Abstract
The excessive production of proinflammatory cytokines plays a significant role in the pathogenesis of severe malaria. Mammalian macrophage migration inhibitory factor (MIF) (mMIF) is an immune mediator that promotes a sustained proinflammatory response by inhibiting the glucocorticoid-mediated downregulation of inflammation. In addition, Plasmodium parasites also encode a homologue of mammalian MIF that is expressed in asexual-stage parasites. We used the Plasmodium yoelii murine model to study the potential role of parasite-encoded MIF in the pathogenesis of malaria. Antibodies raised against purified, non-epitope-tagged P. yoelii MIF (PyMIF) were used to localize expression in trophozoite- and schizont-stage parasites and demonstrate extracellular release. In vitro, recombinant PyMIF was shown to actively induce the chemotaxis of macrophages but did not induce or enhance tumor necrosis factor alpha (TNF-α) production from peritoneal macrophages. To examine the role of parasite-derived PyMIF in vivo, two transgenic parasite lines that constitutively overexpress PyMIF were generated, one in a nonlethal P. yoelii 17X background [Py17X-MIF(+)] and the other in a lethal P. yoelii 17XL background [Py17XL-MIF(+)]. Challenge studies with transgenic parasites in mice showed that the increased expression of PyMIF resulted in a reduction in disease severity. Mice infected with Py17X-MIF(+) developed lower peak parasitemia levels than controls, while malaria-associated anemia was unaltered. Infection with Py17XL-MIF(+) resulted in a prolonged course of infection and a reduction in the overall mortality rate. Combined, the data indicate that parasite-derived MIF does not contribute significantly to immunopathology but, through its chemotactic ability toward macrophages, may attenuate disease and prolong infection of highly virulent parasite isolates.
Malaria, caused by protozoan Plasmodium spp., is a significant global health problem, with about 3.3 billion people at risk of infection worldwide (30, 33). The disease can range from a mild, uncomplicated febrile illness to severe, life-threatening syndromes, including cerebral malaria and severe malarial anemia (41). Studies with human subjects show that severe malaria often resembles a sepsis-like syndrome, in that the excessive production of proinflammatory cytokines such as tumor necrosis factor alpha (TNF-α), interleukin-1 (IL-1), IL-6, and gamma interferon (IFN-γ) can contribute to pathology (19, 20, 34). A fine balance in the regulation of the inflammatory response by modulatory anti-inflammatory cytokines such as IL-10 and transforming growth factor β (TGF-β) is required to control parasite replication while limiting immunopathology (25, 46). Data from studies of the rodent malaria parasites Plasmodium yoelii, Plasmodium chabaudi, and Plasmodium berghei also showed that the timing and magnitude of TNF-α, IFN-γ, TGF-β, and IL-10 production markedly influence disease severity and infection outcome (40, 44, 45). One key immunomodulator that likely influences the overall balance between proinflammatory and anti-inflammatory responses during malaria is macrophage migration inhibitory factor (MIF) (21).
Mammalian MIF (mMIF), one of the earliest cytokines to be discovered, is homotrimeric in structure and functions as an upregulator of the proinflammatory cascade (14). mMIF is expressed in several immune cell types, including activated T cells, monocytes/macrophages, and eosinophils as well as in cells and tissues of the neuroendocrine system, lung, skin, and gastrointestinal tract (9, 14). mMIF exerts its action mainly by inhibiting glucocorticoid-mediated anti-inflammatory responses (13). A mMIF receptor complex that includes CD74, the major histocompatibility complex (MHC) class II invariant chain, along with CD44, a second surface glycoprotein, has been identified (39, 54). mMIF binds directly to CD74, while CD44 serves as the intracellular signaling component. The binding of mMIF to its receptor complex enables the activation of extracellular signal-regulated kinase (ERK) signaling cascades and the upregulation of the proinflammatory response. Mammalian MIF has been shown to be important in the pathology of several inflammatory conditions, including endotoxic shock, rheumatoid arthritis, atherosclerosis, and cancer (7, 8, 14). Studies of malaria in human subjects and in animal models have also shown a role for host MIF. Previously reported studies of P. chabaudi indicated a pathogenic role for mMIF specifically in the development of severe malarial anemia (42, 43). mMIF knockout (KO) mice infected with P. chabaudi showed increased survival and reduced anemia during infection. Studies of human subjects in areas where malaria is endemic point to a similar pathogenic role for mMIF. Increased mMIF production was associated with heightened inflammatory responses in cases of cerebral malaria and placental malaria (16-18, 35). Recent data suggest that polymorphisms in the mammalian mif promoter associated with elevated levels of mMIF production may increase the risk for developing severe malaria (5, 61). It is important to point out, however, that several studies of uncomplicated and severe P. falciparum malaria assign a protective role for mMIF. In some cases, high circulating levels of mMIF were associated with reduced anemia and with milder episodes of malaria in children (3, 4, 6). An explanation for the apparent discrepancies is not obvious. One factor not specifically considered in those previous studies is that all genomes of Plasmodium parasites examined to date also contain mif homologues (2, 29).
Studies of P. berghei-, P. falciparum-, and P. yoelii-encoded MIF revealed a 116-amino-acid protein with ∼30% amino acid sequence identity with mMIF (2, 22, 24, 52). Collectively, the data suggest that plasmodial MIF (pMIF) is structurally similar to mMIF and possesses some but probably not all activities normally attributed to mMIF. As such, the hypothesis that parasite-encoded MIF could enhance proinflammatory responses and contribute to the severity of malaria is reasonable. Infection of mice with P. berghei mif (pbmif) knockout parasites led to earlier and increased reticulocytosis. However, the development of cerebral complications in C57BL/6 mice and hyperparasitemia and severe anemia in BALB/c mice did not differ upon infection with P. berghei wild-type or pbmif knockout parasites (2). While in vitro studies suggested that it is likely that pMIF modulates host immune responses, additional in vivo studies are needed to evaluate its effect on infection outcome (2, 22). Here, we describe the functional properties of recombinant, non-epitope-tagged, P. yoelii MIF (PyMIF) and provide evidence that disease severity is reduced upon infection with nonlethal and lethal P. yoelii transgenic parasites engineered for the increased expression of PyMIF.
MATERIALS AND METHODS
Experimental animals and parasites.
Five- to six-week-old male BALB/cByJ and C57BL/6J mice were purchased from the Jackson Laboratories (Bar Harbor, ME). All animals were housed in the Animal Care Facility of the Drexel University College of Medicine under specific-pathogen-free conditions. Lethal P. yoelii 17XL and nonlethal P. yoelii 17X parasites were originally obtained from William P. Weidanz (University of Wisconsin, Madison, WI) and maintained as cryopreserved stabilates. For challenge studies, mice were infected intraperitoneally (i.p.) with either 1 × 105 or 1 × 106 P. yoelii parasitized red blood cells (RBCs) (pRBCs), as indicated. Infections were monitored by enumerating pRBCs in thin smears of tail blood stained with Giemsa stain and expressed as percent parasitemia, calculated as follows: (number of pRBCs/total number of RBCs) × 100. Hemoglobin concentrations were monitored during infection by using Drabkin's reagent and procedures described previously (43). According to the policy of the Institutional Animal Care and Use Committee of the Drexel University College of Medicine regarding P. yoelii 17XL infections, mice were sacrificed when parasitemia exceeded 50%, and infections were scored as lethal.
RNA isolation and quantitative real-time PCR.
RBCs were collected from P. yoelii-infected mice when parasitemia was between 15 and 25%. Following saponin lysis of erythrocytes and leukocytes, intact P. yoelii parasites were resuspended in Trizol reagent (Invitrogen, Carlsbad, CA), and total RNA was extracted, precipitated, and purified by using the RNeasy RNA isolation kit (Qiagen, Valencia, CA). Similarly, RNA was isolated from ring-, trophozoite-, and schizont-stage P. yoelii parasites following density gradient centrifugation on a discontinuous Percoll gradient (GE Life Sciences, Piscataway, NJ) as previously described (53).
Real-time PCR was used to quantitate the levels of PyMIF RNA using P. yoelii mif (pymif) forward primer 5′-TCAAAATGCGTTATCCGAAAT-3′ and pymif reverse primer 5′-ACTTCCAGAAAATCGGAGGTT-3′ as previously described (48). Assays were performed by using an Applied Biosystems 7900HT real-time PCR system (Applied Biosystems, Foster City, CA), and data were analyzed by using the Applied Biosystems sequence detection system (v2.2.1). Serial dilutions of the input cDNA (12 ng/well to 600 pg/well) were used to generate a standard curve for pymif as well as a 60S ribosomal protein L23 gene (PY07409 [http://PlasmoDB.org]) as a reference. All reactions were carried out in triplicate. The relative expression of pymif normalized to the endogenous reference was calculated based on the threshold cycle of product detection.
Expression and purification of recombinant PyMIF.
The recombinant PyMIF protein (rPyMIF) was expressed in Escherichia coli using pET plasmid vectors and a T7 RNA polymerase expression system (12). The complete coding region of pymif was PCR amplified from cDNA clone MS36, obtained from a sequenced P. yoelii 17XL blood-stage cDNA library (15, 23), using forward primer 5′-GATATCATATGATGCCTTGCTGCGAATTAATAACAAAC-3′ and reverse primer 5′-GAATTCTCGAGTTAGCCAAATAGTGAACCACTAAAA-3′ (NdeI and XhoI restriction sites used for subcloning into the pET-30a plasmid [Novagen, Madison, WI] are underlined). The sequence of the pymif cDNA insert was confirmed and shown to be identical to the deposited GenBank sequence under accession number DQ494171 (52).
E. coli BL21(DE3)pLysS cells (Novagen) were transformed with the pET-30a-pymif construct. Bacteria from induced cultures were harvested and resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 10 mM EDTA) containing 1 mg/ml lysozyme (Sigma-Aldrich, St. Louis, MO). A 40% to 60% ammonium sulfate fraction of the soluble lysate containing rPyMIF was then recovered and separated by preparative isoelectric focusing using the Bio-Rad Rotofor system with 1% Bio-Lyte ampholyte (pH range, 3 to 10) according to the manufacturer's instructions (Bio-Rad Laboratories, Hercules, CA). The fractions containing rPyMIF were then subjected to hydrophobic interaction chromatography using Phenyl Sepharose 6 Fast Flow (high substitution) beads (Amersham Biosciences, Piscataway, NJ) using a stepwise gradient of 1.5 M ammonium sulfate to 0.2 M ammonium sulfate. Eluted rPyMIF was dialyzed into 25 mM Tris-HCl (pH 8.0)-100 mM NaCl. Any residual endotoxin was removed by passage over polymyxin B beads (Detoxi-Gel; Pierce Chemical Company, Rockford, IL). The protein concentration was determined by a bicinchoninic acid (BCA) protein assay (Pierce), and purity was assessed by staining with Coomassie blue following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 4-to-15% gradient gels. Endotoxin levels measured by using a Limulus amebocyte lysate test (Cambrex Bioscience, Walkersville, MD) were <0.01 ng per microgram of rPyMIF. The yield of purified rPyMIF was approximately 1 mg per liter of induced bacterial culture.
Analysis of the native PyMIF protein.
To generate high-titer polyclonal antisera against PyMIF, BALB/cByJ mice (n = 5) were immunized subcutaneously three times at 3-week intervals with 25 μg of purified rPyMIF formulated with 25 μg of Quil A as an adjuvant (Accurate Chemical and Scientific Corp., Westbury, NY). Control animals were immunized with Quil A alone. Sera were collected 2 weeks following the final immunization and pooled. In subsequent assays, serum collected from mice immunized with Quil A alone was used as a negative control and is referred to as normal mouse serum (NMS).
P. yoelii 17X proteins were metabolically labeled with [35S]methionine and [35S]cysteine (EasyTag Express, 1,000 Ci/mmol; Perkin-Elmer, Waltham, MA) as previously described (11). The cell-free culture supernatant was collected and stored at −80°C until further use. Pelleted RBCs were solubilized in buffer containing 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 5 mM EDTA, 1% Triton X-100, and 0.5% deoxycholate. Mouse anti-rPyMIF sera were used to immunoprecipitate PyMIF from 1 × 106 cpm of metabolically labeled proteins from the culture supernatant and parasite-associated fractions. Antigen-antibody complexes were collected by using protein G-agarose beads (Invitrogen) and separated by SDS-PAGE on 15% polyacrylamide gels. Precipitated PyMIF was visualized by autoradiography on fixed gels treated with Amplify fluorographic reagent (GE Life Sciences).
Total P. yoelii 17X antigen was prepared, separated by SDS-PAGE on 15% polyacrylamide gels, and used for immunoblot assays as previously described (12). Similar immunoblot assays were completed by using purified rPyMIF and recombinant human MIF (R&D Systems, Inc.). Blots were blocked and probed with NMS or polyclonal mouse anti-rPyMIF serum diluted 1:1,000 in Tris-buffered saline (TBS) containing 1% bovine serum albumin (BSA) and 0.1% Tween 20. Bound antibodies were detected by chemiluminescence using horseradish peroxidase-conjugated rabbit anti-mouse IgG(H+L) (1:2,000; Zymed, South San Francisco, CA) and SuperSignal West Pico substrate (Pierce). Equivalent loading of parasite lysates in each lane was monitored with immunoblots probed with rabbit antisera specific for PyMSP-8, a trophozoite and merozoite membrane protein of P. yoelii (12). Densitometric analysis was performed by using the uncalibrated OD function of ImageJ software (v1.42q) (National Institutes of Health, Bethesda, MD).
Indirect immunofluorescence assay.
P. yoelii 17XL pRBCs were washed in phosphate-buffered saline (PBS) (pH 7.4) and fixed in suspension by overnight incubation at 4°C with 4% formaldehyde and 0.0075% glutaraldehyde (57). pRBCs were then washed, permeabilized by incubation with 0.1% Triton X-100 for 10 min at room temperature, and treated for an additional 10 min with 0.1 mg/ml sodium borohydride. Following incubation with PBS containing 3% bovine serum albumin to reduce nonspecific binding, cells were incubated with mouse anti-rPyMIF sera (1:1,000) overnight at 4°C. Primary antibody was detected by using goat anti-mouse IgG(H+L)-fluorescein isothiocyanate (FITC) (1:400) (Invitrogen, CA). Parasite nuclei were stained by using SlowFade Gold containing 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Images (×1,000 magnification) were obtained by using an Olympus BX60 fluorescent microscope (Olympus America, Inc., Melville, NY) and a Spot RT Slider digital camera system (Diagnostic Instruments, Sterling Heights, MI).
Assays of rPyMIF-elicited TNF-α production and migration by peritoneal macrophages.
C57BL/6J mice were injected i.p. with 1 ml of 3% Brewer's thioglycolate medium (Sigma-Aldrich). Five days later, peritoneal exudate cells were harvested by lavage with 10 ml cold Hanks’ buffered salts solution (HBSS). Macrophages were adhered to tissue culture plates by incubation in RPMI medium containing 10% fetal bovine serum (Gibco), 20 mM HEPES (pH 7.0), 0.15% sodium bicarbonate, and penicillin (100 units/ml)-streptomycin (100 μg/ml) (Invitrogen) for 1 h at 37°C in 5% CO2.
To test for the ability to elicit a proinflammatory response, peritoneal macrophages (1 × 105 cells/well) were cultured with medium alone or with rPyMIF (10 ng/ml to 500 ng/ml) in the presence or absence of 1 ng/ml of lipopolysaccharide (LPS) (Sigma-Aldrich). Culture supernatants were harvested following 24 h and 48 h of incubation at 37°C in 5% CO2. The concentration of TNF-α in each culture supernatant was determined by using BD Biosciences OptEIA cytokine enzyme-linked immunosorbent assay (ELISA) kits (BD Biosciences, San Jose, CA) according to the manufacturer's protocol. In control experiments, rPyMIF was pretreated with 2 μg of recombinant E. coli methionine amino peptidase (reMAP) (R&D Systems, Minneapolis, MN) for 15 min at 37°C to remove the N-terminal methionine residue.
For chemotaxis assays, peritoneal macrophages (1 × 105 to 2 × 105 cells) were loaded into the upper chamber of a 5-μm Chemo-Tx plate (Neuroprobe, Gaithersburg, MD). Chemoattractants, including monocyte chemoattractant protein 1 (MCP-1) (50 ng/ml) (R&D Biosystems), formyl-methionine-leucine-phenylalanine peptide (f-MLP) (10−7 M) (Sigma-Aldrich), and rPyMIF (50 ng/ml to 1 μg/ml), were loaded into the lower chambers. The transwell plates were incubated for 3 h at 37°C in 5% CO2. Macrophages that had migrated through the filter were dislodged by using 2 mM EDTA. Following staining with trypan blue (Fluka/Sigma-Aldrich), migrated cells were counted by light microscopy. Data are expressed as percent chemotaxis, calculated as follows: (number of macrophages in the lower chamber/number of input macrophages in the upper chamber) × 100. In control experiments, rPyMIF was heat inactivated by incubation at 65°C for 30 min prior to testing in the chemotaxis assay.
Infection-induced antibodies to PyMIF.
Two groups of BALB/cByJ mice (five per group) were infected i.p. with 1 × 105 P. yoelii 17X pRBCs, and parasitemia was monitored. One week following the clearance of parasites from circulation, primary infection sera were collected from the first group of mice. The second group of mice was rechallenged i.p. with 1 × 107 P. yoelii 17X parasitized RBCs 4 weeks later. Parasitemia in rechallenged animals remained subpatent. Four weeks later, mice were infected a third time as described above with 1 × 107 P. yoelii 17X pRBCs. One week after this rechallenge, sera from the tertiary infection were collected for analysis of antibody responses. The level of anti-PyMIF antibodies present in serially diluted (1:100 to 1:3,200) sera from primary and tertiary P. yoelii 17X infection was measured by ELISA as previously described (48).
Generation of PyMIF transgenic parasites.
Plasmid pPbGFPcon was obtained from Andrew Waters (Leiden University Medical Center, Leiden, Netherlands). The vector contains a gfp expression cassette under the control of a constitutive P. berghei ef-1aα (Pbef-1aα) promoter, flanked by sequences to target insertion into the c- and/or d-ssu-rrna (d-small subunit-rRNA) loci by a single-crossover event (28). P. berghei ssu-rrna genes are 96% identical to P. yoelii ssu-rrna genes. Plasmid pPY00068-OE contains a similar expression cassette with the same Pbef-1aα promoter but is designed to replace the pydhfr locus by a double-crossover event. Both plasmids contain the pyrimethamine-resistant Toxoplasma gondii dhfr-ts (Tg-dhfr-ts) gene as a selectable marker. A BamHI pymif cDNA fragment was cloned into gel-purified pPbGFPcon after the excision of the gfp gene fragment to generate pPyMIF(rrna). Similarly, pymif cDNA was cloned into plasmid pPY00068-OE as a BamHI fragment after the excision of the PY00068 gene to generate pPyMIF(dhfr). The correct integration of the transgene into both plasmids was confirmed by sequencing. Prior to the transfection of P. yoelii 17X, pPyMIF(rrna) was linearized by digestion with ApaI and SacII. Prior to the transfection of P. yoelii 17XL, pPyMIF(dhfr) was digested with KpnI and SacII. Transgenic P. yoelii 17X and P. yoelii 17XL parasites with a similar Pbef-1aα promoter-gfp expression cassette and the Tg-dhfr-ts gene in the c/d-ssu-rrna loci were used as controls.
Transfection of P. yoelii parasites was carried out as described previously for P. berghei, with minor modifications (36). P. yoelii pRBCs were collected, washed, and resuspended in 50 ml of RPMI 1640 medium containing 25 mM HEPES (pH 7.0), 7.5% NaHCO3, penicillin (100 units/ml)-streptomycin (100 μg/ml) (Invitrogen), and 20% fetal bovine serum (Gibco). The cell suspension was placed into a 250-ml conical flask, flushed with a blood-gas mix (5% CO2, 5% O2, and 90% N2), and capped tightly. The flask was incubated at 37°C for 4 h with slow shaking to keep the cells in suspension. Late-stage parasites were isolated by density gradient centrifugation through a 60% Percoll solution. Schizont-enriched pRBCs were washed, resuspended in transfection buffer (Amaxa Nucleofactor parasite transfection kit 2; Walkersville, MD), and electroporated with 10 μg of linearized plasmid using the Amaxa Nucleofactor electroporation device (program U33), according to the manufacturer's protocol. Transfected pRBCs were injected intravenously (i.v.) into naïve BALB/cByJ mice. Beginning 24 h later, mice were treated i.p. with pyrimethamine (60 μg/mouse/day; Sigma) for four consecutive days. When parasitemia with drug-resistant P. yoelii parasites became patent, stabilates were prepared and cryopreserved.
The integration of the pymif expression cassette and the Tg-dhfr-ts gene was confirmed by PCR, and Py17X-MIF(+) and Py17XL-MIF(+) transgenic parasite lines were selected for cloning. For cloning, BALB/cByJ mice (three to four per group) were infected i.v. with 25, 10, or 1 P. yoelii transgenic parasite. Pyrimethamine was administered as described above, and drug-resistant parasites from the lowest infective dose were selected as clones. The integration of the pymif expression cassette at the targeted loci was characterized by diagnostic PCR and Southern blot analysis (see Fig. S1 to S4 in the supplemental material).
Statistical analysis.
The statistical significance of differences in mean peak parasitemias and degrees of anemia was calculated by an unpaired Student's t test (two-tailed). The significance of differences in survival between groups was determined by a Mantel-Haenszel log rank test (GraphPad Prism 4.0; GraphPad Software Inc., San Diego, CA). P values of <0.05 were considered significant.
RESULTS
Expression of P. yoelii MIF.
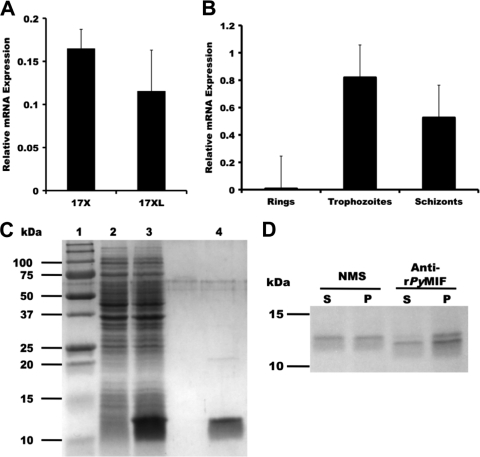
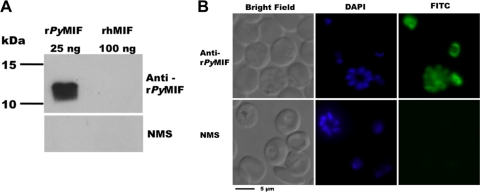
To begin to investigate pMIF, the sequence and expression profile of the mif gene of P. yoelii were determined. Analysis of a P. yoelii 17XL mif cDNA clone confirmed the sequence and the intron-exon structure reported previously by Shao et al. (52). By reverse transcription (RT)-PCR analysis, pymif was found to be expressed by both lethal P. yoelii 17XL and nonlethal P. yoelii 17X strains (Fig. 1A). The expression of pymif was highest in trophozoites, with minimal expression seen in ring-stage parasites (Fig. 1B). Non-epitope-tagged recombinant PyMIF was expressed in E. coli and purified from a soluble bacterial lysate by ammonium sulfate fractionation, preparative isoelectric focusing, and hydrophobic interaction chromatography (Fig. 1C). Polyclonal mouse serum raised against rPyMIF was used to immunoprecipitate metabolically labeled P. yoelii proteins. A specific protein of ∼12 kDa was identified in the parasite-associated fraction of P. yoelii proteins as well as in the fraction of proteins released into the culture supernatant during short-term culture (Fig. 1D). The polyclonal anti-rPyMIF antibody was parasite specific and showed no cross-reactivity with mMIF by immunoblot assay, even when loaded in excess concentrations (Fig. 2A). Indirect immunofluorescence assays with rPyMIF-specific antibodies confirmed the RT-PCR results, with the expressed protein being detected primarily in trophozoites and schizonts (Fig. 2B). Little staining of the erythrocyte cytoplasm was noted. These data are consistent with the conclusion that pymif is expressed as blood-stage parasites mature through the trophozoite and schizont stages and is released extracellularly upon schizont rupture, where it is available to interact with host immune cells.
FIG. 1.
Native and recombinant PyMIF. (A and B) The expression of the pymif gene was measured by quantitative RT-PCR in P. yoelii 17X and P. yoelii 17XL blood-stage parasites (A) and in the ring, trophozoite, and schizont stages of P. yoelii 17X (B), isolated by density gradient centrifugation. pymif mRNA levels are expressed relative to those of PY07409, a 60S P. yoelii ribosomal protein gene. All reactions were carried out in triplicate. (C) Coomassie blue-stained SDS-polyacrylamide gel containing E. coli lysates of uninduced (lane 2) and induced (lane 3) cells expressing rPyMIF and purified rPyMIF (lane 4). (D) Immunoprecipitation of metabolically labeled P. yoelii proteins released into culture supernatants (S) or remaining parasite associated (P), using anti-rPyMIF and normal mouse sera.
FIG. 2.
(A) Mouse anti-rPyMIF antibodies do not recognize mammalian MIF. Immunoblots of 15% polyacrylamide gels containing rPyMIF (25 ng) and recombinant human MIF (100 ng) were probed with anti-rPyMIF sera or normal mouse sera (NMS) diluted 1:1,000. Bound antibody was detected by chemiluminescence using rabbit anti-mouse IgG conjugated to horseradish peroxidase (1:10,000). (B) Localization of PyMIF in P. yoelii-infected erythrocytes. PyMIF was detected in parasitized erythrocytes by indirect immunofluorescence using P. yoelii 17XL parasites fixed in 4% formaldehyde and 0.0075% glutaraldehyde using anti-rPyMIF (top row) or normal mouse (bottom row) sera. Bound antibody was detected with goat anti-mouse IgG conjugated to FITC. Parasite nuclei were stained with DAPI.
In vitro assay of functional activity of rPyMIF.
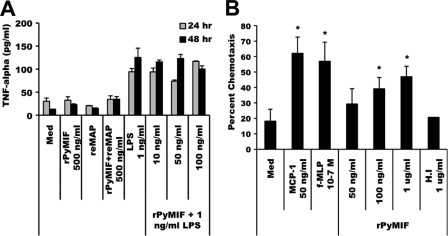
The ability of mMIF to enhance proinflammatory responses is one of its key functions as a modulator of host immune responses. To determine if pMIF possesses this same activity, rPyMIF was tested for its ability to induce and/or enhance TNF-α production by macrophages. Mouse peritoneal macrophages were cultured with rPyMIF in the presence or absence of LPS. The concentrations of TNF-α in culture supernatants after 24 h and 48 h of incubation were determined by ELISA. As shown in Fig. 3A, rPyMIF alone, at a concentration of 500 ng/ml, did not induce significant TNF-α release from peritoneal macrophages upon prolonged incubation compared to medium-alone controls. This was in contrast to incubation with LPS, which readily stimulated the release of TNF-α by macrophages after 24 h of culture at a concentration of only 1 ng/ml. The culture of peritoneal macrophages with LPS (1 ng/ml) combined with increasing concentrations of rPyMIF (10 to 100 ng/ml) did not increase TNF-α release over that observed with LPS alone after 24 h or 48 h of incubation. In mMIF, the N-terminal methionine is cleaved to yield a mature protein with an N-terminal proline residue (56). Sequencing of the N terminus of rPyMIF showed that the initiating methionine residue was retained in the bacterial product. To determine if this affected the ability of rPyMIF to stimulate TNF-α production, rPyMIF was treated with recombinant E. coli methionine amino peptidase (reMAP) to excise the N-terminal methionine residue. No difference was seen in TNF-α production when macrophages were incubated with reMAP-treated PyMIF alone (Fig. 3A) or in combination with LPS (data not shown). These data suggest that unlike its mammalian counterpart, pMIF itself does not elicit significant TNF-α production by macrophages or contribute to the ability of macrophages to enhance or sustain an ongoing response to a proinflammatory stimulus.
FIG. 3.
Functional activity of rPyMIF. (A) TNF-α release. Thioglycolate-elicited peritoneal macrophages (1 × 105 cells/well) were cultured in the presence of LPS alone (1 ng/ml), rPyMIF alone (500 ng/ml), or LPS (1 ng/ml) plus increasing concentrations of rPyMIF (10 to 100 ng/ml). In control experiments, rPyMIF was pretreated with recombinant methionine amino peptidase (reMAP) to remove the N-terminal methionine residue. Culture supernatants were harvested after 24 h or 48 h, and the TNF-α concentration was measured by ELISA. Basal levels of TNF-α release were determined from cells cultured in medium (Med) alone. (B) Chemotaxis. Murine peritoneal macrophages (1 × 105 to 2 × 105 cells/well) were loaded into the top well of a transwell chamber, and increasing concentrations of rPyMIF were loaded into the lower chambers. The migration of macrophages across the transwell membrane after a 3-h incubation was quantitated and expressed as percent chemotaxis. MCP-1 and f-MLP served as positive controls. Heat-inactivated (H.I) rPyMIF and medium alone were used as negative controls. An asterisk indicates significant differences in values (P < 0.05) relative to those of medium alone.
A second hallmark activity associated with mMIF is the ability to induce the chemotaxis of macrophages (31). Peritoneal macrophages in the upper chamber of a transwell assay plate were tested for the ability to migrate into the lower chamber of the transwell containing increasing concentrations of rPyMIF in comparison to known chemoattractants (MCP-1 and f-MLP). As shown in Fig. 3B, rPyMIF induced the migration of macrophages in a dose-dependent manner. Heat denaturation of rPyMIF reduced the chemotaxis of macrophages to background levels, suggesting that the conformation of rPyMIF is important for activity. These data suggest that like its mammalian counterpart, pMIF does possess the ability to induce the migration of macrophages along a concentration gradient.
Antibody response to PyMIF during blood-stage infection.
To investigate the contribution of PyMIF to the outcome of infection in vivo, two sets of experiments were initially completed. First, the antibody response to PyMIF upon infection of naïve BALB/c mice with nonlethal P. yoelii 17X was evaluated. Serum collected from mice 1 week following the resolution of P. yoelii 17X infection contained detectable levels of anti-PyMIF antibodies, as measured by ELISA (Fig. 4). Furthermore, this antibody response was markedly boosted upon repeated reinfection, demonstrating the immunogenicity of PyMIF during the natural course of blood-stage malaria. This increase in the production of PyMIF-specific antibodies coincided with the ability of mice to resist reinfection with homologous parasites, suggesting that anti-PyMIF antibodies may contribute to protective immune responses. A second experiment was completed to investigate this further. BALB/c mice were immunized three times at 3-week intervals with 25 μg of rPyMIF formulated with Quil A as an adjuvant. Upon P. yoelii 17X challenge infection, however, rPyMIF-immunized mice exhibited modest increases in mean peak parasitemia and infection-induced anemia (data not shown). This trend toward a slightly increased parasite burden did not reach statistical significance. The data suggested that anti-PyMIF antibodies are not protective.
FIG. 4.
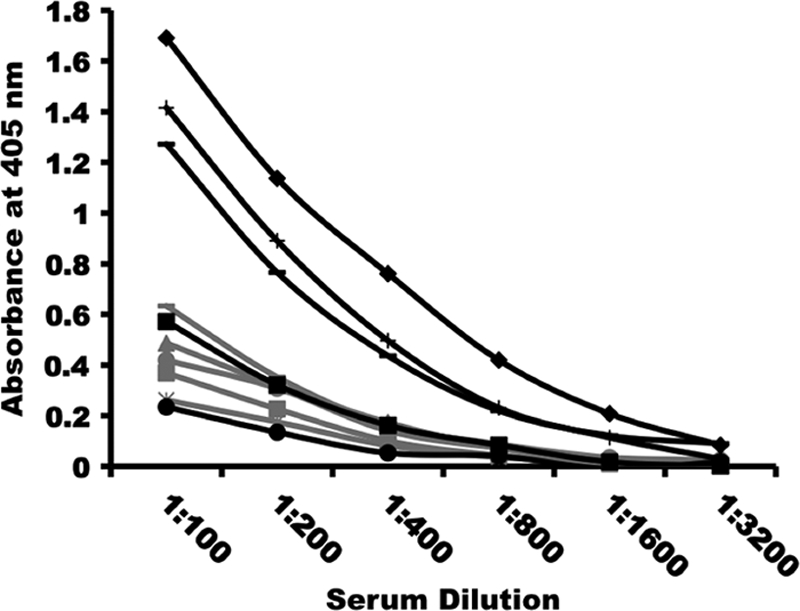
Antibody responses to PyMIF are boosted upon repeated P. yoelii infection. BALB/cByJ mice (n = 5) were infected with 1 × 105 P. yoelii 17X pRBCs. Primary infection sera were collected 30 days postinfection, approximately 1 week following the clearance of blood-stage parasites. A second group of BALB/cByJ mice (n = 5) was infected as described above with 1 × 105 P. yoelii 17X pRBCs. Following the clearance of the primary infection, these mice were rechallenged twice with 1 × 107 P. yoelii 17X pRBCs. Tertiary infection sera were collected 1 week following the clearance of the third P. yoelii challenge infection. Anti-PyMIF antibodies present in primary (gray) and tertiary (black) infection sera were measured by ELISA using rPyMIF-coated plates. Plots depict PyMIF-specific antibody responses for individual mice in both groups.
Overexpression of PyMIF reduces peak parasitemia during nonlethal P. yoelii 17X malaria.
To examine the in vivo influence of parasite-expressed pMIF on the severity of disease more closely, transgenic P. yoelii 17X parasites capable of overexpressing PyMIF were generated. Using a previously described targeting strategy and vector, a second copy of the PyMIF gene under the control of the constitutive P. berghei ef-1aα promoter was targeted for insertion into the c/d-ssu-rrna gene loci (see Fig. S1 in the supplemental material). The integration of the pymif transgene into a cloned P. yoelii 17X line, designated Py17X-MIF(+), was confirmed by diagnostic PCR and Southern blot analysis. Genetic analysis showed that in addition to the endogenous pymif gene, Py17X-MIF(+) parasites contained two copies of the pymif transgene construct integrated in tandem into the c-ssu-rrna gene (Fig. S2A to S2C). The constitutive expression of pymif was confirmed by RT-PCR analysis, with the greatest increase in expression noted for ring-stage parasites (Fig. S2C). By immunoblot analysis and quantitation by densitometry, the PyMIF protein level in Py17X-MIF(+) transgenic parasites was increased approximately 5.5-fold over that of wild-type P. yoelii 17X parasites (Fig. 5).
FIG. 5.
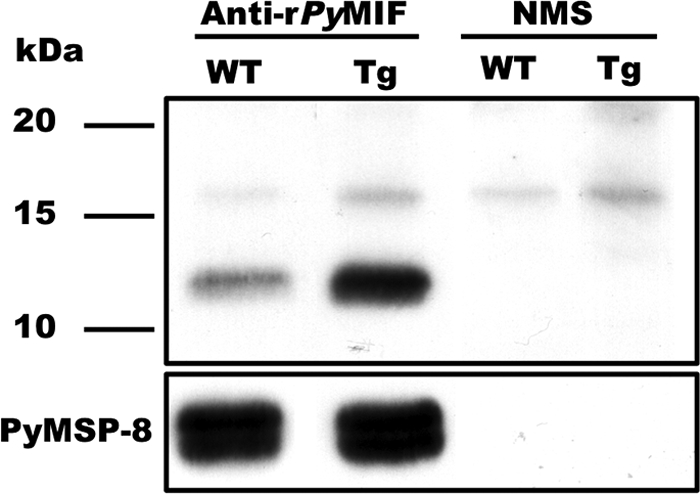
Elevated expression of PyMIF in Py17X-MIF(+) transgenic parasites. Lysates of P. yoelii 17X (wild type [WT]) and Py17X-MIF(+) (transgenic [Tg]) blood-stage parasites were separated by SDS-PAGE, and the levels of PyMIF were compared by immunoblot analysis. Blots were probed with rPyMIF-specific polyclonal sera (1:2,000) or a pool of NMS. The total amount of parasite protein loaded per lane (6 μg) was normalized based on the reactivity of rabbit anti-PyMSP8 on parallel immunoblots. The expression profile of PyMSP8 across blood-stage malaria parasites (53) was similar to that observed for endogenous PyMIF.
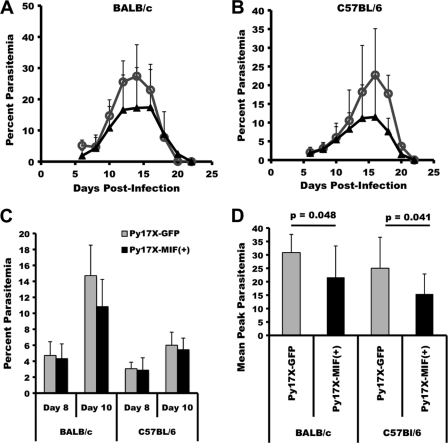
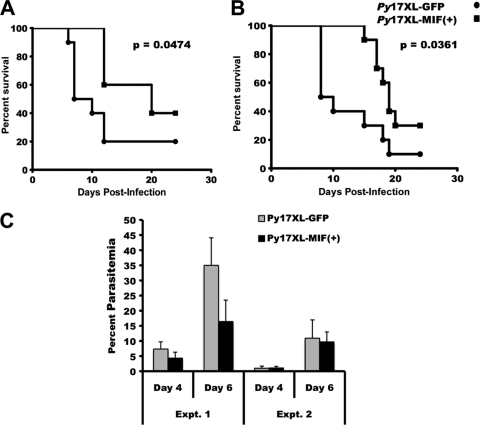
BALB/cByJ and C57BL/6 mice were infected with Py17X-MIF(+) parasites, and the resulting parasitemia and anemia were monitored. P. yoelii 17X transgenic parasites containing a similar Pb-ef-1aα-gfp expression cassette and selectable marker were used for control infections. As shown in Fig. 6, BALB/cByJ and C57BL/6 mice challenged with Py17X-MIF(+) parasites showed a modest decrease in the severity of infection. The mean peak parasitemia for BALB/cByJ mice infected with Py17X-MIF(+) parasites was 21.7% ± 11.7%, compared to 30.9% ± 6.8% for Py17X-green fluorescent protein (GFP)-infected controls (P = 0.048) (Fig. 6D). Similarly, the mean peak parasitemia for Py17X-MIF(+)-infected C57BL/6 mice was 15.5% ± 7.4%, significantly lower than the mean peak parasitemia of 25.0% ± 11.5% observed for Py17X-GFP-infected control mice (P = 0.04) (Fig. 6D). For both strains of mice infected with Py17X-MIF(+) and Py17X-GFP transgenic parasites, no difference in the prepatent period of infection, the slope of the ascending parasitemia curve, day 8 parasitemia, or day 10 parasitemia was noted (Fig. 6A to C). As such, the in vivo growth rates of the two transgenic parasites during the first 8 to 10 days of infection were comparable. As shown in Fig. 7, a moderate, infection-induced anemia developed in both BALB/cByJ and C57BL/6 mice. The anemia observed paralleled the course of infection, with the most significant anemia associated with peak parasitemia and the subsequent return to baseline levels with the suppression of parasite growth. However, neither the severity of anemia nor the period of anemia differed between mice infected with Py17X-MIF(+) and those infected with Py17X-GFP. Somewhat unexpectedly, these data indicate that the increased expression of pMIF during blood-stage malaria does not increase the severity of disease but may in fact attenuate parasite virulence.
FIG. 6.
Overexpression of PyMIF reduces peak parasitemia during nonlethal P. yoelii 17X malaria. (A and B) Groups of BALB/cByJ (A) and C57BL/6 (B) mice (10 animals/group) were infected i.p. with 1 × 105 Py17X-GFP (○) or Py17X-MIF(+) (▴) parasites and monitored for parasitemia. The mean percent parasitemia (± standard deviation [SD]) in each group is plotted versus days postinfection. (C) The mean percent parasitemias (±SD) on day 8 and day 10 of infection were comparable for Py17X-GFP- and Py17X-MIF(+)-infected mice. (D) Mean peak parasitemia (±SD) was significantly reduced (P < 0.05 by an unpaired Student's t test) in Py17X-MIF(+)-infected mice relative to Py17X-GFP-infected controls irrespective of the mouse genetic background.
FIG. 7.
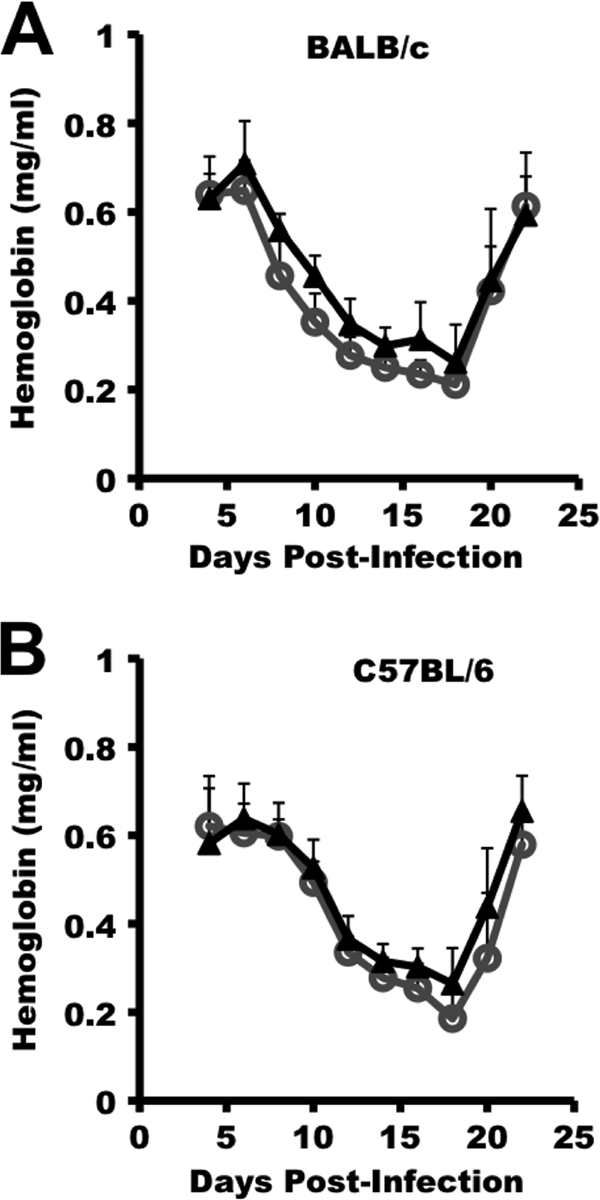
Overexpression of PyMIF does not alter P. yoelii 17X malaria-associated anemia. Groups of BALB/cByJ (A) and C57BL/6 (B) mice (10 animals/group) were infected i.p. with 1 × 105 Py17X-GFP (○) or Py17X-MIF(+) parasites (▴) (Fig. 4), and blood hemoglobin levels were monitored throughout the course of infection. No significant difference in infection-associated anemia was observed between Py17X-GFP- and Py17X-MIF(+)-infected mice irrespective of the mouse genetic background (P > 0.05 by an unpaired Student's t test).
Overexpression of PyMIF reduces the severity of P. yoelii 17XL malaria.
To extend these studies and test the hypothesis that the expression of pMIF has the potential to reduce the severity of blood-stage malaria, a similar pymif transgenic parasite line was constructed by using P. yoelii 17XL, a virulent, typically lethal strain of malaria parasites. In this case, the Pb-ef-1aα-pymif transgene was targeted for insertion at the pydhfr locus (see Fig. S3 in the supplemental material). As described above, the integration of the pymif transgene into a cloned P. yoelii 17XL line, designated Py17XL-MIF(+), was confirmed by diagnostic PCR and Southern blot analysis. Genetic analysis of Py17XL-MIF(+) parasites showed the presence of an intact, endogenous copy of the pymif gene and the integration of the Pb-ef-1aα-pymif construct at the 5′ end of the pydhfr gene locus via a single-crossover event instead of the expected double crossover (Fig. S4A to S4C). The increased expression of pymif Py17XL-MIF(+) parasites was assessed at both the RNA and protein levels. Quantitative RT-PCR analysis indicated an increase in the level of expression of approximately 3.6-fold (data not shown), which was comparable to the increase of 3.7-fold observed at the protein level measured by immunoblot analysis (Fig. 8).
FIG. 8.
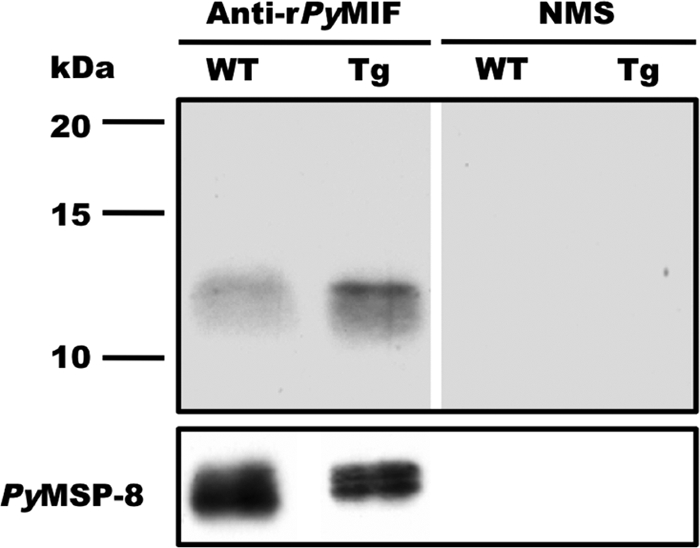
Elevated expression of PyMIF in Py17XL-MIF(+) transgenic parasites. Lysates of P. yoelii 17XL (WT) and Py17XL-MIF(+) (Tg) blood-stage parasites were separated by SDS-PAGE, and the levels of PyMIF were compared by immunoblot analysis. Blots were probed with rPyMIF-specific polyclonal sera (1:2,000) or a pool of NMS. The total amount of parasite protein loaded per lane (3 μg) was normalized based on the reactivity of rabbit anti-PyMSP8 on parallel immunoblots. The expression of PyMSP8 across blood-stage malaria parasites (53) was similar to that observed for endogenous PyMIF.
BALB/cByJ mice were infected with 1 × 106 Py17XL-MIF(+) parasites. For comparison, control mice were challenged with a P. yoelii 17XL transgenic line containing a similar Pb-ef-1aα-gfp expression cassette and selectable marker. In two independent experiments, mice challenged with Py17XL-MIF(+) parasites showed a significant delay in time to death as well as higher survival rates than those infected with control parasites (Fig. 9A and B). The median survival times for Py17XL-MIF(+)-infected mice in the two experiments were 19 and 20 days, respectively, in comparison to 9 and 8.5 days, respectively, for Py17XL-GFP controls. In the first experiment, the level of parasitemia in Py17XL-MIF(+)-infected mice was slightly lower than that in Py17XL-GFP-infected controls during the first week of infection (Fig. 9C). However, this was not the case in the second experiments, where parasitemia levels in the two groups were comparable through day 6 of infection (Fig. 9C). The surviving mice in both experiments cleared their infection completely by day 24 postinfection. These data further support the conclusion that the increased expression of pMIF by blood-stage malaria parasites can alter the course of infection and reduce overall mortality.
FIG. 9.
Overexpression of PyMIF reduces the severity of P. yoelii 17XL malaria. (A and B) Groups of BALB/cByJ mice (10 animals/group) were challenged with 1 × 106 Py17XL-GFP (•) or Py17XL-MIF(+) (▪) parasites. The percent survival plotted against days postinfection from two independent experiments is shown. The significance of the differences in survival times between groups was calculated by using the Mantel-Haenszel log rank test, with a P value of <0.05 considered to be significant. (C) The mean percent parasitemias (±SD) on day 4 and day 6 of infection were reduced in Py17XL-MIF(+)- versus Py17XL-GFP-infected mice in experiment 1 but were comparable in experiment 2.
DISCUSSION
Mammalian MIF has been shown to be an important regulator of proinflammatory responses. Remarkably, MIF appears to be evolutionarily conserved across diverse organisms (14), including nematode (58) and protozoan (37) parasites. The presence of mif homologues in plasmodial parasites has generated interest in assessing its contribution to uncontrolled proinflammatory responses typically associated with the severe complications of malaria. Initial studies of the P. berghei (2, 24), P. falciparum (2, 22), and P. yoelii (52) MIF proteins indicated the potential of pMIF to interact with the host immune system. However, a pathogenic role for pMIF during malaria has not been established. We sought to provide data on the role of pMIF in enhancing disease severity using P. yoelii transgenic parasites, engineered for an increased expression of PyMIF. Unexpectedly, our results suggest that pMIF may modulate host immune responses to attenuate parasite virulence.
Previous in vitro studies suggested that pMIF may not possess all activities normally attributed to mMIF (2, 22, 52). However, those studies relied on analyses of recombinant pMIF that contained an N- or C-terminal epitope tag. To eliminate the influence of the epitope tag on in vitro assays of function and/or on the production of pMIF-specific antisera, we produced and purified non-epitope-tagged, recombinant P. yoelii MIF. Consistent with previous work, our studies showed pMIF to be expressed to some degree throughout the erythrocytic cycle, with peak levels present in late-trophozoite- and schizont-stage parasites. As with P. falciparum and P. berghei MIF, we readily detected PyMIF in the supernatants of short-term, asynchronous cultures of P. yoelii blood-stage parasites. In contrast to previous data, we did not detect significant quantities of PyMIF in the erythrocyte cytosol by indirect immunofluorescence assays of pRBCs. As pMIF lacks recognizable signal sequences and/or PEXEL motifs, we believe that our data are consistent with the conclusion that pMIF is released primarily extracellularly upon schizont rupture. We cannot, however, exclude the possibility that the sensitivity of our assay was insufficient to detect low quantities of pMIF delivered to the erythrocyte cytoplasm via an active export pathway.
Several functional assays for mMIF activity have been described. Mammalian MIF has been shown to possess both tautomerase and oxidoreductase activities in vitro, although the physiological relevance of these activities is not clear (27, 38, 50). Results to date suggested that recombinant pMIF may possess similar but significantly reduced enzyme activity relative to that of mMIF (2, 52). Our studies of rPyMIF showed minimal d-dopachrome tautomerase activity even after treatment with reMAP to remove the initiating methionine and provide the required catalytic N-terminal proline residue (data not shown). More relevant to biologically important activities of mMIF is the ability to elicit and/or enhance the release of proinflammatory cytokines by macrophages in vitro. Previously reported assays of His-tagged recombinant P. falciparum MIF (rPfMIF) showed that it did not stimulate the release IL-8, TNF-α, or IL-12 from human monocytes or enhance the response of these cells to LPS (22). However, previous studies of Brugia malayi-encoded MIF noted that the presence of an epitope tag at either the N or C terminus of the recombinant protein inhibited its activity (47). In our hands, coculturing of peritoneal macrophages with nonfused rPyMIF did not result in the release of TNF-α. The production of TNF-α was not enhanced when macrophages were stimulated with both rPyMIF and LPS. It remains possible that the conformation of our rPyMIF molecule may not fully mimic the native structure, limiting its interaction with specific receptors and explaining the lack of cytokine stimulation. However, our data with PyMIF and mouse macrophages are consistent with data from a previous report of PfMIF and human monocytes (22). Given the inability to directly induce or enhance cytokine release in vitro, we expect that pMIF does not significantly modulate the proinflammatory response of macrophages in vivo during the course of blood-stage malaria. Direct effects of pMIF on NK cell, T-cell, and/or B-cell activation remain to be tested.
One of the earliest recognized attributes of mMIF was its ability to attract macrophages and other cells of the immune system to sites of inflammation. When tested in a conventional transwell chemotaxis assay with peritoneal murine macrophages, our rPyMIF clearly induced the in vitro migration of macrophages in a dose-dependent manner. This finding is consistent with a previously reported finding that rPfMIF could to some degree affect the movement of human monocytes in chemotaxis and migration inhibition assays (22). Recently, the ability of mMIF to bind to two chemokine receptors, CXCR2 and CXCR4, was demonstrated, providing a mechanism by which immune cells are recruited. In addition its ability to bind to CD74, the traditional mMIF receptor (39), it will be of interest to determine if pMIF also interacts with CXCR2/4 to trigger macrophage recruitment (10). Combined, the data on pMIF suggest that its chemotactic activity toward macrophages is particularly relevant to its potential function in vivo.
Initially reported results from studies of the influence of pMIF on the outcome of P. berghei and P. yoelii malaria were probably unanticipated. Infections of C57BL/6 mice with P. berghei mif knockout parasites still resulted in the development of cerebral complications, with mortality comparable to that of infection with wild-type P. berghei parasites. Similarly, infections of BALB/c mice with the P. berghei mif KO line resulted in high levels of parasitemia and severe anemia comparable to those of control infections (2). With the exception of an earlier reticulocytosis, no significant reduction in disease severity was associated with the loss of pMIF in these P. berghei models of severe malaria. On the other hand, the systemic administration of rPyMIF to P. yoelii 17XL-infected mice on days 4 and 5 postchallenge prolonged survival, although all treated mice eventually succumbed (52). In our own initial studies, immunization of mice with rPyMIF to induce high titers of neutralizing antibodies prior to P. yoelii challenge did not reduce the severity of disease. Considering these data, we began to question whether or not pMIF was a likely mediator of malaria-associated immunopathology.
To assess the effects of parasite-produced pMIF on infection outcome, we choose to generate transgenic lines of nonlethal P. yoelii 17X and lethal P. yoelii 17XL parasites that overexpress pymif. Both Py17X-MIF(+) and Py17XL-MIF(+) parasites exhibited constitutive and moderately elevated levels of expression of PyMIF at both the RNA and protein levels. Importantly, the presence of the transgene construct did not appear to affect the Py17X-MIF(+) growth rate through day 8 of infection. With a P. yoelii asexual replication cycle of 18 to 20 h, this indicates that there was no alteration in the replication rate through at least the first 9 to 10 asexual cycles in vivo. Infection of BALB/cByJ mice, C57BL/c mice, and CB6F1/J mice (data not shown) with Py17X-MIF(+), however, resulted in a reduction in mean peak parasitemia relative to control infections. No significant difference in malaria-associated anemia was observed between mice infected with Py17X-MIF(+) and those infected with control parasites. This apparent pMIF-dependent reduction in the severity of P. yoelii malaria was confirmed with the lethal model. Relative to control parasites, mice were able to resist infection with Py17XL-MIF(+) to a significant degree, as evidenced by a prolonged period of infection and a reduction in the overall mortality rate. During malaria, the spleen is essential for the control of infection and the development of protective immune responses (1, 32, 59). Our preliminary studies of splenic mRNA levels of TNF-α, IL-6, IL-12, murine MIF, IL-10, and TGF-β showed no differences between Py17XL-MIF(+)-infected and Py17XL-GFP-infected mice. This finding is consistent with our in vitro data showing little pMIF-dependent modulation of cytokine production by macrophages. We did observe a ∼2-fold increase in splenic IFN-γ mRNA levels in Py17XL-MIF(+)-infected mice on days 4 and 6 postchallenge, which may have contributed to the increased survival (data not shown). Considering the ability of rPyMIF to attract macrophages in vitro, the recruitment of macrophages to the spleen in response to increased levels of PyMIF, and any associated enhancement of phagocytic and/or antigen-processing and presentation ability, may have contributed to the observed reduction in disease severity. Sponaas et al. (55) recently showed that a population of CD11bhigh Ly6C+ monocytes migrates to the spleen during an acute P. chabaudi infection and contributes to the suppression of blood-stage malaria. This population of monocytes exhibited enhanced capacities to present antigens to CD4+ T cells, to phagocytose pRBCs, and to produce inducible nitric oxide synthase (iNOS) and reactive oxygen intermediates. The chemotactic response of this subpopulation of monocytes to pMIF will be of interest to evaluate.
What advantage would malaria parasites gain through the pMIF-dependent manipulation of host immune responses to reduce blood parasitemia levels? A hint may come from studies of MIF homologues that are present in other parasites of humans. The nematodes Wucheria bancrofti and Brugia malayi, the etiological agents of lymphatic filariasis, encode MIF homologues (26, 47, 60). Likewise, the protozoan Leishmania parasites that cause self-limiting cutaneous infections as well as potentially fatal visceral disease also encode MIF homologues (37). Studies of B. malayi MIF (Bm-MIF) and Leishmania major MIF (Lm-MIF) indicate that they are similar in structure and function to mMIF, including their chemotactic activity toward macrophages. However, in the presence of IL-4 induced by B. malayi parasites, Bm-MIF promotes the recruitment and development of alternatively activated macrophages (49). These macrophages and the Th2-associated anti-inflammatory environment may aid in the establishment of more chronic nematode infections. It has been proposed that the antiapoptotic activity associated with Lm-MIF binding to CD74 on macrophages may promote the intracellular survival of Leishmania amastigotes and sustain infection (37). While the release of pMIF by blood-stage malaria parasites may reduce parasite burden to some degree, the possibility that it may concurrently promote persistent, chronic infection is intriguing. If so, such an effect would enhance the likelihood of transmitting parasites to the mosquito vector and, ultimately, a new host.
Additional study of pMIF will be required to further define its role during the course of human malaria. It may also be necessary to reevaluate some of the data on the importance of mMIF during blood-stage malaria. At the moment, there are opposing views. Some reports indicated a pathogenic role for mMIF during P. falciparum malaria (35, 43), while others showed that higher levels of mMIF production correlate with protection from severe disease (3-6). The presence of pMIF in the sera of P. falciparum malaria patients has been reported but has not been factored into this analysis (51). This is further complicated by data on pMIF-induced signaling cascades suggesting that host and plasmodial homologues are synergistic at low concentrations but are antagonistic at higher concentrations (52). If it is the combined action of mMIF and pMIF that influences the severity of P. falciparum malaria, careful consideration must be given to the known variability in mMIF production by individual patients and the possibility for variation in pMIF production by different P. falciparum strains.
Supplementary Material
Acknowledgments
This work was supported by NIH NIAID grants AI069147 and AI035661 and by the Drexel University College of Medicine.
Editor: J. H. Adams
Footnotes
Published ahead of print on 13 September 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Alves, H. J., W. Weidanz, and L. Weiss. 1996. The spleen in murine Plasmodium chabaudi adami malaria: stromal cells, T lymphocytes, and hematopoiesis. Am. J. Trop. Med. Hyg. 55:370-378. [DOI] [PubMed] [Google Scholar]
- 2.Augustijn, K. D., R. Kleemann, J. Thompson, T. Kooistra, C. E. Crawford, S. E. Reece, A. Pain, A. H. G. Siebum, C. J. Janse, and A. P. Waters. 2007. Functional characterization of the Plasmodium falciparum and P. berghei homologues of macrophage migration inhibitory factor. Infect. Immun. 75:1116-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awandare, G. A., J. B. Hittner, P. G. Kremsner, D. O. Ochiel, C. C. Keller, J. B. Weinberg, I. A. Clark, and D. J. Perkins. 2006. Decreased circulating macrophage migration inhibitory factor (MIF) protein and blood mononuclear cell MIF transcripts in children with Plasmodium falciparum malaria. Clin. Immunol. 119:219-225. [DOI] [PubMed] [Google Scholar]
- 4.Awandare, G. A., P. G. Kremsner, J. B. Hittner, C. C. Keller, I. A. Clark, J. B. Weinberg, and D. J. Perkins. 2007. Higher production of peripheral blood macrophage migration inhibitory factor in healthy children with a history of mild malaria relative to children with a history of severe malaria. Am. J. Trop. Med. Hyg. 76:1033-1036. [PubMed] [Google Scholar]
- 5.Awandare, G. A., J. J. Martinson, T. Were, C. Ouma, G. C. Davenport, J. M. Ong'echa, W. Wang, L. Leng, R. E. Ferrell, R. Bucala, and D. J. Perkins. 2009. MIF (macrophage migration inhibitory factor) promoter polymorphisms and susceptibility to severe malarial anemia. J. Infect. Dis. 200:629-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Awandare, G. A., Y. Ouma, C. Ouma, T. Were, R. Otieno, C. C. Keller, G. C. Davenport, J. B. Hittner, J. Vulule, R. Ferrell, J. M. Ong'echa, and D. J. Perkins. 2007. Role of monocyte-acquired hemozoin in suppression of macrophage migration inhibitory factor in children with severe malarial anemia. Infect. Immun. 75:201-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayoub, S., M. J. Hickey, and E. F. Morand. 2008. Mechanisms of disease: macrophage migration inhibitory factor in SLE, RA and atherosclerosis. Nat. Clin. Pract. Rheumatol. 4:98-105. [DOI] [PubMed] [Google Scholar]
- 8.Bach, J. P., B. Rinn, B. Meyer, R. Dodel, and M. Bacher. 2008. Role of MIF in inflammation and tumorigenesis. Oncology 75:127-133. [DOI] [PubMed] [Google Scholar]
- 9.Baugh, J. A., and R. Bucala. 2002. Macrophage migration inhibitory factor. Crit. Care Med. 30:S27-S35. [PubMed] [Google Scholar]
- 10.Bernhagen, J., R. Krohn, H. Lue, J. L. Gregory, A. Zernecke, R. R. Koenen, M. Dewor, I. Georgiev, A. Schober, L. Leng, T. Kooistra, G. Fingerle-Rowson, P. Ghezzi, R. Kleemann, S. R. McColl, R. Bucala, M. J. Hickey, and C. Weber. 2007. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat. Med. 13:587-596. [DOI] [PubMed] [Google Scholar]
- 11.Burns, J., Jr., W. Majarian, J. Young, T. Daly, and C. Long. 1989. A protective monoclonal antibody recognizes an epitope in the carboxyl-terminal cysteine-rich domain in the precursor of the major merozoite surface antigen of the rodent malarial parasite, Plasmodium yoelii. J. Immunol. 143:2670-2676. [PubMed] [Google Scholar]
- 12.Burns, J. M., Jr., C. C. Belk, and P. D. Dunn. 2000. A protective glycosylphosphatidylinositol-anchored membrane protein of Plasmodium yoelii trophozoites and merozoites contains two epidermal growth factor-like domains. Infect. Immun. 68:6189-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calandra, T., J. Bernhagen, C. N. Metz, L. A. Spiegel, M. Bacher, T. Donnelly, A. Cerami, and R. Bucala. 1995. MIF as a glucocorticoid-induced modulator of cytokine production. Nature 377:68-71. [DOI] [PubMed] [Google Scholar]
- 14.Calandra, T., and T. Roger. 2003. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat. Rev. Immunol. 3:791-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlton, J. M., S. V. Angiuoli, B. B. Suh, T. W. Kooij, M. Pertea, J. C. Silva, M. D. Ermolaeva, J. E. Allen, J. D. Selengut, H. L. Koo, J. D. Peterson, M. Pop, D. S. Kosack, M. F. Shumway, S. L. Bidwell, S. J. Shallom, S. E. van Aken, S. B. Riedmuller, T. V. Feldblyum, J. K. Cho, J. Quackenbush, M. Sedegah, A. Shoaibi, L. M. Cummings, L. Florens, J. R. Yates, J. D. Raine, R. E. Sinden, M. A. Harris, D. A. Cunningham, P. R. Preiser, L. W. Bergman, A. B. Vaidya, L. H. van Lin, C. J. Janse, A. P. Waters, H. O. Smith, O. R. White, S. L. Salzberg, J. C. Venter, C. M. Fraser, S. L. Hoffman, M. J. Gardner, and D. J. Carucci. 2002. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 419:512-519. [DOI] [PubMed] [Google Scholar]
- 16.Chaisavaneeyakorn, S., N. Lucchi, C. Abramowsky, C. Othoro, S. C. Chaiyaroj, Y. P. Shi, B. L. Nahlen, D. S. Peterson, J. M. Moore, and V. Udhayakumar. 2005. Immunohistological characterization of macrophage migration inhibitory factor expression in Plasmodium falciparum-infected placentas. Infect. Immun. 73:3287-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaisavaneeyakorn, S., J. M. Moore, C. Othoro, J. Otieno, S. C. Chaiyaroj, Y. P. Shi, B. L. Nahlen, A. A. Lal, and V. Udhayakumar. 2002. Immunity to placental malaria. IV. Placental malaria is associated with upregulation of macrophage migration inhibitory factor in intervillous blood. J. Infect. Dis. 186:1371-1375. [DOI] [PubMed] [Google Scholar]
- 18.Clark, I., M. Awburn, R. Whitten, C. Harper, N. G. Liomba, M. Molyneux, and T. Taylor. 2003. Tissue distribution of migration inhibitory factor and inducible nitric oxide synthase in falciparum malaria and sepsis in African children. Malar. J. 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark, I., A. Budd, L. Alleva, and W. Cowden. 2006. Human malarial disease: a consequence of inflammatory cytokine release. Malar. J. 5:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark, I. A., L. M. Alleva, A. C. Budd, and W. B. Cowden. 2007. Understanding the role of inflammatory cytokines in malaria and related diseases. Travel Med. Infect. Dis. 6:67-81. [DOI] [PubMed] [Google Scholar]
- 21.Clark, I. A., and W. B. Cowden. 2003. The pathophysiology of falciparum malaria. Pharmacol. Ther. 99:221-260. [DOI] [PubMed] [Google Scholar]
- 22.Cordery, D. V., U. Kishore, S. Kyes, M. J. Shafi, K. R. Watkins, T. N. Williams, K. Marsh, and B. C. Urban. 2007. Characterization of a Plasmodium falciparum macrophage-migration inhibitory factor homologue. J. Infect. Dis. 195:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daly, T. M., C. A. Long, and L. W. Bergman. 2001. Interaction between two domains of the P. yoelii MSP-1 protein detected using the yeast two-hybrid system. Mol. Biochem. Parasitol. 117:27-35. [DOI] [PubMed] [Google Scholar]
- 24.Dobson, S. E., K. D. Augustijn, J. A. Brannigan, C. Schnick, C. J. Janse, E. J. Dodson, A. P. Waters, and A. J. Wilkinson. 2009. The crystal structures of macrophage migration inhibition factor (MIF) from Plasmodium falciparum and Plasmodium berghei. Protein Sci. 18:2578-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dodoo, D., F. M. Omer, J. Todd, B. D. Akanmori, K. A. Koram, and E. M. Riley. 2002. Absolute levels and ratios of proinflammatory and anti-inflammatory cytokine production in vitro predict clinical immunity to Plasmodium falciparum malaria. J. Infect. Dis. 185:971-979. [DOI] [PubMed] [Google Scholar]
- 26.Falcone, F. H., P. N. Loke, X. Zang, A. S. MacDonald, R. M. Maizels, and J. E. Allen. 2001. A Brugia malayi homolog of macrophage migration inhibitory factor reveals an important link between macrophages and eosinophil recruitment during nematode infection. J. Immunol. 167:5348-5354. [DOI] [PubMed] [Google Scholar]
- 27.Fingerle-Rowson, G., D. R. Kaleswarapu, C. Schlander, N. Kabgani, T. Brocks, N. Reinart, R. Busch, A. Schutz, H. Lue, X. Du, A. Liu, H. Xiong, Y. Chen, A. Nemajerova, M. Hallek, J. Bernhagen, L. Leng, and R. Bucala. 2009. A tautomerase-null macrophage migration-inhibitory factor (MIF) gene knock-in mouse model reveals that protein interactions and not enzymatic activity mediate MIF-dependent growth regulation. Mol. Cell. Biol. 29:1922-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franke-Fayard, B., H. Trueman, J. Ramesar, J. Mendoza, M. van der Keur, R. van der Linden, R. E. Sinden, A. P. Waters, and C. J. Janse. 2004. A Plasmodium berghei reference line that constitutively expresses GFP at a high level throughout the complete life cycle. Mol. Biochem. Parasitol. 137:23-33. [DOI] [PubMed] [Google Scholar]
- 29.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M.-S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. A. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenwood, B. M. 2008. Malaria: progress, perils, and prospects for eradication. J. Clin. Invest. 118:1266-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gregory, J. L., E. F. Morand, S. J. McKeown, J. A. Ralph, P. Hall, Y. H. Yang, S. R. McColl, and M. J. Hickey. 2006. Macrophage migration inhibitory factor induces macrophage recruitment via CC chemokine ligand 2. J. Immunol. 177:8072-8079. [DOI] [PubMed] [Google Scholar]
- 32.Grun, J. L., C. A. Long, and W. P. Weidanz. 1985. Effects of splenectomy on antibody-independent immunity to Plasmodium chabaudi adami malaria. Infect. Immun. 48:853-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hay, S. I., C. A. Guerra, P. W. Gething, A. P. Patil, A. J. Tatem, A. M. Noor, C. W. Kabaria, B. H. Manh, I. R. F. Elyazar, S. Brooker, D. L. Smith, R. A. Moyeed, and R. W. Snow. 2009. A world malaria map: Plasmodium falciparum endemicity in 2007. PLoS Med. 6:e1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunt, N. H., and G. E. Grau. 2003. Cytokines: accelerators and brakes in the pathogenesis of cerebral malaria. Trends Immunol. 24:491-499. [DOI] [PubMed] [Google Scholar]
- 35.Jain, V., S. McClintock, A. Nagpal, A. Dash, J. Stiles, V. Udhayakumar, N. Singh, and N. Lucchi. 2009. Macrophage migration inhibitory factor is associated with mortality in cerebral malaria patients in India. BMC Res. Notes 2:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janse, C. J., J. Ramesar, and A. P. Waters. 2006. High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat. Protoc. 1:346-356. [DOI] [PubMed] [Google Scholar]
- 37.Kamir, D., S. Zierow, L. Leng, Y. Cho, Y. Diaz, J. Griffith, C. McDonald, M. Merk, R. A. Mitchell, J. Trent, Y. Chen, Y.-K. A. Kwong, H. Xiong, J. Vermeire, M. Cappello, D. McMahon-Pratt, J. Walker, J. Bernhagen, E. Lolis, and R. Bucala. 2008. A Leishmania ortholog of macrophage migration inhibitory factor modulates host macrophage responses. J. Immunol. 180:8250-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleemann, R., H. Rorsman, E. Rosengren, R. Mischke, N. T. Mai, and J. Bernhagen. 2000. Dissection of the enzymatic and immunologic functions of macrophage migration inhibitory factor. Full immunologic activity of N-terminally truncated mutants. Eur. J. Biochem. 267:7183-7193. [DOI] [PubMed] [Google Scholar]
- 39.Leng, L., C. N. Metz, Y. Fang, J. Xu, S. Donnelly, J. Baugh, T. Delohery, Y. Chen, R. A. Mitchell, and R. Bucala. 2003. MIF signal transduction initiated by binding to CD74. J. Exp. Med. 197:1467-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, C., L. A. Sanni, F. Omer, E. Riley, and J. Langhorne. 2003. Pathology of Plasmodium chabaudi chabaudi infection and mortality in interleukin-10-deficient mice are ameliorated by anti-tumor necrosis factor alpha and exacerbated by anti-transforming growth factor beta antibodies. Infect. Immun. 71:4850-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mackintosh, C. L., J. G. Beeson, and K. Marsh. 2004. Clinical features and pathogenesis of severe malaria. Trends Parasitol. 20:597-603. [DOI] [PubMed] [Google Scholar]
- 42.Martiney, J. A., B. Sherry, C. N. Metz, M. Espinoza, A. S. Ferrer, T. Calandra, H. E. Broxmeyer, and R. Bucala. 2000. Macrophage migration inhibitory factor release by macrophages after ingestion of Plasmodium chabaudi-infected erythrocytes: possible role in the pathogenesis of malarial anemia. Infect. Immun. 68:2259-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDevitt, M. A., J. Xie, G. Shanmugasundaram, J. Griffith, A. Liu, C. McDonald, P. Thuma, V. R. Gordeuk, C. N. Metz, R. Mitchell, J. Keefer, J. David, L. Leng, and R. Bucala. 2006. A critical role for the host mediator macrophage migration inhibitory factor in the pathogenesis of malarial anemia. J. Exp. Med. 203:1185-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Omer, F. M., J. B. de Souza, and E. M. Riley. 2003. Differential induction of TGF-beta regulates proinflammatory cytokine production and determines the outcome of lethal and nonlethal Plasmodium yoelii infections. J. Immunol. 171:5430-5436. [DOI] [PubMed] [Google Scholar]
- 45.Omer, F. M., and E. M. Riley. 1998. Transforming growth factor beta production is inversely correlated with severity of murine malaria infection. J. Exp. Med. 188:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Othoro, C., A. A. Lal, B. Nahlen, D. Koech, A. S. Orago, and V. Udhayakumar. 1999. A low interleukin-10 tumor necrosis factor-alpha ratio is associated with malaria anemia in children residing in a holoendemic malaria region in western Kenya. J. Infect. Dis. 179:279-282. [DOI] [PubMed] [Google Scholar]
- 47.Pastrana, D. V., N. Raghavan, P. FitzGerald, S. W. Eisinger, C. Metz, R. Bucala, R. P. Schleimer, C. Bickel, and A. L. Scott. 1998. Filarial nematode parasites secrete a homologue of the human cytokine macrophage migration inhibitory factor. Infect. Immun. 66:5955-5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petritus, P. M. and J. M. Burns, Jr. 2008. Suppression of lethal Plasmodium yoelii malaria following protective immunization requires antibody-, IL-4-, and IFN-gamma-dependent responses induced by vaccination and/or challenge infection. J. Immunol. 180:444-453. [DOI] [PubMed] [Google Scholar]
- 49.Prieto-Lafuente, L., W. F. Gregory, J. E. Allen, and R. M. Maizels. 2009. MIF homologues from a filarial nematode parasite synergize with IL-4 to induce alternative activation of host macrophages. J. Leukoc. Biol. 85:844-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosengren, E., P. Aman, S. Thelin, C. Hansson, S. Ahlfors, P. Bjork, L. Jacobsson, and H. Rorsman. 1997. The macrophage migration inhibitory factor MIF is a phenylpyruvate tautomerase. FEBS Lett. 417:85-88. [DOI] [PubMed] [Google Scholar]
- 51.Shao, D., Z. Han, Y. Lin, L. Zhang, X. Zhong, M. Feng, Y. Guo, and H. Wang. 2008. Detection of Plasmodium falciparum derived macrophage migration inhibitory factor homologue in the sera of malaria patients. Acta Trop. 106:9-15. [DOI] [PubMed] [Google Scholar]
- 52.Shao, D., X. Zhong, Y. F. Zhou, Z. Han, Y. Lin, Z. Wang, L. Bu, L. Zhang, X. D. Su, and H. Wang. 2009. Structural and functional comparison of MIF ortholog from Plasmodium yoelii with MIF from its rodent host. Mol. Immunol. 47:726-737. [DOI] [PubMed] [Google Scholar]
- 53.Shi, Q., A. Cernetich-Ott, M. M. Lynch, and J. J. M. Burns. 2006. Expression, localization, and erythrocyte binding activity of Plasmodium yoelii merozoite surface protein-8. Mol. Biochem. Parasitol. 149:231-241. [DOI] [PubMed] [Google Scholar]
- 54.Shi, X., L. Leng, T. Wang, W. Wang, X. Du, J. Li, C. McDonald, Z. Chen, J. W. Murphy, E. Lolis, P. Noble, W. Knudson, and R. Bucala. 2006. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 25:595-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sponaas, A. M., A. P. Freitas do Rosario, C. Voisine, B. Mastelic, J. Thompson, S. Koernig, W. Jarra, L. Renia, M. Mauduit, A. Potocnik, and J. Langhorne. 2009. Migrating monocytes recruited to the spleen play an important role in control of blood stage malaria. Blood 114:5522-5531. [DOI] [PubMed] [Google Scholar]
- 56.Swope, M., H.-W. Sun, P. Blake, and E. Lolis. 1998. Direct link between cytokine activity and a catalytic site for macrophage migration inhibitory factor. EMBO J. 17:3534-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tonkin, C. J., G. G. van Dooren, T. P. Spurck, N. S. Struck, R. T. Good, E. Handman, A. F. Cowman, and G. I. McFadden. 2004. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol. Biochem. Parasitol. 137:13-21. [DOI] [PubMed] [Google Scholar]
- 58.Vermeire, J. J., Y. Cho, E. Lolis, R. Bucala, and M. Cappello. 2008. Orthologs of macrophage migration inhibitory factor from parasitic nematodes. Trends Parasitol. 24:355-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiss, L. 1989. Mechanisms of splenic control of murine malaria: cellular reactions of the spleen in lethal (strain 17XL) Plasmodium yoelii malaria in BALB/c mice, and the consequences of pre-infective splenectomy. Am. J. Trop. Med. Hyg. 41:144-160. [DOI] [PubMed] [Google Scholar]
- 60.Zang, X., P. Taylor, J. M. Wang, D. J. Meyer, A. L. Scott, M. D. Walkinshaw, and R. M. Maizels. 2002. Homologues of human macrophage migration inhibitory factor from a parasitic nematode. Gene cloning, protein activity, and crystal structure. J. Biol. Chem. 277:44261-44267. [DOI] [PubMed] [Google Scholar]
- 61.Zhong, X.-B., L. Leng, A. Beitin, R. Chen, C. McDonald, B. Hsiao, R. D. Jenison, I. Kang, S.-H. Park, A. Lee, P. Gregersen, P. Thuma, P. Bray-Ward, D. C. Ward, and R. Bucala. 2005. Simultaneous detection of microsatellite repeats and SNPs in the macrophage migration inhibitory factor (MIF) gene by thin-film biosensor chips and application to rural field studies. Nucleic Acids Res. 33:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.