Abstract
Edwardsiella tarda is a Gram-negative enteric pathogen that causes hemorrhagic septicemia in fish and both gastrointestinal and extraintestinal infections in humans. A type III secretion system (T3SS) was recently shown to contribute to pathogenesis, since deletions of various T3SS genes increased the 50% lethal dose (LD50) by about 1 log unit in the blue gourami infection model. In this study, we report EseG as the first identified effector protein of T3SS. EseG shares partial homology with two Salmonella T3SS effectors (SseG and SseF) over a conserved domain (amino acid residues 142 to 192). The secretion of EseG is dependent on a functional T3SS and, in particular, requires the chaperone EscB. Experiments using TEM-1 β-lactamase as a fluorescence-based reporter showed that EseG was translocated into HeLa cells at 35°C. Fractionation of infected HeLa cells demonstrated that EseG was localized to the host membrane fraction after translocation. EseG is able to disassemble microtubule structures when overexpressed in mammalian cells. This phenotype may require a conserved motif of EseG (EseG142-192), since truncated versions of EseG devoid of this motif lose their ability to cause microtubule destabilization. By demonstrating the function of EseG, our study contributes to the understanding of E. tarda pathogenesis. Moreover, the approach established in this study to identify type III effectors can be used to identify and characterize more type III and possible type VI effectors in Edwardsiella.
Edwardsiella tarda is a Gram-negative bacterium associated with septicemia and fatal infections in many animals, including fish and humans (18, 36, 48). In humans, it causes gastrointestinal infections as well as extraintestinal infections such as myonecrosis, bacteremia, septic arthritis, and wound infections (18). Using a comparative proteomics approach, our group reported the presence of two secretion systems, namely, a type III secretion system (T3SS) and a type VI secretion system (T6SS), which are vital for E. tarda pathogenesis (44, 45, 46, 54).
T3SSs are contact-dependent translocation systems that have been found in many Gram-negative bacteria of animals, commensals, and plant-symbiotic rhizobia (3, 31). They have many components, such as secretion and translocon apparatuses, effectors, regulators, and chaperones. The protein secretion machinery directs the secretion and translocation of many bacterial effectors into host cells. The concerted action of these effectors stimulates or interferes with host cellular processes, thereby dictating the terms of bacterium-host cell interactions (8). In Salmonella spp., more than 30 effectors are secreted by two different T3SSs. These effectors are involved in several diverse functions, such as forced entry into epithelial cells, intracellular replication inside vacuoles, and suppression of cellular immune responses (16, 29, 50). In Yersinia species, six Yop effectors have been identified, and they have been found to disrupt vital signaling cascades that are required for innate immunity (20, 41). T3SS effectors are generally less conserved than other components of T3SSs, and they perform unique functions adapted to a pathogen's virulence strategy.
The T3SS facilitates the survival of E. tarda in phagocytes and HEp-2 cells (19, 33, 34, 45) and contributes to virulence in vivo (45). The deletion of various single T3SS genes, such as escC, eseB, eseD, or escA, increased the 50% lethal dose (LD50) by approximately 1 log unit in blue gourami (49, 55). EscC has been shown to act as the chaperone for EseB and EseD, whereas EscA is the chaperone for EseC (49, 55). EseB, EseC, and EseD, which are secreted in large quantities by E. tarda, are homologous to Salmonella sp. SseB, SseC, and SseD, respectively (45). The SseB, SseC, and SseD proteins are secreted by the T3SS of Salmonella pathogenicity island 2 (SPI-2) and predominantly assemble into complexes (SseBCD) that function as a translocon for effector proteins (32). EseB, EseC, and EseD can also form a protein complex (EseBCD) after secretion (55), suggesting that the EseBCD complex functions as a translocon component, rather than as T3SS effectors. To our knowledge, no T3SS effectors have been identified in E. tarda yet.
The E. tarda T3SS has almost a full set of genes that are homologous to those of SPI-2 (45). A bioinformatics analysis of E. tarda T3SS proteins revealed that EseG bears some similarity to two Salmonella effectors, SseG and SseF. SseG is an effector of SPI-2 that targets intracellular Salmonella to the Golgi network, where the close association of bacterial cells allows Salmonella to multiply efficiently (40). SseF, another SPI-2 effector, is involved in positioning and maintaining the Salmonella-containing vacuoles (SCV) in a juxtanuclear position in infected epithelial cells (1). The translocated Salmonella effector proteins SseF and SseG interact with each other and are required for Salmonella to establish an intracellular replication niche (12).
In this study, we used a modified TEM-1 β-lactamase system (TEM) to show that EseG is an E. tarda T3SS effector that is localized in the host membrane fraction after translocation. A functional study showed that EseG interacts with α-tubulin and destabilizes the microtubule through a conserved domain. The ΔeseG mutant showed slightly reduced virulence in blue gourami.
MATERIALS AND METHODS
Bacterial strains and cell cultures.
The bacterial strains and plasmids used in the present study are described in Table 1. E. tarda strains were grown in tryptic soy broth (TSB; BD Biosciences) or Dulbecco modified Eagle medium (DMEM; Invitrogen) at 25°C, while Escherichia coli strains were cultured in Luria-Bertani broth (LB; BD Biosciences) at 37°C. Cultivation of bacteria in DMEM was carried out under a 5% (vol/vol) CO2 atmosphere. When required, the medium was supplemented with appropriate antibiotics at the following concentrations: ampicillin, 100 μg/ml; colistin, 12.5 μg/ml; gentamicin, 100 μg/ml; tetracycline, 15 μg/ml; kanamycin, 50 μg/ml. For the induction of T3SS proteins, E. tarda PPD130/91 was grown in DMEM at 25°C, 35°C, or 37°C. Extracellular proteins (ECP) were prepared from E. tarda cultures according to the protocol established previously (54). HEK293A and HeLa cells were grown at 37°C in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS) under a 5% (vol/vol) CO2 atmosphere.
TABLE 1.
List of strains and plasmids
| Strain or plasmid | Description and/or genotypea | Reference or source |
|---|---|---|
| Strains | ||
| E. tarda | ||
| PPD130/91 | Wild type; Kms Colr Amps; LD50 of 105.0 | 26 |
| ΔesaN | PPD130/91 with in-frame deletion of esaN | This study |
| ΔeseG | PPD130/91 with in-frame deletion of eseG | This study |
| ΔescB | PPD130/91 with in-frame deletion of escB | This study |
| ΔescB/escB | ΔescB with pACYC-escB | This study |
| ΔeseG/eseG | ΔeseG with pACYC-eseG | This study |
| ΔesaN/esaN | ΔesaN with pACYC-esaN | This study |
| E. coli | ||
| DH5α | α complementation | Stratagene |
| MC1061 λpir | (λpir) thi thr-1 leu6 proA2 his-4 argE2 lacY1 galK2 ara14 xyl5 supE44 λpir | 39 |
| S17-1 λpir | RK2 tra regulon, pir, host for pir-dependent plasmids | 42 |
| BL21(DE3) | E. coli B F−ompThsdS(rB− mB−) dcm+ Tetrgal (DE3) endA Hte | Novagen |
| Plasmids | ||
| pGEM-Teasy | Ampr | Promega |
| pACYC184 | Tetr Cmr | Amersham |
| pACYC-escB | pACYC184 with wild-type escB | This study |
| pACYC-eseG | pACYC184 with wild-type eseG | This study |
| pACYC-esaN | pACYC184 with wild-type esaN | This study |
| pCX341 | pBR322 derivative, used to fuse EseG to TEM-1 β-lactamase | 6 |
| pCX-eseG | eseG fused to TEM-1 | This study |
| pRE112 | pGP704 suicide plasmid, pir dependent; CmroriToriVsacB | 13 |
| pRE-ΔesaN | pRE112 containing esaN fragment with aa 21 to 421 deleted | This study |
| pRE-ΔeseG | pRE112 containing eseG fragment with aa 47 to 335 deleted | This study |
| pRE-ΔescB | pRE112 containing escB fragment with aa 2 to 109 deleted | This study |
| pEGFP-N1 | Kmr | BD Biosciences Clontech |
| pEGFP-eseG1-88 | pEGFP-N1 with eseG encoding aa 1-88 | This study |
| pEGFP-eseG89-339 | pEGFP-N1 with eseG encoding aa 89-339 | This study |
| pEGFP-eseG | pEGFP-N1 with full-length eseG | This study |
| pCDNA-3.1+ | CMV promoter; Ampr Neor | Invitrogen |
| pCDNA-2HA-eseG | pCDNA3.1 with 2HA tag at the N terminus of EseG | This study |
| pCDNA-eseG | pCDNA3.1 with full-length eseG | This study |
| pCDNA-eseG89-339 | pCDNA3.1 with eseG encoding aa 89-339 | This study |
| pCDNA-eseG89-339,Δ142-192 | pCDNA3.1 with eseG encoding aa 89-141 and 193-339 | This study |
Km, kanamycin; Col, colistin; Amp, ampicillin; Cm, chloramphenicol; Tet, tetracycline; CMV, cytomegalovirus. Superscripts: r, resistance; s, sensitivity.
Construction of deletion mutants, complementation, and plasmids.
Nonpolar esaN, eseG, and escB deletion mutants were generated by sacB-based allelic exchange (13) as described previously (56). For example, two PCR fragments were generated from PPD130/91 genomic DNA for the construction of the ΔeseG mutant. Primer pairs eseG-for plus eseG-int-rev and eseG-int-for plus eseG-rev were used. The resulting products were a 633-bp fragment containing the upstream region of eseG and a 690-bp fragment containing the downstream region of eseG. A 16-bp overlap in the sequences permitted the amplification of a 1,323-bp product during a second PCR with primers eseG-for and eseG-rev, each of which introduced a KpnI restriction enzyme site. The resulting PCR product, containing a deletion from amino acid (aa) 47 to aa 335 of EseG, was ligated into the suicide vector pRE112 (13) and transformed into E. coli SM10 λpir. Single-crossover mutants were obtained by conjugal transfer of the resulting plasmid into E. tarda PPD130/91. Deletion mutants were screened on 10% sucrose-tryptic soy agar (TSA) plates. Mutants were verified by PCR, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blot analysis. No mutants showed growth defects when cultured in TSB or DMEM. All primers used for the construction of mutants are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Designation | Nucleotide sequence |
|---|---|
| esaN-for | ATGGTACC GTTCATAGGCTGTCTCCACGGT |
| esaN-int-rev | TCCCACCCTGCCGTCG CCGCTGAGCGCCACCCTCA |
| esaN-int-for | CGACGGCAGGGTGGGACAGTCGA |
| esaN-rev | ATGGTACC AGATTCTGATCCAGCCAGATA |
| esaN-com-for | AGAATTCGAACGCGAGCTGTTCACCAC |
| esaN-com-rev | AAGTACTGCGTAGCTGCGTCAGCAT |
| eseG-for | ATGGTACC TGGCTGGAGACGAAGAAGGG |
| eseG-int-rev | GATGATCAACGGCGTTTACAGCTTTGCCTGACC |
| eseG-int-for | AACGCCGTTGATCATCATAGGCTC |
| eseG-rev | ATGGTACCCGCTATTGCTGCCGCTAACC |
| eseG-com-for | AGAATTCGCAGCAGCAGCAGGAGTAT |
| eseG-com-rev | AAGTACTGTATGGGAGGAGGGGGTCA |
| escB-for | AAGGTACCAAAGACTTTCAGGCCAAGCAGG |
| escB-int-rev | GCACTCCGCCATGTGAAACATGGGGGTCTCCAAATGATATGATATATT |
| escB-int-for | TTTCACATGGCGGAGTGCCTGCT |
| escB-rev | TTGGTACCCAGGGTGAGCACTTTGGCGAC |
| escB-com-for | AAGAATTCATTAAAGAGGAGAAATTAACCATGACGTCCGCACCGCTCT |
| escB-com-rev | AAAGTACTTATTAGGGTTGATTAAGCGTA |
| pEGFP-eseGF | AAAAGCTTATGCCTTGGATGCCACGAT |
| pEGFP-eseGR | AAGGATCCGCAAAGCTGTAGCGTCGCG |
| pEGFP-eseG88R | AAGGATCCGCCCCCGGCGCTCGGCTGCG |
| pEGFP-eseG89F | AAAAGCTTATGTCAGCGCCCCTGGACCAGC |
| escB-For | AAGGATCCATGACGTCCGCACCGCTCT |
| escB-Rev | TTAAGCTTGGGTTGATTAAGCGTATCCAG |
| pCDNA-2HA-eseG-For | AAGCCCGGGCAATGCCTTGGATGCCACGAT |
| pCDNA-eseG-For | AAAAGCTTGCCACCATGGCACCTTGGATGCCACGATTGCG |
| pCDNA-eseG-Rev-stop | AAGGATCCTCAGGCAAAGCTGTAGCGTCG |
| pCDNA-eseG89-For | AAGAATTCGCCACCATGGCATCAGCGCCCCTGGACCAGC |
| pCDNA-eseG142-int-Rev | GAGCCCGGTGATCAGTGCGGC |
| pCDNA-eseG193-int-for | GCCGCACTGATCACCGGGCTCTGGCTGGGAAAAGTCAGCG |
Site-directed mutagenesis of eseG was performed using overlap extension PCR (17). For complementation of the ΔescB mutant, the low-copy-number plasmid pACYC184 was used, and the wild-type gene was inserted into the EcoRI and ScaI sites of a chloramphenicol gene. The recombinant plasmid was screened by tetracycline and was transferred into PPD130/91. The primers used are listed in Table 2 as eseG-com-for, eseG-com-rev, escB-com-for, escB-com-rev, esaN-com-for, and esaN-com-rev.
Secretion of EseG.
To examine the secretion of EseG, overnight cultures of the E. tarda wild-type strain PPD130/91, the ΔesaN and ΔeseG mutants, and the ΔeseG/eseG and ΔesaN/esaN complemented strains were diluted 1:200 in DMEM with 5 μg/ml tetracycline. Bacteria were then grown without shaking for 20 h at 25°C under 5% CO2. Each bacterial culture was centrifuged. The supernatant (labeled as ECP) was filtered (pore size, 0.22 μm; Millipore) and was then concentrated by an Amicon Ultra-15 centrifugal filtration device with a 10,000-molecular-weight cutoff filter (Millipore). The bacterial pellet was resuspended in 200 μl phosphate-buffered saline (PBS), sonicated, and then centrifuged. The supernatant was labeled as total bacterial proteins (TBP). One hundred fifty micrograms of each TBP, or 6.0 μg of each ECP, was boiled for 5 min in the SDS sample buffer before each protein mixture was subjected to denaturing PAGE on a NuPAGE 4 to 12% gradient gel (Invitrogen). Antigen-antibody complexes were visualized with the SuperSignal West Pico chemiluminescent substrate (Pierce), followed by exposure on Lumi-Film chemiluminescent detection film (Roche). Proteins were separated by PAGE and transferred to a nitrocellulose membrane. The membrane was cut into two parts; the upper part was probed with a mouse anti-DnaK monoclonal antibody (Stressgen) at 1:2,000 and horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG at 1:5,000 (Sigma), and the lower part was probed with a rabbit anti-EseG antibody (monopeptide antibodies were raised in rabbits against keyhole limpet hemocyanin-conjugated peptides of EseG [EseG aa 93 to 106; DQLRAQAQEAEQKA] by BioGenes [Berlin, Germany]) diluted at 1:2,000 and HRP-conjugated goat anti-rabbit IgG at 1:3,000 (Santa Cruz).
Fluorescence microscopy for observation of EseG translocation.
HeLa cells were seeded into wells of a 24-well plate (with coverslips at the bottom) at 1 × 105 cells per well in DMEM for 18 h at 35°C in a 5% CO2 incubator. Wild-type PPD130/91, the ΔesaN mutant with pCX-eseG, and PPD130/91 with pCX341 were inoculated into TSB, supplemented with tetracycline, and grown at 35°C without shaking the night before infection. HeLa cells were washed twice with PBS, and bacteria were applied to HeLa cells at a multiplicity of infection (MOI) of 100 in DMEM without serum. Three hours later, HeLa cell monolayers were washed twice with PBS. The infection then proceeded for another 5 h in DMEM. Pictures were taken under a Zeiss Axiovert inverted microscope by using a Zeiss AxioCam MRc digital camera.
Fractionation of infected HeLa cells.
Infected HeLa cells were fractionated as described by Coombes et al. (9) with minor modifications. Briefly, HeLa cells were seeded at 5 × 106 per 100-mm-diameter culture dish 1 day before the infection. Before infection, overnight E. tarda cultures were diluted 1:20 into fresh TSB, grown for 3 h without shaking, and then applied to HeLa cells at an MOI of 100 in DMEM. Culture dishes were maintained at 35°C in a 5% CO2 incubator for 3 h. The culture dishes were then washed 3 times with prewarmed PBS, and the medium was replaced with prewarmed DMEM with 5% FBS and 30 μg/ml gentamicin. Infections were allowed to proceed for another 5 h before harvest and resuspension in 300 μl homogenization buffer (HB) (250 mM sucrose, 3 mM imidazole, 0.5 mM EDTA [pH 7.4]). The cells were homogenized by mechanical lysis using a 1-ml syringe with a 22-gauge needle. The lysate was spun at 3,000 × g for 15 min at 4°C, and the supernatant was transferred to a small plastic ultracentrifuge tube and spun at 41,000 × g in an ultracentrifuge (Beckman) for 30 min at 4°C. The supernatant (cytosolic fraction) was mixed with 60 μl 5× SDS sample buffer containing 5% β-mercaptoethanol. The pellet (membrane fraction) was resuspended in a mixture of 240 μl HB and 60 μl 5× SDS sample buffer containing 5% β-mercaptoethanol and was boiled for 5 min. Fractions (40 μl) were transferred to a polyvinylidene difluoride (PVDF) membrane for Western blot analysis. The membrane was probed with the rabbit anti-EseG antiserum at 1:2,000 and with a polyclonal anti-EvpP antiserum (54) at 1:3,000. The membrane was stripped with 8.0 M guanidine hydrochloride and 10 mM dithiothreitol and was reprobed with a monoclonal anti-β-tubulin antibody (Sigma) at 1:2,000 and a monoclonal anti-calnexin antibody (Stressgen) at 1:2,000.
Virulence in blue gourami.
Healthy naïve blue gourami (Trichogaster trichopterus [Pallas]) were infected with E. tarda as described previously by Ling et al. (26). Fish mortalities were recorded over a period of 7 days after intramuscular injections near the dorsal fins. The LD50s were calculated by the method of Reed and Muench (37). For each strain, the LD50 estimation was performed at least three times.
Transfection of HEK293A cells and immunofluorescence microscopy.
HEK293A cells were seeded on glass coverslips in 6-well tissue culture plates for 24 h before transfection at a density of 3 × 105 cells/well. Cells were transiently transfected with 6 μl Fugene 6 (Roche) using 2.0 μg DNA optimized according to the manufacturer's instructions. Cells were subsequently incubated at 37°C under 5% CO2 for 18 h prior to fixation and antibody staining. For staining, transfected cells were fixed with 4% paraformaldehyde in PBS, permeabilized with 0.1% Triton X-100 in PBS, and stained as described by Abrahams and coworkers (1). Antibodies were used at the following dilutions: mouse monoclonal anti-α-tubulin antibody (Sigma), 1:5,000; rabbit anti-EseG antiserum, 1:300; goat anti-mouse IgG (Alexa 594; Molecular Probes), 1:200; and goat anti-rabbit IgG (Alexa 488; Molecular Probes), 1:200. For the quantification of microtubule destabilization, 120 positively transfected cells were scored for the disappearance of microtubule structure in at least three independent experiments.
Immunoprecipitation (IP) and Western blot analysis.
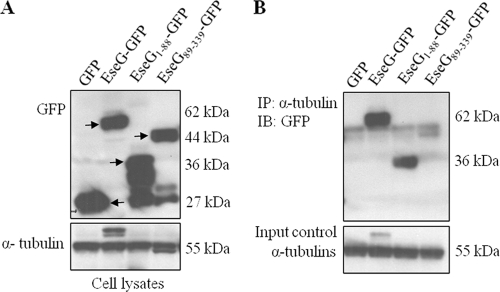
HEK293A cells were grown in 100-mm-diameter culture dishes transfected with 24 μg pCDNA-EseG by using 20 μl Lipofectamine 2000 reagent (Invitrogen). After 20 h, cells were washed with cold PBS and were scraped. Cells were then washed three times with cold PBS and were lysed for 30 min with a modified radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1% NP-40, 10% glycerol, 50 mM NaF, 0.4 mM EDTA, 10 mM sodium pyrophosphate) containing protease inhibitors (Complete; Roche). Homogenates were centrifuged, and supernatants were incubated overnight at 4°C, with gentle rotation, with IgG(κ) or an anti-α-tubulin antibody bound to protein A-agarose (Invitrogen). After incubation, immunoprecipitates were washed extensively with ice-cold modified RIPA buffer. Proteins bound to the agarose were eluted by heating at 95°C for 4 min in SDS-PAGE sample buffer. Eluted proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with IgG(κ) or an anti-α-tubulin antibody, followed by detection by chemiluminescence. A mouse monoclonal antibody against green fluorescent protein (GFP) (Molecular Probes), a mouse monoclonal anti-α-tubulin antibody (Sigma), and mouse IgG(κ) (Sigma), all at 1:2,000, were used.
Quantitative 2HA-EseG immunoprecipitation and mass spectrometry.
Binding partners for EseG were identified exactly as described previously for SopB (38). Briefly, unlabeled HEK293 cells were transfected with EseG tagged with 2 hemagglutinin molecules (2HA-EseG) for 24 h and were then mixed with an equal number of nontransfected cells labeled with 13C6-Arg and 2H4-Lys. EseG was immunoprecipitated and prepared for mass spectrometry as described previously (38). Digested immunoprecipitates were analyzed with an LTQ Orbitrap XL system (5). The protein was identified, and quantitative ratios were calculated, exactly as described previously (38).
Pulldown assay and Western blotting.
EscB-His6 was purified according to a batch purification protocol (Qiagen). E. tarda with pCX-eseG and E. tarda with pCX341 were subcultured separately into DMEM and were induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 5 h before harvesting. Cell pellets were sonicated and then centrifuged to collect supernatants. His-tagged EscB beads were then incubated with cell lysates for 2 h at 4°C with gentle rotation. Beads were washed with the washing buffer 7 times, and proteins bound to beads were resolved by a NuPAGE 4 to 12% Bis-Tris gel (Invitrogen). Proteins were transferred to a PVDF membrane and were probed with a mouse anti-β-lactamase antibody at 1:2,000 (QED Bioscience Inc.) and with HRP-conjugated goat anti-mouse IgG at 1:5,000 (Sigma).
Statistical analysis.
All data are presented as means ± standard deviations (SD). Comparisons between groups were tested by one-way analysis of variance (ANOVA), and the least significant difference (LSD) was used for multiple comparisons.
RESULTS
Sequence analysis of EseG.
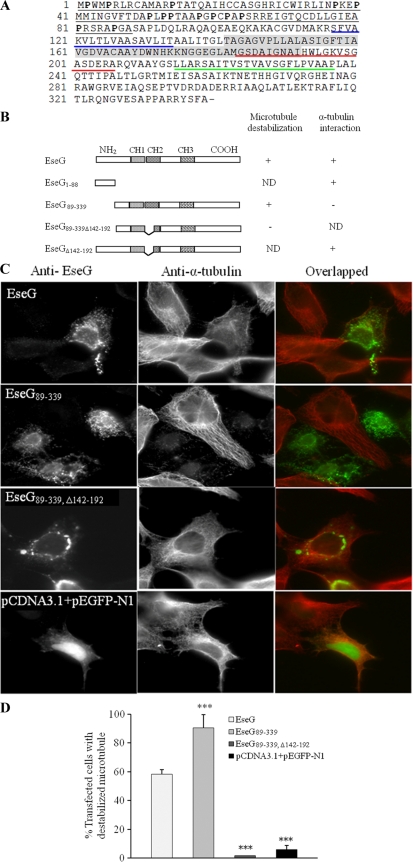
EseG is a 339-amino-acid protein encoded inside the E. tarda T3SS gene cluster (45). It possesses nine positive charges (http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html) (7) and is classified as an unstable protein according to the instability index calculated by a computer model (http://www.expasy.org/tools/protparam.html). Amino acid sequence comparison shows that EseG is very similar to a hypothetical protein from Edwardsiella ictaluri (ABC60070) (70% identity and 79% similarity). It also shows homology to hypothetical proteins from other bacteria, such as ABN61461 of Shewanella baltica OS155 (24.4% identity and 45.9% similarity) and ZP04613838 of Yersinia rohdei ATCC 43380 (22.9% identity and 41.9% similarity). Interestingly, EseG also shares homology with SseF (CAA12191) and SseG (CAA12192), effector proteins secreted by a T3SS of Salmonella enterica serovar Typhimurium (12, 23, 24), suggesting that EseG may also function as an effector protein of the T3SS in E. tarda. In addition, EseG and its homologs, especially SseF and SseG, share a conserved region (Fig. 1), which is used by SseF to recruit dynein to the SCV (1) and by SseG to target the Golgi complex and to allow Salmonella to multiply vigorously (40). We used this information to examine a possible function of this conserved fragment in EseG.
FIG. 1.
Amino acid sequence alignment of a conserved region in E. tarda EseG (AAX76916) and its homologs Salmonella serovar Typhimurium SseF and SseG (CAA12191 and CAA12192, respectively) and hypothetical proteins of E. ictaluri (ABC60070), S. baltica OS155 (ABN61461), and Yersinia intermedia ATCC 29909 (ZP00833567). Asterisks denote identical residues; colons, conserved substitutions; periods, similar amino acids. Boldface indicates residues of EseG that are identical in all sequences compared.
EseG is secreted in a T3SS-dependent manner.
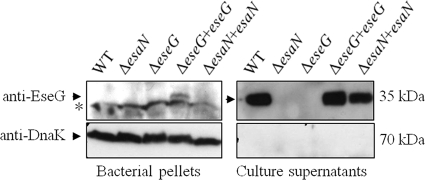
The E. tarda wild-type strain PPD130/91, a T3SS-deficient ΔesaN mutant, a ΔeseG mutant, and two complemented strains (the ΔesaN/esaN and ΔeseG/eseG strains) were examined for the secretion of EseG. escN, a homolog of esaN in E. coli, is an ATPase that energizes the transportation of T3SS effectors and translocon components. Mutation in escN has been shown to abolish the secretion or translocation of T3SS substrates (2). Equal amounts of extracellular proteins (ECP) and total bacterial proteins (TBP) from these E. tarda strains were collected separately and subjected to Western blot analyses using anti-EseG and anti-DnaK antibodies. In TBP fractions, EseG was detected only in the complemented ΔeseG mutant (Fig. 2). However, in ECP fractions, EseG was detected in the wild type and in complemented ΔesaN and ΔeseG mutants. EseG was not detected in the bacterial pellets of the wild type or the complemented ΔesaN mutant. A possible explanation may be that EseG was rapidly secreted once it was synthesized by the chromosome. Thus, EseG was detected in a bacterial pellet only when the mutant was complemented with eseG in the low-copy-number plasmid pACYC184. DnaK was not detected in any ECP, showing that detection of EseG is not due to leakage from bacterial pellets. These results indicate that the secretion of EseG is dependent on a functional T3SS.
FIG. 2.
EseG is secreted into the culture supernatant in a T3SS-dependent manner. Western blot analysis of EseG was conducted with bacterial pellets (TBP) and culture supernatants (ECP) from identical numbers of organisms of wild-type E. tarda PPD130/91, its ΔesaN and ΔeseG mutants, and the ΔeseG/eseG and ΔesaN/esaN complemented strains. The Western blot was probed with anti-EseG and an antibody against DnaK, a bacterial cytosolic marker. EseG was not detected in the ECP of the ΔesaN or ΔeseG background, and DnaK was not detected in any ECP, showing that detection of EseG is not due to leakage from bacterial pellets. The asterisk indicates the nonspecific band detected in the bacterial pellet.
EseG is translocated into HeLa cells at 35°C and is localized in the host cell membrane.
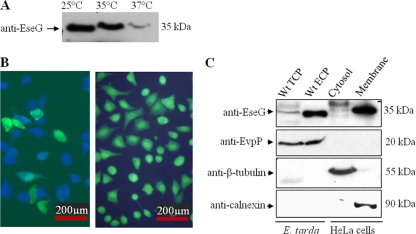
Effector translocation was detected directly in live host cells (mammalian cells) by using the fluorescent β-lactamase substrate CCF2-AM (6, 25). When CCF2-AM permeates host cells, it exhibits fluorescence resonance energy transfer (FRET), resulting in a green fluorescence signal. Cleavage of the substrate by an effector-β-lactamase molecule disrupts FRET, resulting in a shift to a blue fluorescence signal (6). Considering that E. tarda is also a human pathogen (19) and that the secretion of EseG decreased drastically at 37°C (Fig. 3A), we chose to culture E. tarda, and infect HeLa cells, at 35°C, the condition under which EseG was maintained at a high level (Fig. 3A). Under a fluorescence microscope, most HeLa cells appeared blue, indicating that the translocation of EseG-TEM into these cells had taken place (Fig. 3B, left). In contrast, almost all HeLa cells infected with E. tarda expressing pCX341 (empty vector) appeared green (Fig. 3B, right). Taken together, our results suggested that EseG is a T3SS effector.
FIG. 3.
EseG is translocated into the cytosol of the host cell and is localized in its membrane fraction. (A) EseG is secreted in a temperature-dependent manner. ECP were prepared from identical amounts of the wild-type E. tarda strain PPD130/91 cultured in DMEM at 25°C, 35°C, or 37°C and were then probed with anti-EseG for Western blot analysis. (B) Demonstration of the translocation of the E tarda effector EseG into live HeLa cells using TEM-1 fusions and fluorescence microscopy. HeLa cells were infected with wild-type E. tarda strains expressing EseG-TEM (left) or TEM-1 only (right). Eight hours after infection, HeLa cells were washed and loaded with CCF2-AM. Translocation of the effector-TEM into host cells causes the cleavage of the CCF2-AM product, resulting in the emission of a blue fluorescence signal. Uncleaved CCF2 emits green fluorescence. Bars, 200 μM. (C) EseG is localized at the HeLa membrane fraction. HeLa cells were infected with wild-type E. tarda PPD130/91 and were subjected to subcellular fractionation by differential centrifugation. Fractions were analyzed by Western blot analyses using anti-EseG, anti-EvpP, anti-β-tubulin, and anti-calnexin antibodies.
To localize EseG in infected host cells, HeLa cells were infected with the E. tarda wild-type strain PPD130/91 and were subjected to subcellular fractionation. Calnexin and β-tubulin were used as subcellular markers of membrane and cytosolic proteins, respectively, in HeLa cells. EvpP, a T6SS-dependent secreted protein (54), was found in the E. tarda pellet (TCP) and E. tarda ECP but not in the infected HeLa cell cytosolic or membrane fraction (Fig. 3C), indicating that these fractions were not contaminated by the bacteria. Our data showed that EseG is localized in the HeLa cell membrane fraction (Fig. 3C).
Role of EseG in E. tarda pathogenesis.
To characterize further the effects of eseG and esaN on the virulence of E. tarda, we determined the LD50s of nonpolar deletion mutants devoid of eseG or esaN in vivo (Table 3). In this study, LD50 refers to the minimum lethal dose of E. tarda required to kill half of the blue gourami tested within 7 days. The ΔesaN mutant was modestly attenuated—its LD50 was about 1 log unit higher than that of the wild type, and the difference was statistically significant (Table 3). In contrast, the ΔeseG mutant showed only slight attenuation, with an LD50 about 0.17 to 0.35 log unit higher than that of the wild type, and the difference was not statistically significant (Table 3).
TABLE 3.
Characterization of wild-type E. tarda PPD130/91 and its mutants
| Strain | Length of WTa protein (aa) | Codons deleted (aa) | LD50 | Pb |
|---|---|---|---|---|
| PPD130/91 | 104.91 ± 0.15 | NA | ||
| ΔesaN mutant | 438 | 21-421 | 105.94 ± 0.19 | <0.001 |
| ΔeseG mutant | 339 | 47-335 | 105.01 ± 0.10 | >0.05 |
| ΔescB mutant | 163 | 2-109 | NDc | ND |
WT, wild type.
Obtained using one-way ANOVA and LSD. All P values refer to the wild-type LD50 (n, ≥3). NA, not applicable.
ND, not determined.
EscB is the chaperone of EseG and is involved in the stabilization of EseG.
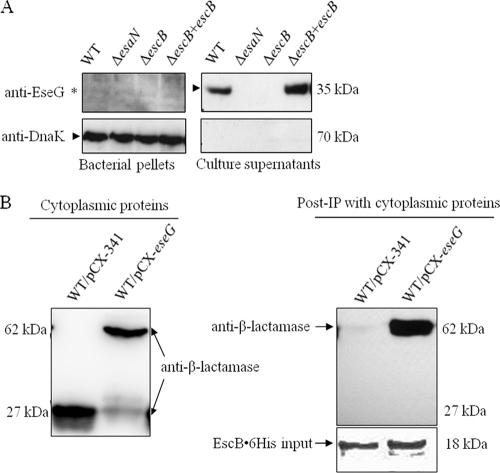
The escB gene is located upstream of eseG and is transcribed in the same direction as eseG in the E. tarda PPD130/91 T3SS gene cluster (45). The start codon of EseG overlaps with EscB. EscB is small (18 kDa) and acidic (pI 4.64), characteristics common to most chaperones (10). Furthermore, it has a tetratricopeptide repeat domain, as identified by a Pfam scan (http://pfam.janelia.org/). This domain is also present in the LcrH/SycD-like chaperone (35). To test whether EscB is required for the secretion of EseG, E. tarda ECP were harvested from equal numbers of organisms of the wild type, the ΔesaN and ΔescB mutants, and the ΔescB/escB complemented strain and were probed with anti-EseG and anti-DnaK antibodies. As shown in Fig. 4A, EseG was not detected in the ECP of the ΔescB or ΔesaN mutant; however, it was detected in the ECP when the ΔescB mutant was complemented with the low-copy-number plasmid pACYC-escB. Thus, EsaN and EscB are required for the secretion of EseG.
FIG. 4.
EscB is the chaperone of EseG. (A) EseG was not secreted in the absence of EscB, and its secretion was restored by EscB trans-complementation. ECP from equal amounts of E. tarda PPD130/91, ΔesaN, ΔescB, and ΔescB/escB organisms were probed with anti-EseG and anti-DnaK antibodies. The asterisk indicates the nonspecific band detected in the bacterial pellet. (B) EscB interacts with EseG-TEM. His-tagged EscB beads were used to immunoprecipitate identical amounts of cell lysates prepared from E. tarda PPD130/91/pCX-341 and PPD130/91/pCX-eseG. Proteins pulled down by the His-tagged EscB were separated by SDS-PAGE and subjected to Western blot analysis with anti-His and anti-β-lactamase antibodies.
Chaperone proteins usually bind to their cognate substrate directly (47, 49, 55). Since EscB likely functions as a chaperone of EseG, we performed in vitro binding assays to test whether EscB binds to EseG directly. His-tagged EscB was expressed and purified to pull down cell lysates of E. tarda PPD130/91 harboring pCX-eseG (WT/pCX-eseG) or pCX341 (WT/pCX-341). Under IPTG induction conditions, WT/pCX-eseG and WT/pCX-341 expressed EseG-TEM and TEM, respectively (Fig. 4B). Our results showed that EscB could pull down EseG-TEM but not TEM (Fig. 4C), suggesting that EscB and EseG interact with each other in the cytosol of E. tarda.
EseG interacts with α-tubulin.
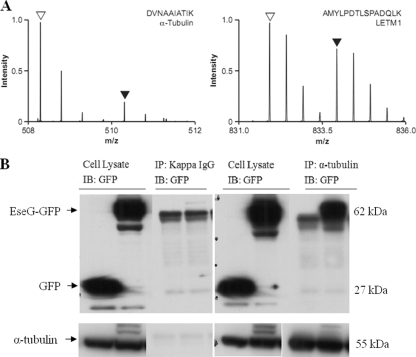
In order to investigate host interaction partners of EseG, two approaches were used. First, SILAC (stable isotope labeling with amino acids in cell culture)-based quantitative proteomics, as previously used to identify host targets of the Salmonella type III effector protein SopB (38), was used to analyze immunoprecipitates of pCDNA3.1-2HA-EseG-transfected cells (expressing HA-tagged EseG) versus nontransfected cells (Fig. 5A). Second, immunoprecipitation of cell lysates prepared from pEGFP-N1- or pEGFP-eseG-transfected 293A cells, followed by mass spectrometric analysis of additional bands obtained from EseG, was performed (data not shown). The SILAC result identified at least eight peptides from tubulin, all showing the same pattern. Together, these two approaches identified α-tubulin, β-tubulin, and dynein as possible interaction partners of EseG. The interactions between EseG and α-tubulin were further confirmed by IP using a monoclonal anti-α-tubulin antibody (Fig. 5B, right). IgG(κ) was used as a negative control for the IP experiment to show that it did not pull down α-tubulin (Fig. 5B, left).
FIG. 5.
EseG interacts with α-tubulin. (A) SILAC 2HA-EseG immunoprecipitations. Anti-HA was used to immunoprecipitate cell lysates from nontransfected cells (stable isotope-labeled cells) and HA-EseG-overexpressing cells (unlabeled cells). A peptide from α-tubulin (left) is present predominantly in the unlabeled, specific form (open arrowhead), while a peptide from LETM1 and EF-hand domain-containing protein 1 (right) is present in roughly equal amounts in the unlabeled (open arrowhead) and labeled (filled arrowhead) forms. The SILAC result showed that more than eight peptides, all showing the same pattern, were identified from tubulin. (B) EseG interacts with α-tubulin. HEK293A cells were transfected with either pEGFP-N1 or pEGFP-EseG, followed by solubilization to produce cell lysates. These cell lysates were immunoprecipitated (IP) with IgG(κ) (as a control) (left) or with an anti-α-tubulin antibody (right). EseG-GFP fusion proteins were detected by an anti-GFP antibody (immunoblotting [IB]). The doublet background bands are likely heavy chains of IgG produced by IP.
The N-terminal fragment of EseG containing 88 amino acid residues is important for its interaction with α-tubulin.
To identify the region(s) of EseG that interacts with α-tubulin, a series of truncated EseG proteins fused at the N terminus of GFP were constructed and overexpressed in HEK293A cells by transfection. These included EseG-GFP, EseG1-88-GFP, EseG89-339-GFP (251 residues of the C-terminal end), and GFP alone (negative control). As shown in Fig. 6, GFP was detected in all cell lysates, with molecular weights consistent with the predicted sizes of fusion proteins representing the respective constructs. Multiple bands were detected for EseG1-88-GFP, possibly due to the unstable nature of the N-terminal end of EseG. EseG1-88-GFP and EseG-GFP could be pulled down by the anti-α-tubulin antibody (Fig. 6B). These results suggested that the N-terminal fragment of EseG consisting of 88 amino acid residues is necessary for its interaction with α-tubulin.
FIG. 6.
N-terminal residues (aa 1 to 88) of EseG are important for interaction with α-tubulin. (A) HEK293A cells were transfected with pEGFP-N, pEGFP-EseG, pEGFP-EseG1-88, or pEGFP-EseG89-339, followed by solubilization to produce cell lysates. EseG-GFP fusion proteins exhibited different molecular sizes (indicated by arrows), depending on the size of each construct (upper panel). α-Tubulin was used as a loading control (lower panel). (B) EseG was immunoprecipitated (IP) with an anti-α-tubulin antibody. GFP fusion proteins were detected by an anti-GFP antibody (upper panel), and α-tubulin was used as a loading control (lower panel). EseG1-88-GFP and EseG-GFP were pulled down by the anti-α-tubulin antibody (indicated by arrows).
Functional dissection of EseG.
Subcellular fractionation revealed the presence of translocated EseG in the membrane fractions of host cells (Fig. 3C). Sequence analyses of EseG found three hydrophobic regions that may act as transmembrane domains (Fig. 7A). In addition, the N-terminal region is rich in proline residues, with 14 prolines in the first 88 residues (Fig. 7A). The conserved fragment (EseG142-192) that has similarity with Salmonella effectors SseF and SseG covered part of the CH2 region (the second transmembrane domain). To gain further insight into the potential functions of these domains, a series of GFP-tagged (constructs in pEGFP-N1) and nontagged (pCDNA-3.1+) EseG constructs were prepared. Immunoprecipitation analyses revealed that all truncated EseG constructs that contained the 88-aa N-terminal fragment interacted with α-tubulin (Fig. 6 and 7). Immunofluorescence of nontagged truncated EseG showed that the conserved motif EseG142-192 (Fig. 1) was involved in the microtubule destabilization activity of EseG (Fig. 7).
FIG. 7.
Functional dissection of the E tarda T3SS effector EseG. (A) Sequence analysis of EseG. Hydrophobic regions CH1, CH2, and CH3 are underlined in blue, red, and green, respectively. Amino acid residues of the N-terminal region (residues 1 to 88) of EseG interacting with α-tubulin are underlined. Prolines in the N-terminal region are in boldface. The conserved fragment (EseG142-192) that has similarity with Salmonella effectors SseF and SseG is shaded. (B) Schematic representation of truncated polypeptides derived from EseG. The ability of each polypeptide to destabilize microtubules and to interact with α-tubulin is indicated next to its schematic structure. Various truncations of GFP-tagged EseG were constructed for the α-tubulin interaction studies, whereas untagged truncated EseG proteins in pCDNA3.1 were used for microtubule destabilization studies (panels C and D). ND, not determined. (C) The conserved hydrophobic segment in EseG is essential for EseG-mediated phenotypes. HEK293A cells were transiently transfected with EseG, EseG89-339, EseG89-339, Δ142-192, or pCDNA3.1 with pEGFP-N1 (empty-vector control). EseG and microtubules are labeled with an anti-EseG antibody (green) and an anti-α-tubulin antibody (red), respectively. (D) Measurement of microtubule destabilization in HEK293A cells transfected with EseG, EseG89-339, EseG89-339, Δ142-192, or an empty vector (negative control). One hundred twenty transfected cells were counted in at least three independent experiments. Data are presented as means ± SD. Bars with three asterisks are significantly different from the EseG bar (P ≤ 0.001). P values were obtained using one-way ANOVA and the least-significant-difference (LSD) method. All values refer to the wild type (n ≥ 3).
EseG disassembles microtubules in vivo.
To investigate whether EseG is colocalized with α-tubulin, several truncated versions of EseG without tags were constructed in the pCDNA3.1+ vector and were expressed in HEK293A cells, namely, pCDNA-eseG, pCDNA-eseG89-339, pCDNA-eseG89-339, Δ142-192, and empty vectors (Fig. 7C). HEK 293A cells were then stained with anti-EseG (green) and anti-α-tubulin (red) antibodies. As shown in Fig. 7C, HEK293A cells transfected with full-length EseG, or with EseG89-339, showed destabilization of microtubule structures. In contrast, HEK293A cells transfected with an empty vector showed normal filamentous microtubules that could be stained by the anti-α-tubulin antibody. Similar phenotypes were observed in both HeLa and Cos-7 cells (data not shown). Interestingly, removal of the conserved fragment (residues 142 to 192) resulted in a complete loss of microtubule destabilization activity. EseG89-339, Δ142-192, which did not contain the conserved domain, was, instead, redistributed in a punctate form, mainly around the nucleus (Fig. 7C).
The level of microtubule destabilization was then measured in HEK293A cells after transfection with different truncated versions of EseG. The percentages of transfected cells that showed microtubule destabilization are presented in Fig. 7D. Expression of EseG in HEK293A cells caused 58.3% of transfected cells to show destabilized microtubules, whereas the removal of an 88-residue N-terminal fragment (pCDNA-eseG89-339) resulted in 90.5% of transfected cells exhibiting microtubule destabilization. Most importantly, deletion of residues 142 to 192 (pCDNA-eseG89-339, Δ142-192) resulted in a total loss of microtubule destabilization; the rate of microtubule destabilization dropped to a level (1.67%) approximately the same as that of the vector control (Fig. 7D). These results suggest that the conserved domain of EseG (amino acid residues 142 to 192) is required to destabilize microtubule networks.
DISCUSSION
E. tarda belongs to the family Enterobacteriaceae and is closely related to other enteric pathogens, such as pathogenic Escherichia coli strains and pathogenic Yersinia and Salmonella species. These enteric pathogens and pathogenic E. tarda strains can cause many diseases in humans, such as typhoid fever, diarrheal diseases, and bubonic plague. One of the virulence factors shared by these pathogens is the T3SS. The study of T3SS effectors in E. tarda will help us to understand not only the infection pathway of this pathogen but also the general pathogenic features of these effectors in enteric diseases. To dissect the pathogenicity of E. tarda, the previously established fish infection model, blue gourami, was used (26). This model helps us to identify the important virulence factors of E. tarda by comparing the LD50s of various mutants with that of the wild type. For example, when one of the two T3SS-regulatory genes, esrB and esrC, was deleted in E. tarda PPD130/91, the LD50s of the resulting mutants were 3 log units higher than that of the wild type (43, 56). Although it has been speculated that Edwardsiella effectors may be responsible for the intracellular growth of the bacteria in HEp-2 cells (34) and for their multiplication in murine and fish macrophages (33, 45), no T3SS effectors of E. tarda have been identified yet. In this study, we have identified the first effector of E. tarda, EseG. The role of EseG in the pathogenesis of E. tarda was further investigated in tissue culture models and in fish.
Establishing a protocol for studying the translocation of E. tarda T3SS effectors into host cells.
TEM-1 β-lactamase was developed as a fluorescence-based reporter to directly identify effectors that are translocated into mammalian host cells by extracellular pathogens (6, 25). Previously, it has been shown that E. tarda is an intracellular pathogen, capable of penetrating and replicating inside cultured HEp-2 cells, HeLa cells, and murine macrophages (19, 27, 33, 34). In this study, we modified the TEM-based system to identify a T3SS effector (EseG) from E. tarda.
Wild-type E. tarda PPD130/91 is a fish isolate whose T3SS is functionally active at lower temperatures, such as 25°C, but not at a higher temperatures, such as 37°C (44). To study the translocation of EseG into host cells, we initially cultured E. tarda expressing a EseG-TEM fusion protein at 25°C and used these bacteria to infect a fish cell line, epithelioma papillosum of carp. All cells infected with E. tarda appeared green (data not shown), suggesting that either EseG-TEM was not translocated into cells of epithelioma papillosum of carp or the catalytic activity of TEM was inhibited in epithelioma papillosum of carp cells at 25°C. To circumvent the limitation caused by these possibilities, HeLa cells were chosen for TEM experiments. We grew E. tarda and HeLa cells at 35°C, a condition under which the T3SS of E. tarda was functionally active (Fig. 3A). Also, HeLa cell growth was not affected (4). As expected, we observed the translocation of EseG-TEM into HeLa cells 8 h after infection. Thus, this modified TEM-based protocol was shown to be suitable for the identification of effectors secreted by the T3SS, and possibly the T6SS, of Edwardsiella.
Cellular functions of EseG.
Full-length EseG was able to destabilize microtubule structures when overexpressed in HeLa cells (Fig. 7). This phenotype is reminiscent of those caused by a group of T3SS effectors, such as enteropathogenic E. coli (EPEC) effectors EspG and EspG2 and the Shigella effector VirA (14, 28). EspG, EspG2, and VirA belong to the EspG protein family (http://pfam.janelia.org/family?acc=PF06872). These three proteins share a striking homology at the amino acid level (14, 28). VirA can affect the structure of microtubule networks when transiently expressed in host cells, and EspG and EspG2 trigger the destruction of microtubule networks beneath adherent EPEC (28, 52). Either EspG or EspG2 can rescue the invasive ability of a Shigella virA mutant, indicating that these effector proteins are functionally equivalent to those in Shigella (14). Like EspG family proteins, EseG can also trigger microtubule rearrangement mediated by its microtubule destabilization activity. Despite the fact that EseG shows functional phenotypes similar to those of the EspG family proteins, there is no amino acid homology between them. It is possible that EseG has a microtubule-inhibitory mechanism different from those of EspG, EspG2, and VirA. Alternatively, EseG and the EspG family may share a similar 3-dimensional structural motif that determines their common microtubule-inhibitory activity.
Mutations of either one of the homologs of eseG in Salmonella (sseF and sseG) have been reported to render Salmonella less able to replicate in epithelial cells (1, 40). SseF and SseG help to maintain the SCV in a juxtanuclear, Golgi network-associated localization, which is required for the intracellular proliferation of Salmonella (1, 40). Furthermore, EseG, like SseG and SseF, is localized to the host cell membrane fraction (Fig. 3C) (1). Since EseG shares homology with SseF and SseG (Fig. 1), it is likely that EseG may have a function similar to that of SseF and SseG, i.e., to enhance the proliferation of E. tarda in host cells. In future work, we will explore whether the localization of the ΔeseG mutant in host cells differs from that of the wild type and whether ΔeseG bacteria have a decreased ability to replicate in host cells.
Further comparison of the amino acid sequences of EseG, SseF, and SseG revealed a conserved domain in all three proteins (Fig. 1), namely, EseG142-192, SseF85-135, and SseG90-133. SseF85-135 is located in the second transmembrane domain of SseF (residues 128 to 179), which is involved in the positioning of intracellular Salmonella and the recruitment of dynein to SCV (1). SseG90-133 is located in a Golgi complex-targeting domain of SseG (residues 88 to 142) that allows Salmonella to multiply near the Golgi network (40). EseG142-192 might be required for EseG to destabilize microtubule structures, as evidenced by the observation that the microtubule destabilization ability of EseG dropped to the same level as that of the negative control when EseG142-192 was deleted (Fig. 7). Furthermore, since SseF and SseG share similarities with EseG in this conserved domain, it will be interesting to determine whether SseF and SseG can also destabilize microtubules using this domain. Functional study of EseG may help to reveal novel functions of SseF and SseG in Salmonella virulence.
EseG interacts with α-tubulin. The first 88 N-terminal residues of EseG (EseG1-88) are important for this interaction (Fig. 6 and 7). Two properties of EseG might account for this interaction. First, these 88 N-terminal residues contain 14 proline residues (Fig. 7A), which appear to be critical in protein-protein interactions. It has been reported that several protein-interacting domains prefer ligand sequences that are rich in proline residues, for example, Src homology 3 (SH3) and WW domains (21). Second, these 88 residues include 5 residues with net positive charges, which are important for protein-protein interactions. Considering that EseG destabilizes microtubule structures through the conserved domain EseG142-192, while the first 88 N-terminal residues of EseG are important for the interaction between EseG and α-tubulin (Fig. 6 and 7), we speculate that EseG142-192 interacts with another protein, or that it exhibits an unknown enzyme-like activity, to destabilize microtubule structures. Alternatively, the deletions in EseG142-192 may interfere with the proper cellular localization of EseG, resulting in the loss of its microtubule destabilization activity. However, we cannot rule out the possibility that the disruption in the microtubule destabilization function of EseG142-192 may arise from the effect of its improper folding. The fact that the ability to bind α-tubulin and the ability to destabilize microtubule structures are not mediated by the same domain (or enzyme-like activity) is not unique to EseG. The Shigella effector VirA also interacts with α- and β-tubulin heterodimers but induces microtubule destabilization through an α-tubulin-specific cysteine protease-like activity (51, 52, 53). Interestingly, VirA is not a protease, nor does it directly sever or destabilize microtubules (11, 15).
In conclusion, we have demonstrated the various cellular activities of EseG, such as its interaction with α-tubulin, destabilization of microtubules, and contribution to the virulence of E. tarda in vivo. Our ongoing work is to explore whether there is any microtubule destabilization in infected HeLa cells when EseG is translocated into the host cells at a high level and to examine the cellular localization of the translocated EseG. Since microtubules are also involved in initiating the process of bacterial entry (22), intracellular movement, and the spread of bacteria (30), it will be interesting to determine whether E. tarda exploits microtubules through EseG for its entry into, and movement in, fish cells.
Acknowledgments
We are very grateful to Jack E. Dixon for providing equipment, reagents, and helpful advice on part of this research. We thank Dixon lab members Lorena Navarro, Yvonne Lee, Neal Alto, and Seema Mattoo for helpful discussions. Thanks to Mo Li, ZhaoLan Mo, and Chakraborty Smarajit for experimental assistance. Our thanks also go to Shashikant Joshi, Anthony Siame, and Heather Snowball for proofreading of the manuscript.
This research was supported by Biomedical Research Council (BMRC) grants from the Agency for Science, Technology, and Research (A*STAR), Singapore, to K.Y.L. (04/1/21/19/346, 06/1/21/16/431) and Y.-K.M. (07/1/21/19/495), a Natural Science and Engineering Research Council (NSERC) discovery grant (372373-2010) and a Trinity Western startup grant (0488) to K.Y.L., a Canadian Institutes of Health Research (CIHR) operating grant (MOP-77688) to L.J.F., and operating grants to B.B.F. from the CIHR. H.B.Y. is supported by a CIHR fellowship. L.J.F. is the Canada Research Chair in Quantitative Proteomics and a Michael Smith Foundation Scholar. B.B.F. is a Howard Hughes Medical Institute (HHMI) International Research Scholar and the University of British Columbia Peter Wall Distinguished Professor.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 20 September 2010.
REFERENCES
- 1.Abrahams, G. L., P. Müller, and M. Hensel. 2006. Functional dissection of SseF, a type III effector protein involved in positioning the Salmonella-containing vacuole. Traffic 7:950-965. [DOI] [PubMed] [Google Scholar]
- 2.Andrade, A., J. P. Pardo, N. Espinosa, G. Pérez-Hernández, and B. González-Pedrajo. 2007. Enzymatic characterization of the enteropathogenic Escherichia coli type III secretion ATPase EscN. Arch. Biochem. Biophys. 468:121-127. [DOI] [PubMed] [Google Scholar]
- 3.Büttner, D., and U. Bonas. 2006. Who comes first? How plant pathogenic bacteria orchestrate type III secretion. Curr. Opin. Microbiol. 9:193-200. [DOI] [PubMed] [Google Scholar]
- 4.Byrd, M. P., M. Zamora, and R. E. Lloyd. 2005. Translation of eukaryotic translation initiation factor 4GI (eIF4GI) proceeds from multiple mRNAs containing a novel cap-dependent internal ribosome entry site (IRES) that is active during poliovirus infection. J. Biol. Chem. 280:18610-18622. [DOI] [PubMed] [Google Scholar]
- 5.Chan, Q. W., C. G. Howes, and L. J. Foster. 2006. Quantitative comparison of caste differences in honeybee hemolymph. Mol. Cell. Proteomics 5:2252-2262. [DOI] [PubMed] [Google Scholar]
- 6.Charpentier, X., and E. Oswald. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 186:5486-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claros, M. G., and P. Vincens. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241:779-786. [DOI] [PubMed] [Google Scholar]
- 8.Coburn, B., I. Sekirov, and B. B. Finlay. 2007. Type III secretion systems and diseases. Clin. Microbiol. Rev. 20:535-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coombes, B. K., M. J. Lowden, J. L. Bishop, M. E. Wickham, N. F. Brown, N. Duong, S. Osborne, O. Gal-Mor, and B. B. Finlay. 2007. SseL is a Salmonella-specific translocated effector integrated into the SsrB-controlled Salmonella pathogenicity island 2 type III secretion system. Infect. Immun. 75:574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 11.Davis, J., J. W. Wang, J. E. Tropea, D. Zhang, Z. Dauter, D. S. Waugh, and A. Wlodawer. 2008. Novel fold of VirA, a type III secretion system effector protein from Shigella flexneri. Protein Sci. 17:2167-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deiwick, J., S. P. Salcedo, E. Boucrot, S. M. Gillil, T. Henry, N. Petermann, S. R. Waterman, J. P. Gorvel, D. W. Holden, and S. Méresse. 2006. The translocated Salmonella effector proteins SseF and SseG interact and are required to establish an intracellular replication niche. Infect. Immun. 74:6965-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 14.Elliott, S. J., E. O. Krejany, J. L. Mellies, R. M. Robins-Browne, C. Sasakawa, and J. B. Kaper. 2001. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri. Infect. Immun. 69:4027-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Germane, K. L., R. Ohi, M. B. Goldberg, and B. W. Spiller. 2008. Structural and functional studies indicate that Shigella VirA is not a protease and does not directly destabilize microtubules. Biochemistry 47:10241-10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 17.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 18.Janda, J. M., and S. L. Abbott. 1993. Infections associated with the genus Edwardsiella: the role of Edwardsiella tarda in human disease. Clin. Infect. Dis. 17:742-748. [DOI] [PubMed] [Google Scholar]
- 19.Janda, J. M., S. L. Abbott, and L. S. Oshiro. 1991. Penetration and replication of Edwardsiella spp. in HEp-2 cells. Infect. Immun. 59:154-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juris, S. J., F. Shao, and J. E. Dixon. 2002. Yersinia effectors target mammalian signalling pathways. Cell. Microbiol. 4:201-211. [DOI] [PubMed] [Google Scholar]
- 21.Kay, B. K., M. P. Williamson, and M. Sudol. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14:231-241. [PubMed] [Google Scholar]
- 22.Kopecko, D. J., L. Hu, and K. J. Zaal. 2001. Campylobacter jejuni microtubule-dependent invasion. Trends Microbiol. 9:389-396. [DOI] [PubMed] [Google Scholar]
- 23.Kuhle, V., and M. Hensel. 2002. SseF and SseG are translocated effectors of the type III secretion system of Salmonella pathogenicity island 2 that modulate aggregation of endosomal compartments. Cell. Microbiol. 4:813-824. [DOI] [PubMed] [Google Scholar]
- 24.Kuhle, V., D. Jäckel, and M. Hensel. 2004. Effector proteins encoded by Salmonella pathogenicity island 2 interfere with the microtubule cytoskeleton after translocation into host cells. Traffic 5:356-370. [DOI] [PubMed] [Google Scholar]
- 25.Li, M., I. Rosenshine, H. B. Yu, C. Nadler, E. Mills, C. L. Hew, and K. Y. Leung. 2006. Identification and characterization of NleI, a new non-LEE-encoded effector of enteropathogenic Escherichia coli (EPEC). Microbes Infect. 8:2890-2898. [DOI] [PubMed] [Google Scholar]
- 26.Ling, S. H. M., X. H. Wang, L. Xie, T. M. Lim, and K. Y. Leung. 2000. Use of green fluorescent protein (GFP) to track the invasive pathways of Edwardsiella tarda in the in vivo and in vitro fish models. Microbiology 146:7-19. [DOI] [PubMed] [Google Scholar]
- 27.Marques, L. R. M., M. R. F. Toledo, N. P. Silva, M. Magalhäes, and L. R. Trabulsi. 1984. Invasion of HeLa cells by Edwardsiella tarda. Curr. Microbiol. 10:129-132. [Google Scholar]
- 28.Matsuzawa, T., A. Kuwae, S. Yoshida, C. Sasakawa, and A. Abe. 2004. Enteropathogenic Escherichia coli activates the RhoA signaling pathway via the stimulation of GEF-H1. EMBO J. 23:3570-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGhie, E. J., L. C. Brawn, P. J. Hume, D. Humphreys, and V. Koronakis. 2009. Salmonella takes control: effector-driven manipulation of the host. Curr. Opin. Microbiol. 12:117-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer, D. H., J. E. Rose, J. E. Lippmann, and P. M. Fives-Taylor. 1999. Microtubules are associated with intracellular movement and spread of the periodontopathogen Actinobacillus actinomycetemcomitans. Infect. Immun. 67:6518-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mota, L. J., and G. R. Cornelis. 2005. The bacterial injection kit: type III secretion systems. Ann. Med. 37:234-249. [DOI] [PubMed] [Google Scholar]
- 32.Nikolaus, T., J. Deiwick, C. Rappl, J. A. Freeman, W. Schröder, S. I. Miller, and M. Hensel. 2001. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J. Bacteriol. 183:6036-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okuda, J., M. Kiriyama, E. Yamanoi, and T. Nakai. 2008. The type III secretion system-dependent repression of NF-κB activation to the intracellular growth of Edwardsiella tarda in human epithelial cells. FEMS Microbiol. Lett. 283:9-14. [DOI] [PubMed] [Google Scholar]
- 34.Okuda, J., Y. Arikawa, Y. Takeuchi, M. M. Mahmoud, E. Suzaki, K. Kataoka, T. Suzuki, Y. Okinaka, and T. Nakai. 2006. Intracellular replication of Edwardsiella tarda in murine macrophage is dependent on the type III secretion system and induces an up-regulation of anti-apoptotic NF-κB target genes protecting the macrophage from staurosporine-induced apoptosis. Microb. Pathog. 41:226-240. [DOI] [PubMed] [Google Scholar]
- 35.Pallen, M. J., M. S. Francis, and K. Fütterer. 2003. Tetratricopeptide-like repeats in type-III-secretion chaperones and regulators. FEMS Microbiol. Lett. 223:53-60. [DOI] [PubMed] [Google Scholar]
- 36.Plumb, J. A. 1993. Edwardsiella septicaemia, p. 61-79. In V. Inglis, R. J. Roberts, and N. R. Bromage (ed.), Bacterial diseases of fish. Cambridge University Press, Cambridge, England.
- 37.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent end points. Am. J. Hyg. (Lond.) 27:493-497. [Google Scholar]
- 38.Rogers, L. D., A. R. Kristensen, E. C. Boyle, D. P. Robinson, R. T. Ly, B. B. Finlay, and L. J. Foster. 2008. Identification of cognate host targets and specific ubiquitylation sites on the Salmonella SPI-1 effector SopB/SigD. J. Proteomics 71:97-108. [DOI] [PubMed] [Google Scholar]
- 39.Rubirés, X., F. Saigi, N. Piqué, N. Climent, S. Merino, S. Albertí, J. M. Tomás, and M. Regué. 1997. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J. Bacteriol. 179:7581-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salcedo, S. P., and D. W. Holden. 2003. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 22:5003-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao, F. 2008. Biochemical functions of Yersinia type III effectors. Curr. Opin. Microbiol. 11:21-29. [DOI] [PubMed] [Google Scholar]
- 42.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Nat. Biotechnol. 1:784-791. [Google Scholar]
- 43.Srinivasa Rao, P. S., T. M. Lim, and K. Y. Leung. 2003. Functional genomics approach to the identification of virulence genes involved in Edwardsiella tarda pathogenesis. Infect. Immun. 71:1343-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srinivasa Rao, P. S., Y. Yamada, Y. P. Tan, and K. Y. Leung. 2004. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol. Microbiol. 53:573-586. [DOI] [PubMed] [Google Scholar]
- 45.Tan, Y. P., J. Zheng, S. L. Tung, I. Rosenshine, and K. Y. Leung. 2005. Role of type III secretion in Edwardsiella tarda virulence. Microbiology 151:2301-2313. [DOI] [PubMed] [Google Scholar]
- 46.Tan, Y. P., Q. Lin, X. H. Wang, S. Joshi, C. L. Hew, and K. Y. Leung. 2002. Comparative proteomic analysis of extracellular proteins of Edwardsiella tarda. Infect. Immun. 70:6475-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas, N. A., W. Deng, J. L. Puente, E. A. Frey, C. K. Yip, N. C. Strynadka, and B. B. Finlay. 2005. CesT is a multi-effector chaperon and recruitment factor required for the efficient type III secretion of both LEE- and non-LEE-encoded effectors of enteropathogenic Escherichia coli. Mol. Microbiol. 57:1762-1779. [DOI] [PubMed] [Google Scholar]
- 48.Thune, R. L., L. A. Stanley, and R. K. Cooper. 1993. Pathogenesis of gram-negative bacterial infections in warm water fish. Annu. Rev. Fish Dis. 3:37-68. [Google Scholar]
- 49.Wang, B., Z. L. Mo, Y. X. Mao, Y. X. Zou, P. Xiao, J. Li, J. Y. Yang, X. H. Ye, K. Y. Leung, and P. J. Zhang. 2009. Investigation of EscA as a chaperone for the Edwardsiella tarda type III secretion system putative translocon component EseC. Microbiology 155:1260-1271. [DOI] [PubMed] [Google Scholar]
- 50.Waterman, S. R., and D. W. Holden. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 5:501-511. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida, S., and C. Sasakawa. 2003. Exploiting host microtubule dynamics: a new aspect of bacterial invasion. Trends Microbiol. 11:139-143. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida, S., E. Katayama, A. Kuwae, H. Mimuro, T. Suzuki, and C. Sasakawa. 2002. Shigella deliver an effector protein to trigger host microtubule destabilization, which promotes Rac1 activity and efficient bacterial internalization. EMBO J. 21:2923-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida, S., Y. Handa, T. Suzuki, M. Ogawa, M. Suzuki, A. Tamai, A. Abe, E. Katayama, and C. Sasakawa. 2006. Microtubule-severing activity of Shigella is pivotal for intercellular spreading. Science 314:985-989. [DOI] [PubMed] [Google Scholar]
- 54.Zheng, J., and K. Y. Leung. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66:1192-1206. [DOI] [PubMed] [Google Scholar]
- 55.Zheng, J., N. Li, Y. P. Tan, J. Sivaraman, Y.-K. Mok, Z. L. Mo, and K. Y. Leung. 2007. EscC is a chaperone for the Edwardsiella tarda type III secretion system putative translocon components EseB and EseD. Microbiology 153:1953-1962. [DOI] [PubMed] [Google Scholar]
- 56.Zheng, J., S. L. Tung, and K. Y. Leung. 2005. Regulation of a type III and a putative secretion system in Edwardsiella tarda by EsrC is under the control of a two-component system, EsrA-EsrB. Infect. Immun. 73:4127-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]