental PCM (13). To further characterize the function of this protein during the host-pathogen interaction, we generated mitotically stable P. brasiliensis PbHAD32 aRNA strains with significantly reduced levels of PbHAD32 gene and protein expression, as proven by RT-PCR and Western blot analysis. Moreover, the reproducibility of the assays among PbHAD32 aRNA transformants generated with different aRNA oligonucleotides (AS1, AS2, or AS3) also supported the hypothesis that the observed alterations were due to PbHAD32 silencing and not random gene disruption via genomic insertion by ATMT. The knockdown of this hydrolase gene did not affect the viability or vitality of P. brasiliensis and PbHAD32 aRNA strains, as both showed similar growth rates, suggesting that PbHAD32 is not directly involved in cellular processes related to glucose metabolism and yeast cell growth in batch cultures. Interestingly, downregulation of PbHAD32 resulted in yeast morphological alterations but did not influence mycelial or conidial aspects. Specifically, more elongated buds were observed, suggesting a function for this hydrolase in the maintenance of cell shape during growth. However, the specific mechanism(s) by which this downregulation affected the morphological alterations was not elucidated in this study. Further investigations should be addressed in order to identify the molecular mechanisms that participate in these kinds of alterations.
The results observed in the present study lead us to hypothesize that both morphological alterations in yeast cells and the reduced expression of PbHAD32, probably on their surface, were associated with a decreased capacity of PbHAD32 aRNA strains to adhere to human lung epithelial cells. In addition, the absence of detectable levels of cytokine mRNA during interaction with both PbWt and PbHAD32 aRNA yeast cells suggests that the decrease in the adherence capacity is due to reduced expression of PbHAD32 rather than cytokine signaling. Although posing as a relatively passive physical barrier to infection, epithelial cells have been proven to contribute with signaling events during the initial immune response against P. brasiliensis infection (23). In addition, previous studies have demonstrated that P. brasiliensis strains exhibiting enhanced adhesion to host cells in vitro are more virulent (16) and that strains with different yeast cell morphologies are associated with distinct virulence profiles (20, 40). Our data show that a reduction in the level of expression of PbHAD32 also leads to significantly increased survival in mice challenged with PbHAD32 aRNA yeast cells. Moreover, hydrolase expression increased significantly during the first 12 h of the epithelial cell interaction with PbWt yeast cells but not with PbHAD32 aRNA yeast cells, suggesting that it is specifically elicited by host stimulation. Taking into account previous reports showing that this 32-kDa protein is mainly located at the cell wall of P. brasiliensis (14), our data suggest that the decrease in protein levels, most likely at the cell surface, may lead to a reduced capacity to bind to ECM proteins and a decreased capability to evade host defenses by modulation of the initial immune response (15). Nonetheless, after the initial adherence to epithelial cells and endocytosis, we do not discard the relevance that phagocytosis by macrophages has in the production of pro- and/or anti-inflammatory cytokines and dissemination of the fungus. The alteration in the ability to adhere to epithelial cells and a possible lack of modulation of the immune response during infection with PbHAD32 aRNA yeast cells may be assisted by increased phagocytic capacity or enhanced fungicidal mechanisms.
In other clinically relevant dimorphic fungi, interaction with ECM proteins of host cells and subsequent adherence to tissues constitute crucial steps in the establishment of the initial focal infection and dissemination to other organs (17, 18, 22, 25). Protein sequence analysis revealed a conserved homology of this HAD superfamily hydrolase among human fungal pathogens, both dimorphic (H. capsulatum, Coccidioides spp., Blastomyces dermatitidis, and P. marneffei) and filamentous (Aspergillus spp. and F. graminearum), but not with other more distantly related fungi (e.g., Cryptococcus neoformans, Candida spp., and S. cerevisiae). In fact, the high degree of sequence identity among the proteins of the analyzed fungal species coincides with the findings of recent research pointing out significant evolutionary events during comparative genomic analysis that phylogenetically group these fungal human pathogens (36). Altogether, these data further support the relevance of this HAD superfamily hydrolase in the virulence of P. brasiliensis but also open the door for further studies related to the relevance of this protein in other fungal human pathogens. Future studies can now be directed at the biochemical evaluation of the possible substrates of this enzyme and the differential relevance that they may have during adaptation of P. brasiliensis to different niches during its life cycle, either as an environmental saprophytic mold or as a parasitic yeast cell.
Acknowledgments
This work was supported by COLCIENCIAS Colombia (project no. 2213-343-19183), the Corporación para Investigaciones Biológicas, and the Instituto de Biología of the Universidad de Antioquia. The National Doctoral Program of COLCIENCIAS 2008 supported Orville Hernández.
We thank the GIEM Group of the Universidad de Antioquia, Medellin, Colombia, and especially Carlos Peláez for their help in evaluating cell vitality. We thank Fernando Rodrigues from the School of Health Sciences, University of Minho, Braga, Portugal. We thank Oliver Clay for revision and suggestions for the manuscript. We are grateful to Isaura Torres for valuable collaboration during this project.
Editor: G. S. Deepe, Jr.
Footnotes
Published ahead of print on 27 September 2010.
REFERENCES
- 1.Almeida, A. J., J. A. Carmona, C. Cunha, A. Carvalho, C. A. Rappleye, W. E. Goldman, P. J. Hooykaas, C. Leao, P. Ludovico, and F. Rodrigues. 2007. Towards a molecular genetic system for the pathogenic fungus Paracoccidioides brasiliensis. Fungal Genet. Biol. 44:1387-1398. [DOI] [PubMed] [Google Scholar]
- 2.Almeida, A. J., C. Cunha, J. A. Carmona, B. Sampaio-Marques, A. Carvalho, I. Malavazi, H. Y. Steensma, D. I. Johnson, C. Leao, E. Logarinho, G. H. Goldman, A. G. Castro, P. Ludovico, and F. Rodrigues. 2009. Cdc42p controls yeast-cell shape and virulence of Paracoccidioides brasiliensis. Fungal Genet. Biol. 46:919-926. [DOI] [PubMed] [Google Scholar]
- 3.Andreotti, P. F., J. L. Monteiro da Silva, E. C. Teixeira, M. C. Bertolini, C. P. Soares, G. Benard, and M. J. Mendes-Giannini. 2007. Identification of a gene encoding adaptin-like protein in the Paracoccidioides brasiliensis genome by random amplified polymorphic DNA analysis. J. Med. Microbiol. 56:884-887. [DOI] [PubMed] [Google Scholar]
- 4.Barbosa, M. S., S. N. Bao, P. F. Andreotti, F. P. de Faria, M. S. Felipe, L. dos Santos Feitosa, M. J. Mendes-Giannini, and C. M. Soares. 2006. Glyceraldehyde-3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect. Immun. 74:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beijersbergen, A., A. D. Dulk-Ras, R. A. Schilperoort, and P. J. Hooykaas. 1992. Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science 256:1324-1327. [DOI] [PubMed] [Google Scholar]
- 6.Brummer, E., E. Castaneda, and A. Restrepo. 1993. Paracoccidioidomycosis: an update. Clin. Microbiol. Rev. 6:89-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calich, V. L., A. Purchio, and C. R. Paula. 1979. A new fluorescent viability test for fungi cells. Mycopathologia 66:175-177. [DOI] [PubMed] [Google Scholar]
- 8.da Silva Neto, B. R., J. de Fatima da Silva, M. J. Mendes-Giannini, H. L. Lenzi, C. M. de Almeida Soares, and M. Pereira. 2009. The malate synthase of Paracoccidioides brasiliensis is a linked surface protein that behaves as an anchorless adhesin. BMC Microbiol. 9:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.den Dulk-Ras, A., and P. J. Hooykaas. 1995. Electroporation of Agrobacterium tumefaciens. Methods Mol. Biol. 55:63-72. [DOI] [PubMed] [Google Scholar]
- 10.Donofrio, F. C., A. C. Calil, E. T. Miranda, A. M. Almeida, G. Benard, C. P. Soares, S. N. Veloso, C. M. Soares, and M. J. Mendes Giannini. 2009. Enolase from Paracoccidioides brasiliensis: isolation and identification as a fibronectin-binding protein. J. Med. Microbiol. 58:706-713. [DOI] [PubMed] [Google Scholar]
- 11.Garcia, A. M., O. Hernandez, B. H. Aristizabal, L. E. Cano, A. Restrepo, and J. G. McEwen. 2009. Identification of genes associated with germination of conidia to form mycelia in the fungus Paracoccidioides brasiliensis. Biomedica 29:403-412. [PubMed] [Google Scholar]
- 12.Goldman, G. H., E. dos Reis Marques, D. C. Duarte Ribeiro, L. A. de Souza Bernardes, A. C. Quiapin, P. M. Vitorelli, M. Savoldi, C. P. Semighini, R. C. de Oliveira, L. R. Nunes, L. R. Travassos, R. Puccia, W. L. Batista, L. E. Ferreira, J. C. Moreira, A. P. Bogossian, F. Tekaia, M. P. Nobrega, F. G. Nobrega, and M. H. Goldman. 2003. Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: identification of putative homologues of Candida albicans virulence and pathogenicity genes. Eukaryot. Cell 2:34-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez, A., E. Caro, C. Munoz, A. Restrepo, A. J. Hamilton, and L. E. Cano. 2008. Paracoccidioides brasiliensis conidia recognize fibronectin and fibrinogen which subsequently participate in adherence to human type II alveolar cells: involvement of a specific adhesin. Microb. Pathog. 44:389-401. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez, A., B. L. Gomez, S. Diez, O. Hernandez, A. Restrepo, A. J. Hamilton, and L. E. Cano. 2005. Purification and partial characterization of a Paracoccidioides brasiliensis protein with capacity to bind to extracellular matrix proteins. Infect. Immun. 73:2486-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez, A., B. L. Gomez, C. Munoz, B. H. Aristizabal, A. Restrepo, A. J. Hamilton, and L. E. Cano. 2008. Involvement of extracellular matrix proteins in the course of experimental paracoccidioidomycosis. FEMS Immunol. Med. Microbiol. 53:114-125. [DOI] [PubMed] [Google Scholar]
- 16.Hahn, R. C., A. M. Macedo, C. J. Fontes, R. D. Batista, N. L. Santos, and J. S. Hamdan. 2003. Randomly amplified polymorphic DNA as a valuable tool for epidemiological studies of Paracoccidioides brasiliensis. J. Clin. Microbiol. 41:2849-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton, A. J., L. Jeavons, S. Youngchim, and N. Vanittanakom. 1999. Recognition of fibronectin by Penicillium marneffei conidia via a sialic acid-dependent process and its relationship to the interaction between conidia and laminin. Infect. Immun. 67:5200-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton, A. J., L. Jeavons, S. Youngchim, N. Vanittanakom, and R. J. Hay. 1998. Sialic acid-dependent recognition of laminin by Penicillium marneffei conidia. Infect. Immun. 66:6024-6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurita, N., A. Sano, K. I. Coelho, K. Takeo, K. Nishimura, and M. Miyaji. 1993. An improved culture medium for detecting live yeast phase cells of Paracoccidioides brasiliensis. J. Med. Vet. Mycol. 31:201-205. [DOI] [PubMed] [Google Scholar]
- 20.Kurokawa, C. S., C. R. Lopes, M. F. Sugizaki, E. E. Kuramae, M. F. Franco, and M. T. Peracoli. 2005. Virulence profile of ten Paracoccidioides brasiliensis isolates: association with morphologic and genetic patterns. Rev. Inst. Med. Trop. Sao Paulo 47:257-262. [DOI] [PubMed] [Google Scholar]
- 21.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 22.McMahon, J. P., J. Wheat, M. E. Sobel, R. Pasula, J. F. Downing, and W. J. Martin II. 1995. Murine laminin binds to Histoplasma capsulatum. A possible mechanism of dissemination. J. Clin. Invest. 96:1010-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendes-Giannini, M. J., J. L. Monteiro da Silva, J. de Fatima da Silva, F. C. Donofrio, E. T. Miranda, P. F. Andreotti, and C. P. Soares. 2008. Interactions of Paracoccidioides brasiliensis with host cells: recent advances. Mycopathologia 165:237-248. [DOI] [PubMed] [Google Scholar]
- 24.Mendes-Giannini, M. J., C. P. Soares, J. L. da Silva, and P. F. Andreotti. 2005. Interaction of pathogenic fungi with host cells: molecular and cellular approaches. FEMS Immunol. Med. Microbiol. 45:383-394. [DOI] [PubMed] [Google Scholar]
- 25.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Hook. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 26.Pereira, L. A., S. N. Bao, M. S. Barbosa, J. L. da Silva, M. S. Felipe, J. M. de Santana, M. J. Mendes-Giannini, and C. M. de Almeida Soares. 2007. Analysis of the Paracoccidioides brasiliensis triosephosphate isomerase suggests the potential for adhesin function. FEMS Yeast Res. 7:1381-1388. [DOI] [PubMed] [Google Scholar]
- 27.Puccia, R., S. Schenkman, P. A. Gorin, and L. R. Travassos. 1986. Exocellular components of Paracoccidioides brasiliensis: identification of a specific antigen. Infect. Immun. 53:199-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rappleye, C. A., J. T. Engle, and W. E. Goldman. 2004. RNA interference in Histoplasma capsulatum demonstrates a role for alpha-(1,3)-glucan in virulence. Mol. Microbiol. 53:153-165. [DOI] [PubMed] [Google Scholar]
- 29.Restrepo, A., and B. Jimenez. 1980. Growth of Paracoccidioides brasiliensis yeast phase in a chemically defined culture medium. J. Clin. Microbiol. 12:279-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Restrepo, A., J. G. McEwen, and E. Castaneda. 2001. The habitat of Paracoccidioides brasiliensis: how far from solving the riddle? Med. Mycol. 39:233-241. [DOI] [PubMed] [Google Scholar]
- 31.Restrepo, A., M. Salazar, L. Cano, and M. Patino. 1986. A technique to collect and dislodge conidia produced by Paracoccidioides brasiliensis mycelial form. J. Med. Vet. Mycol. 24:247-250. [PubMed] [Google Scholar]
- 32.Restrepo, A., and A. Tobón. 2009. Paracoccidioides brasiliensis, p. 3357-3364. In B. Mandell, J. Bennett, and R. Dolin (ed.), Mandell, Douglas and Bennett's principles and practice of infectious diseases, 7th ed. Churchill Livingston, Elsevier, Philadelphia, PA.
- 33.Restrepo, S., A. Tobon, J. Trujillo, and A. Restrepo. 1992. Development of pulmonary fibrosis in mice during infection with Paracoccidioides brasiliensis conidia. J. Med. Vet. Mycol. 30:173-184. [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1998. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 35.Serrano, R. 1991. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthase, and energetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Sharpton, T. J., J. E. Stajich, S. D. Rounsley, M. J. Gardner, J. R. Wortman, V. S. Jordar, R. Maiti, C. D. Kodira, D. E. Neafsey, Q. Zeng, C. Y. Hung, C. McMahan, A. Muszewska, M. Grynberg, M. A. Mandel, E. M. Kellner, B. M. Barker, J. N. Galgiani, M. J. Orbach, T. N. Kirkland, G. T. Cole, M. R. Henn, B. W. Birren, and J. W. Taylor. 2009. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 19:1722-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Straus, A. H., E. Freymuller, L. R. Travassos, and H. K. Takahashi. 1996. Immunochemical and subcellular localization of the 43 kDa glycoprotein antigen of Paracoccidioides brasiliensis with monoclonal antibodies. J. Med. Vet. Mycol. 34:181-186. [DOI] [PubMed] [Google Scholar]
- 38.Tronchin, G., M. Pihet, L. M. Lopes-Bezerra, and J. P. Bouchara. 2008. Adherence mechanisms in human pathogenic fungi. Med. Mycol. 46:749-772. [DOI] [PubMed] [Google Scholar]
- 39.Vicentini, A. P., J. L. Gesztesi, M. F. Franco, W. de Souza, J. Z. de Moraes, L. R. Travassos, and J. D. Lopes. 1994. Binding of Paracoccidioides brasiliensis to laminin through surface glycoprotein gp43 leads to enhancement of fungal pathogenesis. Infect. Immun. 62:1465-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villar, L. A., and A. Restrepo. 1989. Virulence of a variant of Paracoccidioides brasiliensis that exists in the yeast form at room temperature. J. Med. Vet. Mycol. 27:141-148. [PubMed] [Google Scholar]