Abstract
Yersinia pestis, the causative agent of plague, is one of the most virulent microorganisms known. The outer membrane protein X (OmpX) in Y. pestis KIM is required for efficient bacterial adherence to and internalization by cultured HEp-2 cells and confers resistance to human serum. Here, we tested the contribution of OmpX to disease progression in the fully virulent Y. pestis CO92 strain by engineering a deletion mutant and comparing its ability in mediating pneumonic plague to that of the wild type in two animal models. The deletion of OmpX delayed the time to death up to 48 h in a mouse model and completely attenuated virulence in a rat model of disease. All rats challenged with 1 × 108 CFU of the ompX mutant survived, compared to the 50% lethal dose (LD50) of 1.2 × 103 CFU for the wild-type strain. Because murine serum is not bactericidal for the ompX mutant, the mechanism underlying the delay in time to death in mice was attributed to loss of adhesion/internalization properties but not serum resistance. The rat model, which is most similar to humans, highlighted the critical role of serum resistance in disease. To resolve conflicting evidence for the role of Y. pestis lipopolysaccharide (LPS) and OmpX in serum resistance, ompX was cloned into Escherichia coli D21 and three isogenic derivatives engineered to have progressively truncated LPS core saccharides. OmpX-mediated serum resistance, adhesiveness, and invasiveness, although dependent on LPS core length, displayed these functions in E. coli, independently of other Yersinia proteins and/or LPS. Also, autoaggregation was required for efficient OmpX-mediated adhesiveness and internalization but not serum resistance.
Yersinia pestis, one of the most virulent microorganisms known, has caused three major pandemics of plague. This pathogen is transmitted by the bite of an infected flea (bubonic plague) or by inhalation of airborne bacteria (pneumonic plague) (31). Y. pestis carries a number of pathogenesis genes on the chromosome, but the pCD1 plasmid is essential for virulence. pCD1 encodes a type III secretion system and effector proteins with several functions, including circumventing host innate immunity (8). One mechanism by which Y. pestis evades the immune system involves entry into epithelial cells (9, 32). Although it is not completely understood, Y. pestis adherence and internalization into epithelial cells involve the Psa fimbria (32), the yadBC operon (17), yapE (30), yapC (15), and the outer membrane protein X (OmpX) (26).
OmpX was first described in Enterobacter cloacae (54, 55), but homologues, including PagC, Lom, Rck, and Ail (the attachment-invasion locus protein of Yersinia enterocolitica), have been identified in other Gram-negative bacteria (13, 20-22, 35). Y. pestis KIM OmpX, encoded by the y1324 gene, expressed at 28° and 37°C, is required for efficient bacterial adherence to and internalization by cultured HEp-2 cells and confers resistance to the bactericidal effect of human serum (4, 26). In vitro, deletion of OmpX reduces the autoaggregation phenotype, and pellicle formation is lost (26). OmpX is required for efficient delivery of Yersinia outer proteins (Yops) into human epithelial and monocyte cell lines (14). Additionally, OmpX is one of the most abundant proteins found in the Y. pestis outer membrane (OM) (37), and as such, is likely an important part of that structure.
The OM of Gram-negative bacteria is comprised of an asymmetric lipid bilayer containing lipopolysaccharide (LPS)-phospholipid embedded with numerous proteins. LPS consists of three domains: lipid A (acylated glucosamine residues), core (hetero-oligosaccharide), and an O antigen (a strain-specific, highly variable polysaccharide) (48). The amphiphilic character of LPS is important for the transfer of Omps and their positioning in the bilayer (11). Close LPS-protein interactions are required for protein support, proper folding, and biological activity (16, 27). Additionally, the proper arrangement of certain Omps in the bacterial cell wall is required for bacterial pathogenesis (27).
LPS of Y. pestis, in comparison to those of other Yersinia species, does not contain O antigen and has characteristic tetra-acylated lipid A at the mammalian host temperature (42, 43, 47). This modified form of lipid A results in poor induction of the host Toll-like receptor 4 (TLR4)-mediated innate immune response (36). In addition, a lack of O antigen and full LPS core length are essential for Pla protease activity (27, 56), which is responsible for dissemination of bacteria in the host (29, 50). Another component of Y. pestis LPS, the core fragment, was hypothesized to be responsible for bacterial serum resistance, an essential phenotype of pathogen survival in the bloodstream. It has been shown that Y. pestis strains that have a shortened LPS core, through directed mutagenesis or as a result of spontaneous mutations, are much more sensitive to human serum (3, 24, 25). However, this effect may be indirect, because LPS is known to be required for the structural integrity of numerous Omps (11, 16, 27, 56).
Here we tested the contribution of OmpX to disease progression with the fully virulent Y. pestis CO92 strain by engineering a deletion mutant and comparing its ability in mediating fatal disease to that of the wild type in a mouse model of pneumonic plague. Because murine serum is not bactericidal for Y. pestis ΔompX, experiments with mice cannot fully assess the role of serum resistance in plague pathogenesis, so rats, whose serum is bactericidal, were also used. To resolve the conflicting evidence for Y. pestis LPS and OmpX contributions to serum resistance, OmpX was cloned into Escherichia coli D21 and its three isogenic derivatives engineered to have progressively truncated LPS core saccharides, and OmpX display in the OM, cellular aggregation, attachment to and internalization into host cells, and serum resistance were assessed.
MATERIALS AND METHODS
Media, strains, and plasmids.
The strains and plasmids used in the study are listed in Table 1. E. coli strains were cultured in Luria-Bertani (LB) low-salt medium (EMD). Y. pestis CO92 was cultured under biosafety level 3 (BSL-3) conditions in brain heart infusion (BHI) medium (EMD) in accordance with the requirements and procedures involving the use of select agents and toxins. Antibiotics were used at the following concentrations: ampicillin (Amp), 50 μg ml−1; kanamycin (Kn), 50 μg ml−1. Cefsulodin-Irgasan-novobiocin (CIN) agar, also known as Yersinia selective agar (BD), was used to select for single-crossover recombinants, and LB agar with 5% sucrose and 50 μg ml−1 kanamycin and lacking NaCl was used to select for double-crossover recombinants, employing the sacBR locus.
TABLE 1.
Strains and plasmids used in the study
| Strain or plasmid | Genotype and/or relevant characteristics | Reference or source |
|---|---|---|
| Y. pestis strains | ||
| CO92 | pgm+ pYV+ pMT1+ pPCP1+ | Centers for Disease Control, Fort Collins, CO |
| CO92 ompX+/ompX::aph | pgm+ pYV+ pMT1+ pPCP1+ompX+ with integrated pMHZ3; merodiploid for ompX | This study |
| CO92 ΔompX::aph | pgm+ pYV+ pMT1+ pPCP1+ ΔompX::aph | This study |
| KIM6+ | pgm+ pYV− pMT1+ pPCP1+ | 26 |
| KIM6+ ΔompX::npt | pgm+ pYV− pMT1+ pPCP1+ ΔompX::npt | 26 |
| E. coli K-12 strains | ||
| DH5α | F−endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG φ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK− mK+) | Invitrogen |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(Strr) endA1 | Invitrogen |
| D21 | F−proA23 lac-28 tsx-81 trp-30 his-51 rpsL173(Strr) ampCp-1 | H. G. Boman, Coli Genetic Stock Center, Yale University |
| D21e7 | F−proA23 lac-28 tsx-81 trp-30 his-51 rpsL173(Strr) rfa-1 ampCp-1 | H. G. Boman, Coli Genetic Stock Center, Yale University |
| D21f1 | F−proA23 lac-28 tsx-81 trp-30 his-51 rpsL173(Strr) rfa-21 rfa-1 ampCp-1 | H. G. Boman, Coli Genetic Stock Center, Yale University |
| D21f2 | F−proA23 lac-28 tsx-81 trp-30 his-51 rpsL173(Strr) rfa-31 rfa-1 ampCp-1 | H. G. Boman, Coli Genetic Stock Center, Yale University |
| CC118 λpir | r− m+ λpir+; cloning strain | 12 |
| SM10 λpir | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr λpir+; conjugation strain | 51 |
| Plasmids | ||
| pPCR2.1-TOPO | Ampr Kmr | Invitrogen |
| pOmpX | pPCR2.1-TOPO carrying ompX gene with its native promoter from Y. pestis | This study |
| pTA | ompX− control plasmid derived from pOmpX by deletion of the fragment between EcoRI restriction enzyme-cut sites | This study |
| pMHZ1 | mob+, pir-dependent oriR6K, sacBR Cmr containing y1324ompX gene from Y. pestis | 26 |
| pMHZ3 | pMHZ1 containing ΔompX::aph | This study |
| pGP704 | mob+, pir-dependent oriR6K, Ampr | J. J. Mekalanos, PlasmID, Harvard Medical School |
| pRL250 | sacBR+ | 52 |
| pGP704.L | pGP704 containing sacBR cloned from pRL250 | This study |
| pGP704.O | pGP704.L containing ΔompX::aph; allelic exchange plasmid | This study |
HEp-2 cells (ATCC CCL-23) were grown in 6% CO2 (37°C) in growth medium (GM) (low-glucose Dulbecco's modified Eagle's medium [Gibco] supplemented with 10% [vol/vol] fetal bovine serum [FBS] [HighClone] and 1% [vol/vol] penicillin-streptomycin solution [Gibco]). For the cell association and internalization assays, internalization medium (IM) (GM lacking FBS and antibiotics) was used.
Generation of a ΔompX mutant of Y. pestis CO92.
The pRL250 plasmid was cut with SalI and EcoRV, and the ∼2.6-kb band carrying the sacBR locus was gel purified. The DNA fragment was cloned into pGP704, creating the pGP704.L plasmid, which was transformed into E. coli CC118 λpir. The pMHZ1 plasmid harboring the ompX gene was cut with MfeI and NdeI (sites in ompX), generating a 426-bp deletion. A gene conferring Knr (aminoglycoside 3′-phosphotransferase; aph) was amplified by PCR (forward primer, 5′ATGCCAATTGCGCAAAGAGAAAGCAGGTA3′; reverse primer, 5′CGCGCATATGTTTCAATTCAGAAGAACTCGTC3′) using plasmid pCR2.1-TOPO (Invitrogen) as a template and cloned into pMHZ1 between the MfeI and NdeI sites. The resulting construct, pMHZ3 with ompX disrupted by Knr, was used as a template for PCR with primers complementary to the cloned upstream (45-bp) and downstream (99-bp) regions of the ompX gene and engineered SacI sites (forward primer, 5′GCAGGAGCTCTCATGTCAGATATTTG3′; reverse primer, 5′ATACGAGCTCTAGCCTACCCCTATTAA3′). The generated PCR product (1,351 bp) was cloned into the multiple-cloning site of plasmid pGP704.L, and the resulting construct, pGP704.O, was transformed into Escherichia coli SM10 λpir. A representative clone was mated with Y. pestis CO92 as described previously (26) and counterselected on CIN agar. This merodiploid strain (ompX+ ompX::aph) was generated by a homologous single-crossover recombination. It was maintained and served as (i) a single-copy ompX complementation control and (ii) an isogenic precursor for selecting the ompX::aph disruption. The latter was isolated on LB agar containing sucrose to select for a second crossover event while maintaining selection for the ompX::aph disruption. Sucrose-resistant, Amp-sensitive colonies were tested by PCR (26) for the ompX::aph disruption.
Infection studies.
Animal exposures with Y. pestis CO92 were performed under BSL-3 conditions in accordance with the requirements and procedures involving the use of select agents and toxins. The use of the animals was approved by the University of Idaho Biosafety Committee, and the animals were handled in accordance with the University of Idaho's Animal Care and Use Committee guidelines. Infection studies were performed as previously described (1). Briefly, Y. pestis CO92 strains were grown overnight at 28°C, washed, resuspended in 25% glycerol, and frozen at −80°C. The bacterial numbers in the stock cultures were verified by plate count on LB agar medium, and the pigmentation phenotype of the ompX mutant was confirmed from the original frozen stocks used in the animal challenge experiments by visualizing red colonies on Congo red agar. On the day of the experiment, the cultures were thawed, aliquoted, and diluted in phosphate-buffered saline (PBS) to the desired concentration. Eight- to 10-week-old female BALB/c mice or 6- to 8-week-old Sprague-Dawley rats (Simonsen Labs) were challenged intranasally (10 μl total [5 μl/naris] for mice and 50 μl total [25 μl/naris] for rats). Challenges of 1 50% lethal dose (LD50), 10 LD50, and 100 LD50 established previously for the wild type (1 LD50 = 2 × 104 CFU [1]) were used for mouse infections. Serial 10-fold dilutions (102 to 108 CFU of the ΔompX mutant and 102 to 104 CFU of the wild type) were used in the rat infections. Bacterial numbers in the challenge inocula were confirmed by plate count on LB agar. The animals were monitored for morbidity (ruffled fur and decreased mobility) and mortality through 10 days postchallenge.
Generation of pOmpX and pTA constructs.
The DNA fragment including the Y. pestis KIM6+ ompX gene (y1324) with neighboring upstream and downstream regions was amplified by PCR with the primers indicated above. The resulting 750-bp PCR fragment was cloned into the pPCR2.1-TOPO plasmid, creating the pOmpX plasmid. The pTA plasmid (vector control) was made by deletion of the 766-bp fragment between the EcoRI restriction enzyme cut sites in the pOmpX plasmid and ligation of the backbone. The resulting plasmids were transformed into E. coli strains TOP10, D21, D21e7, D21f1, and D21f2. pOmpX-transformed colonies with strong autoaggregation and pellicle formation were selected and used for further studies.
Proteomic analysis of pOmpX and pTA constructs.
To verify expression of OmpX from the pOmpX vector, whole-cell lysate proteins were separated by SDS-PAGE through 12.5% acrylamide as previously described (26). Proteins extracted from bacteria grown at 37°C (200 rpm) in LB medium to mid-exponential phase (optical density at 600 nm [OD600] of ∼0.6 to 0.8) were used for the separation.
Cell association and internalization assays.
Cell association and internalization assays were performed as described previously with slight modifications (26). Briefly, E. coli cells from mid-exponential phase (as described above) were washed in PBS (0.01 M sodium phosphate, 0.8% NaCl, pH 7.2) and resuspended in IM. Dilutions were made in PBS to determine cell numbers by plate count. HEp-2 cells (1 × 105 per well) in GM were incubated in 24-well plates (6% CO2, 37°C) for 42 h. The cell monolayers were washed thrice with IM, and approximately 107 CFU E. coli was added to each well to produce cocultures with a multiplicity of infection (MOI) of 20 to 50. The plates were centrifuged (5 min, 1,000 × g, 18°C) and incubated for 1 h as described above. Cocultures were either washed nine times to remove unbound bacteria (cell association assay) or washed three times with IM, incubated for another 1.5 h in IM with 500 μg gentamicin per well (Gibco) to kill extracellular bacteria without lysing host cells, and then washed three times with PBS (internalization assay). Trypsin-like enzyme (Tryp-Le Express; Gibco) was added to each well and incubated for 7 min to detach the HEp-2 cells from the culture plates. Triton X-100 (0.025%) was added to release intracellular bacteria, and bacterial numbers were determined by plate count.
Serum resistance assays.
A serum resistance assay was performed as described previously (26). Bacteria grown at 37°C (200 rpm) in LB medium were collected at mid-exponential phase (OD600 of ∼0.6 to 0.8), washed twice in PBS (0.01 M sodium phosphate, 0.8% NaCl, pH 7.2), diluted 100-fold, mixed with an equal volume of normal serum or heat-inactivated serum (HIS) (incubated at 56°C for 30 min to inactivate complement), and incubated at 37°C. Viable bacteria were quantified by plate counts as described above following incubation in serum for 1 h.
OMP purification.
Bacteria were grown as described for the serum resistance assay, collected by centrifugation (6,000 × g at 4°C), and frozen at −80°C. Triton X-100-insoluble outer membrane proteins (OMPs) were extracted as previously described by Biedzka-Sarek et al. (6). To confirm that the OmpX protein was in the OM and not in cytosolic inclusion bodies, the cells were centrifuged and sonicated and the pellet was collected (1 h at 45,000 × g) as described previously (6). Potential protein aggregates in inclusion bodies were removed by a 1-h wash with 5 M urea in PBS at 4°C as described previously (33), followed by 2 h of centrifugation at 45,000 × g to collect the membrane fractions. Proteins were separated by 12.5% SDS-PAGE as described previously (26).
Field emission scanning electron microscopy (FESEM) imaging.
The Hep-2 cells were seeded (105 cells/chamber) in a Lab-Tek four-chamber slide system (Thermo Fisher Scientific) and grown for 48 h. Bacteria were grown, collected, and used for the adhesion assay as described above. Cocultures were incubated for 15 min, washed nine times, and fixed with 2% paraformaldehyde-2.5% glutaraldehyde in 0.1 M cacodylate buffer overnight at 4°C. The next day, samples were rinsed three times (5 min each wash) with 0.1 M cacodylate buffer and postfixed with 2% OsO4 in 0.1 M cacodylate buffer for 2 h room temperature (RT). After three rinses with water the samples were stained with 1% tannic acid for 1 h at RT, rinsed twice with water, and dehydrated with ethanol and hexamethyldisilizane (HMSDS) as the final dehydration step. Samples were sputter coated with gold and analyzed by Quanta 200F (Field Emission Instruments) operating at 30 kV and at a magnification of 16,000.
Statistical analysis.
Animal survival data were analyzed with the log rank test. Point estimates of the LD50 and 95% confidence intervals were determined using a logistic regression model with the logit link function. Data from cell association, internalization, and serum resistance assays were analyzed using either analysis of variance (ANOVA) with Tukey's posttests, the Kruskal-Wallis test with Dunn's posttests, or repeated-measures ANOVA. These analyses were conducted with R or SAS software.
RESULTS
Deletion of ompX extends the time to death in Y. pestis CO92 pneumonic infections.
To assess the biological significance of OmpX in fully virulent Y. pestis in vivo, studies in a mouse model of infection were performed. ompX was deleted and marked by a kanamycin resistance cassette in Y. pestis CO92, and a merodiploid strain was used as a control. Mice were challenged intranasally with 1, 10, and 100 LD50 and observed for 10 days. Challenge with the ΔompX strain at both 10 LD50 (Fig. 1A) and 100 LD50 (Fig. 1B) resulted in significant extensions of time to death. Pneumonic infection with the wild-type strain resulted in death of the animals on day 3 (90% mortality in animals infected with 10 LD50 and 100% in animals infected with 100 LD50), with no fatalities in the group infected with the ΔompX mutant at that time. All mice challenged with the wild type at both doses developed terminal plague by day 4, while the survival among animals challenged with the ΔompX mutant on that day was 90% (10-LD50 group) and 50% (100-LD50 group). One hundred percent mortality of the animals infected with the ΔompX mutant was delayed by 24 h (10-LD50 group) or 48 h (100-LD50 group). The merodiploid ompX+/ΔompX strain restored full virulence and all challenged mice died by day 4, similar to the case for mice challenged with the wild type. These data and experiments using a dose of 1 LD50 (data not shown) were used to calculate the LD50 value for the ΔompX mutant as 2 × 104 CFU (95% confidence interval, (9.7 × 103 to 4.5 × 104 CFU), similar to that of the wild type. However, the pronounced delayed time to death demonstrated that loss of OmpX altered the course of pneumonic plague infection.
FIG. 1.
Deletion of ompX extends the time to death in Y. pestis CO92 pneumonic infections. Groups of BALB/c mice (n = 8/group) were challenged intranasally with 10 LD50 (A) or 100 LD50 (B), doses established previously for WT Y. pestis CO92 (LD50 = 2 × 104 CFU). The animals were monitored for morbidity and mortality through 10 days postchallenge. The graph presents survival of the animals challenged with WT, ΔompX, or ompX+/ΔompX strains. A difference in survival times for animals infected with the ΔompX strain was observed (log rank test, P < 0.001). Data represent combined survival numbers from two experiments performed on separate days.
Deletion of ompX decreases the virulence of Y. pestis CO92.
Our previous work and that of others showed that human serum is bactericidal to the ΔompX (ail) mutant (4, 26). Also, work by Bartra et al. showed that mouse serum is not bactericidal to the ΔompX (ail) mutant (4), and for this reason the mouse infection model cannot be used to fully assess the role of OmpX and serum resistance in plague pathogenesis. To select a more appropriate animal model that would assess OmpX-conferred complement resistance, rat sera were tested. A resistance assay with Y. pestis KIM6+ and its isogenic ΔompX derivative was performed, comparing normal rat serum (NRS) and normal mouse serum (NMS) (Fig. 2). One hour of incubation in NRS resulted in death of 2 orders of magnitude (98%) of the ΔompX strain, and incubation for an additional 1.5 h increased bacterial death by another order of magnitude (99.84%). In contrast, this strain survived incubation in mouse serum for both of these durations, as expected. Similarly, the parental strain was not susceptible to complement killing by mouse or rat serum. These serum assays indicated that rat infections with Y. pestis CO92 may better reflect the role of OmpX in human disease; thus, a rat model of pneumonic infections was employed. Rats were challenged intranasally with 102 to 108 CFU of the ΔompX mutant and 102 to 104 CFU of the wild type and observed for 10 days (Fig. 3). Deletion of ompX caused complete attenuation of the Y. pestis CO92 strain in the rat pneumonic model of infection. One animal infected with 107 CFU died on day 8, but no fatalities were recorded with the challenge dose of 108 CFU. Hence, the LD50 of the mutant was higher than 108 CFU, while the value estimated for the wild type was 1.2 × 103 CFU (95% confidence interval, 4.2 × 102 to 3.2 × 103 CFU). These results indicate the essential role of serum resistance in pneumonic plague. They also underline the significant contribution of OmpX-conferred serum resistance over bacterial adhesion and invasion in the pathogenesis of Y. pestis.
FIG. 2.
Rat but not mouse serum is bactericidal to the ΔompX strain. Y. pestis KIM6+ and its isogenic ΔompX derivative were grown to mid-exponential phase at 28°C; incubated in 50% normal mice serum (NMS), normal rat serum (NRS), or the respective heat-inactivated sera (HIS) for 0, 1, and 2.5 h at 37°C; and plated on LB agar. Graphs represent surviving bacteria after incubation with NMS and NRS as a percentage of the number of bacteria that survived treatment with HIS (100%). A significant effect of the type of serum for the ΔompX strain was observed (repeated-measures ANOVA, P < 0.001). Results are the mean ± standard errors of the means (SEM) from data derived from two assays performed on separate days (n = 4).
FIG. 3.
Deletion of ompX attenuates virulence of Y. pestis CO92 in rat pneumonic infection. Groups of Sprague-Dawley rats (n = 3/group/challenge) were challenged intranasally with 10-fold serial dilutions of the WT Y. pestis CO92 (102 to 104 CFU) or the isogenic ΔompX derivative (102 to 108 CFU). The animals were monitored for morbidity and mortality through 10 days postchallenge. The graph shows survival of the animals challenged with the WT or ΔompX strains. Survival comparisons between the animals infected with the highest number of WT organisms that killed 100% of the animals (104 CFU) and the animals infected with that number of ΔompX organisms were evaluated by the log rank test (P = 0.01). Data represent combined survival numbers from three experiments.
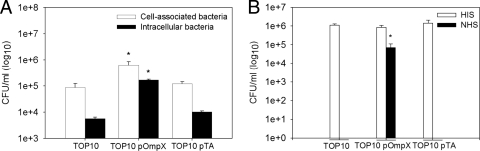
Expression of OmpX in E. coli K-12 TOP10 promotes adherence, internalization, and serum resistance.
The deletion of OmpX protein in Y. pestis KIM6+ leads to the impairment of adherence of bacteria to and internalization into human epithelial cells and loss of resistance to human serum (26). To confirm that these traits are mediated by OmpX protein independently from other Y. pestis proteins or LPS, the y1324 gene with its neighboring upstream and downstream DNA was cloned into plasmid pPCR2.1-TOPO, expressed in the standard cloning strain E. coli TOP10 (Fig. 4), and tested for growth characteristics, cell association, and serum resistance. Expression of OmpX resulted in phenotypic bacterial changes, including flocculent growth and pellicle formation, in LB medium (data not shown). Both characteristics are traits observed in Y. pestis and attributed to OmpX (26). Cell association assays showed that E. coli expressing OmpX had a 5- to 7-fold increase in association (both extra- and intracellular bacteria) with the human epithelial cell line (Hep-2) compared to the control strains (Fig. 5A). An even more pronounced effect was observed with internalized bacteria. There was a 16- to 30-fold increase in the number of OmpX-expressing E. coli bacteria internalized by the epithelial cells compared to control E. coli. This high rate of internalization accounted for nearly 27% of the total cell-associated bacteria that express OmpX, versus 6.5% and 8.5% of the host and plasmid control strains, respectively. We also confirmed that OmpX expressed from the plasmid efficiently mediated resistance to human complement (Fig. 5B). One hour of incubation in normal human serum (NHS) killed all host and plasmid control bacterial cells, while cells expressing OmpX survived. HIS was not lethal to any of the tested strains, which indicated that the bactericidal effect observed with NHS was complement mediated. Our results demonstrated that OmpX expressed in a bacterial host other than Y. pestis mediated adherence to and internalization into epithelial cells and conferred resistance to human serum. Also, our observations confirmed that the product of the cloned fragment was efficiently expressed from the pOmpX plasmid construct and was functional.
FIG. 4.
Generation of pOmpX and pTA constructs for expression of OmpX in E. coli. (A) Schematic representation of the DNA fragment cloned into plasmid pPCR2.1-TOPO. The y1324 gene from Y. pestis KIM6+ with neighboring upstream and downstream regions (750 bp) was cloned into the pPCR2.1-TOPO plasmid, creating the pOmpX plasmid; the pTA plasmid (vector control) was made by deletion of the 766-bp fragment between EcoRI restriction enzyme-cut sites in the pOmpX plasmid and relegation of the backbone. (B) PCR analysis with M13 primers and confirmation of pOmpX and pTA constructs. Lane 1, DNA size marker (Fisher Scientific); lane 2, negative control (no DNA); lane 3, pOmpX; lane 4; pTA. (C) Analysis of OmpX-expressing E. coli TOP10 strains. E. coli Top10, E. coli Top10(pOmpX), and E. coli Top10(pTA) were grown to mid-exponential phase at 37°C and collected by centrifugation. Whole-cell lysate proteins were extracted, separated by SDS-PAGE, and stained with Coomassie blue. Molecular masses were estimated with a prestained Pageruler protein standard (Fermentas). The arrow indicates OmpX protein expressed by the pOmpX-carrying strain.
FIG. 5.
Expression of OmpX in E. coli K-12 TOP10 promotes adherence, internalization, and serum resistance. (A) Cell association. Bacteria were grown to mid-exponential phase at 37°C and coincubated for 1 h with Hep-2 monolayers at 37°C (MOI = 20 to 50). Hep-2 host cells were disrupted and plated on LB agar or incubated with gentamicin for 1.5 h to kill extracellular bacteria before plating. Open bars represent the number of cell-associated bacteria (extracellular and intracellular), and filled bars represent the number of intracellular bacteria. Results are means ± SEM from assays performed in duplicate on two separate days (n = 8). (B) Serum resistance. Bacteria were grown as described above and incubated in 50% normal human serum (NHS) or heat-inactivated serum (HIS) for 1 h at 37°C and plated on LB agar. Results are means ± SEM from two assays performed on separate days (n = 8 for cell association assay; n = 4 for serum resistance assay). The asterisk indicates a difference (ANOVA, P < 0.0001, for cell association assay; Kruskal-Wallis, P < 0.001, for serum resistance assay) between the strain expressing OmpX [TOP10(pOmpX)] and the two controls [cell TOP10 and plasmid control TOP10(pTA) strains).
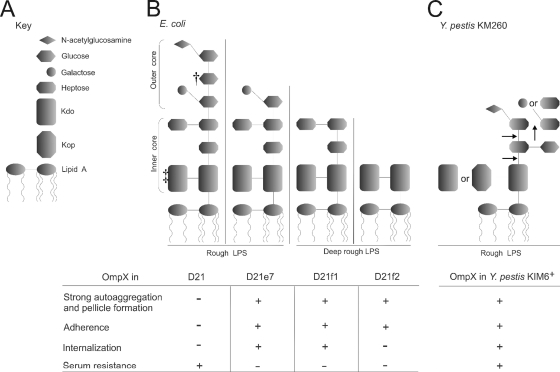
The length of the LPS core determines OmpX-conferred serum resistance.
As shown previously by Knirel et al., Y. pestis mutants possessing different LPS core lengths have altered sensitivity to human serum (24). To investigate the potential effect of LPS core length on OmpX functions, the pOmpX plasmid was transformed into the E. coli D21 strain and its three isogenic derivatives D21e7 (rough), D21f1 (deep rough), and D21f2 (deep rough), each with progressively truncated core saccharide residues (Fig. 6). Each construct was tested for serum resistance. After 1 h of incubation in NHS, all host and OmpX-expressing strains were killed except for the D21(pOmpX) strain (Fig. 7). Only the strain with the full-length LPS core and OmpX expression survived the exposure to NHS (20% survival). All transformed colonies used in the study had enhanced autoaggregation and pellicle formation during mid-exponential growth, with the exception of the parental D21 strain, in which rapid loss of these two traits during culture passage was observed (data not shown). These results confirmed the hypothesis that OmpX-mediated resistance to human serum depends on the LPS core and showed that both the inner and outer core are required for effective resistance to the bactericidal effect of complement in this model.
FIG. 6.
Schematic representation of E. coli and Y. pestis LPS structures and summary of the results. (A) Key to LPS moieties. (B) LPS structures of E. coli K-12 D21 and its isogenic mutants (23, 40, 44, 46, 59). †, some sources indicate galactose (23); ‡, some sources indicate two or three KDO residues (40). (C) LPS core structure of Y. pestis KM260 grown at ambient temperature; arrows indicate truncation of the core residues that resulted in loss of serum resistance (24). The table shows presence (+) or lack (−) of OmpX-mediated phenotypes (autoaggregation and pellicle formation, adhesion to Hep-2 cells, internalization into Hep-2 cells, and human serum resistance) related to a particular strain. Data are a summary of this study and previous work on OmpX of Y. pestis KIM6+ (26).
FIG. 7.
The length of the LPS core determines OmpX-conferred serum resistance. E. coli K-12 D21 (rough LPS) and its three isogenic derivatives D21e7 (rough LPS), D21f1 (deep rough LPS), and D21f2 (deep rough LPS) with or without pOmpX were grown to mid-exponential phase at 37°C, incubated in 50% normal human serum (NHS) or heat-inactivated serum (HIS) for 1 h at 37°C, and plated on LB agar. Results are means ± SEM from data derived from two assays performed on separate days (n = 4). The asterisk (*) indicates the difference (Kruskal-Wallis test, P < 0.001) between E. coli carrying pOmpX and its respective cell control strain after incubation with NHS.
OmpX localizes to the OM in E. coli K-12 D21 strains.
To confirm that the differences in serum resistance between the E. coli D21 strains were not due to a lack of OmpX on the bacterial surface, whole-cell lysates and outer membrane (OM) fractions were extracted and separated by SDS-PAGE (Fig. 8). The results showed that the protein was expressed in all pOmpX-carrying strains (Fig. 8A). Also, the OM preparations indicated the presence of OmpX in the OMs of all pOmpX-carrying strains, with some differences in the protein concentration (Fig. 8B). Higher levels were extracted from the parental D21 strain and the D21f1 mutant than from the other strains. To show that OmpX was present only in the OM and not in cytosolic inclusion bodies that may copurify with OM proteins, extracts were washed with 5 M urea to dissolve the potential aggregates (33), and urea-resistant membrane fractions were collected (Fig. 8C). OmpX protein was present in the urea-insoluble membrane fractions of all OmpX-expressing strains, confirming the proper localizations of the protein in the OM.
FIG. 8.
Analysis of OmpX localization in E. coli K-12 D21 strains. OmpX-expressing E. coli D21, D21e7, D21f1, and D21f2 and their parental strains were grown to mid-exponential phase at 37°C and collected by centrifugation. Whole-cell lysate proteins (A) and Triton X-100-insoluble (B) or 5 M urea-insoluble (C) outer membrane proteins were extracted, separated by SDS-PAGE, and stained with Coomassie blue. Molecular mass markers were estimated with a prestained Pageruler protein standard (Fermentas). Arrows indicate OmpX protein.
The length of the LPS core determines OmpX-mediated internalization and adherence.
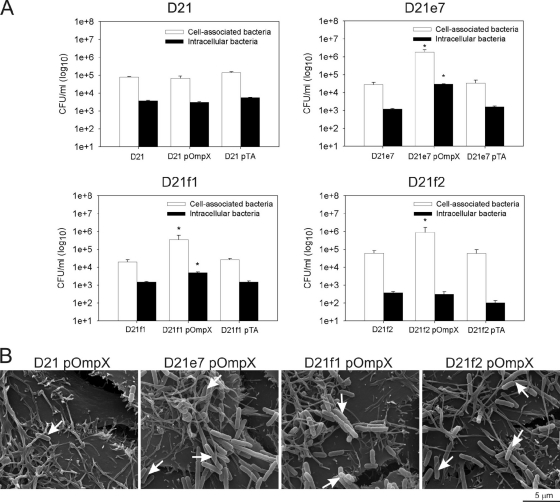
To investigate the role of LPS in other traits conferred by OmpX, the recombinant E. coli strains were analyzed in cell association and invasion assays (Fig. 9A). All of the pOmpX-carrying strains except for the E. coli D21 parental strain showed enhanced cell association, with the highest number of associating cells occurring with D21e7, the mutant strain with the longest LPS core. Comparison of the number of cell-associated CFU showed that approximately 66-fold more D21e7 and 14-fold more D21f1 and D21f2 OmpX-expressing bacteria adhered to the host cells than the respective host and plasmid control strains. The lack of enhanced association with Hep-2 cells of E. coli D21(pOmpX) correlated with loss of autoaggregation in this strain. FESEM imaging confirmed the pattern of a lower level of cell association of D21(pOmpX) and enhanced adherence mediated by OmpX expression in the D21e7, D21f1, and D21f2 strains (Fig. 9B).
FIG. 9.
The length of the LPS core determines OmpX-mediated internalization and adherence. (A) E. coli K-12 D21 (rough LPS) strain and its three isogenic derivatives D21e7 (rough LPS), D21f1 (deep rough LPS), and D21f2 (deep rough LPS) with or without pOmpX were grown to mid-exponential phase at 37°C and coincubated for 1 h with Hep-2 monolayers at 37°C (MOI = 20 to 50). Hep-2 host cells were disrupted and plated on LB agar or incubated with gentamicin for 1.5 h to kill extracellular bacteria before plating. Open bars represent the number of cell-associated bacteria (extracellular and intracellular), and filled bars represent the number of intracellular bacteria. Results are means ± SEM from assays performed in duplicate on two separate days (n = 8). The asterisk (*) indicates differences (ANOVA, P < 0.0001) between the strain expressing OmpX (pOmpX) and cell (D21, D21e7, D21f1, or D21f2) and plasmid (pTA) control strains. (B) Field emission scanning microscopy imaging of OmpX-expressing E. coli associated with Hep-2 cells. Bacteria and eukaryotic cells were grown as described above and cocultured for 15 min at 37°C. After extensive washing, the cells were fixed, treated with tannic acid, OsO4, dehydrated, and gold coated. The samples were analyzed with Quanta 200F (Field Emission Instruments) at a magnification of ×16,000. The bar represents 5 μm; white arrows indicate bacteria associated with Hep-2 cells.
Interestingly, even though D21e7, D21f1, and D21f2 OmpX-expressing strains adhered readily to the host cells, there was a pronounced difference in internalization rate depending on the polysaccharide length of the LPS core (Fig. 9A). The progressive truncation of the core residues resulted in progressive loss of OmpX-mediated internalization. Expression of OmpX in the D21f2 strain with the shortest inner core, containing only 2-keto-3-deoxyoctulosonic acid (KDO) residues, did not enhance the bacterial invasion into Hep-2 cells [P > 0.05 for D21f2(pOmpX) versus D21f2 and D21f2(pTA)]. The mutant strain with three additional heptose residues in the core had in a small, yet significant (P < 0.0001), increase in internalization. Approximately 5-fold more OmpX-expressing D21f1 bacteria invaded epithelial cells than the D21f1 or D21f1(pTA) control strains. The biggest difference in invasion between the OmpX-expressing strains and the controls was seen with the rough mutant D21e7, with an ∼20-fold increase versus the controls. As a consequence of the lack of enhanced adherence, there was no change in internalization of the D21(pOmpX) strain even though the protein was present in the OM and this strain was resistant to human serum (Fig. 7 and 8B and C).
These results confirmed that OmpX, independent from other Y. pestis proteins, conferred bacterial adherence to and internalization into host cells; however, the efficiency of adherence and invasion, similar to serum resistance, was affected by the length of the heterosaccharide content in the LPS core. The results obtained with the D21 strain suggested that autoaggregation of bacterial cells also depended on the display of OmpX in a specific LPS core background and that flocculent growth was critical for effective bacterial adherence.
DISCUSSION
The most important findings of this study were that (i) the loss of a single gene (ompX) caused complete attenuation of Y. pestis by affecting serum resistance and adhesion to and invasion of host cells, (ii) avirulence was dependent on an appropriate animal model, (iii) OmpX conferred its characteristics independently from other Y. pestis proteins or LPS, and (iv) recombinant isogenic strains of E. coli showed that the OmpX-dependent phenotypes were influenced by the LPS core saccharide length. For the first time, the role of OmpX in Y. pestis virulence was comprehensively assessed using a fully virulent strain, pneumonic route of infection, and two animal models to account for differences in serum resistance.
There are conflicting roles for ompX in an intravenous mouse model of plague with a ΔompX mutant in the attenuated Y. pestis strain KIM5. Bartra et al. (4) showed that deletion of ompX does not affect virulence, while Felek et al. (15) showed that the deletion causes a >3,000-fold increase in the LD50 and that fewer bacteria colonize host organs in the early days of infection compared to the wild type. Our mouse model studies using the fully virulent Y. pestis CO92 strain with a pneumonic route of challenge showed a delayed time to death but no decrease in virulence (no increased LD50) of an ompX mutant compared to the wild type. Thus, our studies are in agreement with the findings of Bartra et al. (4). Our assays also confirmed that murine serum is not bactericidal for the ompX mutant (4). Thus, the mechanism underlying the delay in time to death in mice challenged with Y. pestis ΔompX may be attributed to loss of adhesion/internalization properties but not serum resistance. These studies also showed that the role of serum resistance in Y. pestis pathogenesis cannot be tested in mice. To assess the full contribution of OmpX (adherence, internalization, and serum resistance) to virulence, we used a rat model of pneumonic plague. In this model, the Y. pestis ΔompX was completely attenuated (all rats challenged with 1 × 108 CFU survived, compared to the LD50 of 1.2 × 103 CFU of the wild-type strain). Even though the lung is not an organ known for high complement activity (7), the importance of serum resistance in pneumonic plague was highlighted by comparing differences in mortality between mice and rats. Serum resistance contributed to virulence more prominently than bacterial association with host cells.
The bactericidal properties of complement vary by species, and activity often reflects the host's response to infection and efficient bacterial transmission (28). For example, the pattern of serum resistance of different Borrelia spp. correlates with the host that they are able to infect (28). The mechanisms of these differences are not clear, but they involve both bacterial and host components (53). From the host side, the complement regulatory system, rather than components of the membrane attack complex, is suspected (45, 58). Some animal species have a more complex protease inhibitor system regulating the complement cascade; for example, analysis of mouse, rat, and human genomes reveals that the systems are encoded by 199, 183, and 156 genes, respectively (45). Mouse complement is particularly interesting because its bactericidal activity is limited (18, 34). A number of common microorganisms are killed by sera of other species, e.g., human or rat, but remain resistant to murine sera (10, 19, 34, 57). Studies to distinguish complement species specificity and activation on bacterial surfaces identified two critical regulatory proteins: factor H (fH) and C4 binding protein (C4bp) (39). C4bp is involved in regulation of the classical pathway (5), and fH is an inhibitor of an alternative complement pathway (53). Exclusive binding of human fH and C4bp by N. gonorrhoeae ensures its resistance to human serum and reflects restricted infection of the human species (38). Similarly, differential binding of fH obtained from sera of various mammalian species and its correlation with the natural hosts occurs for Borrelia burgdorferi (53). Although the underlying differences between mouse and rat complement that affect Y. pestis are beyond the scope of this work, the findings of Bartra et al. (4) and our findings here confirm that species specificity for the complement alternative pathway also applies to Y. pestis ΔompX.
From the bacterial side, the mechanism of serum resistance in Y. pestis has been controversial. Previously we showed that deletion of ompX leads to a pronounced increased sensitivity to human serum (26). However, Knirel et al. (24) showed that Y. pestis loss of serum resistance was due to mutations affecting LPS core structure. To address these two observations, we first determined that E. coli OmpX-expressing cells conferred the Y. pestis phenotypes of serum resistance, autoaggregation, and invasiveness. This indicated that these traits were independent of other Y. pestis factors. This is consistent with the observations of Bartra et al. (4), who also showed that E. coli expressing OmpX was serum resistant. We next examined OmpX-dependent serum resistance in three isogenic strains of E. coli differing only in the length of the LPS core region. Deep rough strains D21e7 (having the KDO moiety only) and D21f1 (having KDO with three heptose residues) and a rough strain D21e7 (with additional glucose and branched galactose) were sensitive to human serum. This is in contrast to the fully serum-resistant E. coli parental strain expressing OmpX. These experiments clearly show that serum resistance is OmpX dependent, but, significantly, this resistance is influenced by the molecular composition of the LPS core structure. This interaction between OmpX and LPS composition explains the loss of serum resistance conferred by both by a mutation in ompX (26) and mutations affecting LPS (24).
The deletion of ompX in Y. pestis impairs adhesiveness and invasiveness of the bacterium (26). A decrease in cell attachment after ompX deletion in Y. pestis and a gain of adhesiveness in E. coli AAEC185 expressing OmpX were confirmed by Felek and Krukonis (14). Similar to the case for serum resistance, we showed that OmpX-conferred adhesiveness and invasion were dependent on the LPS background in which the protein was expressed. While all of the E. coli OmpX-expressing LPS mutant strains readily adhered to Hep-2 cells, the parental D21 strain did not, even though OmpX was present in the OM and the strain was serum resistant. Lack of cell association correlated with the fact that the OmpX-mediated autoaggregation phenotype was not maintained in this strain. Also, progressive truncation of the LPS core resulted in a proportional decrease in internalization of OmpX-expressing E. coli cells, with the most pronounced effect in strain D21f2 (possessing only KDO residues of the core inner portion), which had invasion levels similar to those of the OmpX-negative control strain. LPS core mediates internalization of some bacteria (41), and alterations in its structure can lead to impairment of bacterial invasion (60). Our data indicated that this was also the case for the E. coli K-12 D21 mutant strains (lower invasion level of strains with shorter LPS cores). However, introduction of OmpX significantly increased invasiveness of strains having low levels of internalization (D21f1 and D21e7). If internalization was driven only by LPS, this would not be observed. If LPS alone conferred these properties, one would expect to see the levels of internalized bacteria increasing as the length of the LPS core increased, but with no difference between OmpX-expressing and nonexpressing strains. Hence, we concluded that OmpX function as an invasin is influenced by the LPS core structure.
In summary, this work showed for the first time that the Y. pestis ompX gene is a novel essential virulence factor in a subset of mammalian hosts where OmpX-dependent serum resistance is required. The loss of ompX had little consequence in mice (where serum resistance is independent of OmpX expression), with no change in LD50. This is in stark contrast to the complete attenuation of a Y. pestis CO92 ompX deletion mutant in rats (where serum resistance is dependent on OmpX expression), an animal model more similar to human disease (2, 49). Finally, OmpX-mediated serum resistance, as well as host cell adhesion and internalization, is dependent on the LPS core structure, which contributes to the OM environment.
Acknowledgments
This work was supported by the National Institutes of Health (grants P20 RR15587, P20 RR016454, and U54AI57141) and the Idaho Agricultural Experimental Station.
We appreciate advice and technical assistance with electron microscopy provided by Valerie Roberts and technical assistance provided by Dylan Sinclair.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 13 September 2010.
REFERENCES
- 1.Airhart, C. L., H. N. Rohde, G. A. Bohach, C. J. Hovde, C. F. Deobald, S. S. Lee, and S. A. Minnich. 2008. Induction of innate immunity by lipid A mimetics increases survival from pneumonic plague. Microbiology 154:2131-2138. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, D. M., N. A. Ciletti, H. Lee-Lewis, D. Elli, J. Segal, K. L. DeBord, K. A. Overheim, M. Tretiakova, R. R. Brubaker, and O. Schneewind. 2009. Pneumonic plague pathogenesis and immunity in Brown Norway rats. Am. J. Pathol. 174:910-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anisimov, A. P., S. V. Dentovskaya, G. M. Titareva, I. V. Bakhteeva, R. Z. Shaikhutdinova, S. V. Balakhonov, B. Lindner, N. A. Kocharova, S. N. Senchenkova, O. Holst, G. B. Pier, and Y. A. Knirel. 2005. Intraspecies and temperature-dependent variations in susceptibility of Yersinia pestis to the bactericidal action of serum and to polymyxin B. Infect. Immun. 73:7324-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartra, S. S., K. L. Styer, D. M. O'Bryant, M. L. Nilles, B. J. Hinnebusch, A. Aballay, and G. V. Plano. 2008. Resistance of Yersinia pestis to complement-dependent killing is mediated by the Ail outer membrane protein. Infect. Immun. 76:612-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berggard, K., E. Johnsson, F. R. Mooi, and G. Lindahl. 1997. Bordetella pertussis binds the human complement regulator C4BP: role of filamentous hemagglutinin. Infect. Immun. 65:3638-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biedzka-Sarek, M., R. Venho, and M. Skurnik. 2005. Role of YadA, Ail, and lipopolysaccharide in serum resistance of Yersinia enterocolitica serotype O:3. Infect. Immun. 73:2232-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolger, M. S., D. S. Ross, H. Jiang, M. M. Frank, A. J. Ghio, D. A. Schwartz, and J. R. Wright. 2007. Complement levels and activity in the normal and LPS-injured lung. Am. J. Physiol. 292:L748-L759. [DOI] [PubMed] [Google Scholar]
- 8.Brubaker, R. R. 2003. Interleukin-10 and inhibition of innate immunity to Yersiniae: roles of Yops and LcrV (V antigen). Infect. Immun. 71:3673-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowan, C., H. A. Jones, Y. H. Kaya, R. D. Perry, and S. C. Straley. 2000. Invasion of epithelial cells by Yersinia pestis: evidence for a Y. pestis-specific invasin. Infect. Immun. 68:4523-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cybulska, J., and J. Jeljaszewicz. 1966. Bacteriostatic activity of serum against staphylococci. J. Bacteriol. 91:953-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Cock, H., K. Brandenburg, A. Wiese, O. Holst, and U. Seydel. 1999. Non-lamellar structure and negative charges of lipopolysaccharides required for efficient folding of outer membrane protein PhoE of Escherichia coli. J. Biol. Chem. 274:5114-5119. [DOI] [PubMed] [Google Scholar]
- 12.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupont, M., E. De, R. Chollet, J. Chevalier, and J. M. Pages. 2004. Enterobacter aerogenes OmpX, a cation-selective channel mar- and osmo-regulated. FEBS Lett. 569:27-30. [DOI] [PubMed] [Google Scholar]
- 14.Felek, S., and E. S. Krukonis. 2009. The Yersinia pestis Ail protein mediates binding and Yop delivery to host cells required for plague virulence. Infect. Immun. 77:825-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felek, S., M. B. Lawrenz, and E. S. Krukonis. 2008. The Yersinia pestis autotransporter YapC mediates host cell binding, autoaggregation and biofilm formation. Microbiology 154:1802-1812. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson, A. D., W. Welte, E. Hofmann, B. Lindner, O. Holst, J. W. Coulton, and K. Diederichs. 2000. A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins. Structure 8:585-592. [DOI] [PubMed] [Google Scholar]
- 17.Forman, S., C. R. Wulff, T. Myers-Morales, C. Cowan, R. D. Perry, and S. C. Straley. 2008. yadBC of Yersinia pestis, a new virulence determinant for bubonic plague. Infect. Immun. 76:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gondwe, E. N., M. E. Molyneux, M. Goodall, S. M. Graham, P. Mastroeni, M. T. Drayson, and C. A. MacLennan. 2010. Importance of antibody and complement for oxidative burst and killing of invasive nontyphoidal Salmonella by blood cells in Africans. Proc. Natl. Acad. Sci. U. S. A. 107:3070-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanski, C., M. Naumann, A. Grutzkau, G. Pluschke, B. Friedrich, H. Hahn, and E. O. Riecken. 1991. Humoral and cellular defense against intestinal murine infection with Yersinia enterocolitica. Infect. Immun. 59:1106-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heffernan, E. J., J. Harwood, J. Fierer, and D. Guiney. 1992. The Salmonella typhimurium virulence plasmid complement resistance gene rck is homologous to a family of virulence-related outer membrane protein genes, including pagC and ail. J. Bacteriol. 174:84-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heffernan, E. J., S. Reed, J. Hackett, J. Fierer, C. Roudier, and D. Guiney. 1992. Mechanism of resistance to complement-mediated killing of bacteria encoded by the Salmonella typhimurium virulence plasmid gene rck. J. Clin. Invest. 90:953-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heffernan, E. J., L. Wu, J. Louie, S. Okamoto, J. Fierer, and D. G. Guiney. 1994. Specificity of the complement resistance and cell association phenotypes encoded by the outer membrane protein genes rck from Salmonella typhimurium and ail from Yersinia enterocolitica. Infect. Immun. 62:5183-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Junkes, C., A. Wessolowski, S. Farnaud, R. W. Evans, L. Good, M. Bienert, and M. Dathe. 2008. The interaction of arginine- and tryptophan-rich cyclic hexapeptides with Escherichia coli membranes. J. Pept. Sci. 14:535-543. [DOI] [PubMed] [Google Scholar]
- 24.Knirel, Y. A., S. V. Dentovskaya, O. V. Bystrova, N. A. Kocharova, S. N. Senchenkova, R. Z. Shaikhutdinova, G. M. Titareva, I. V. Bakhteeva, B. Lindner, G. B. Pier, and A. P. Anisimov. 2007. Relationship of the lipopolysaccharide structure of Yersinia pestis to resistance to antimicrobial factors. Adv. Exp. Med. Biol. 603:88-96. [DOI] [PubMed] [Google Scholar]
- 25.Knirel, Y. A., S. V. Dentovskaya, S. N. Senchenkova, R. Z. Shaikhutdinova, N. A. Kocharova, and A. P. Anisimov. 2006. Structural features and structural variability of the lipopolysaccharide of Yersinia pestis, the cause of plague. J. Endotoxin Res. 12:3-9. [DOI] [PubMed] [Google Scholar]
- 26.Kolodziejek, A. M., D. J. Sinclair, K. S. Seo, D. R. Schnider, C. F. Deobald, H. N. Rohde, A. K. Viall, S. S. Minnich, C. J. Hovde, S. A. Minnich, and G. A. Bohach. 2007. Phenotypic characterization of OmpX, an Ail homologue of Yersinia pestis KIM. Microbiology 153:2941-2951. [DOI] [PubMed] [Google Scholar]
- 27.Kukkonen, M., M. Suomalainen, P. Kyllonen, K. Lahteenmaki, H. Lang, R. Virkola, I. M. Helander, O. Holst, and T. K. Korhonen. 2004. Lack of O-antigen is essential for plasminogen activation by Yersinia pestis and Salmonella enterica. Mol. Microbiol. 51:215-225. [DOI] [PubMed] [Google Scholar]
- 28.Kurtenbach, K., S. De Michelis, S. Etti, S. M. Schafer, H. S. Sewell, V. Brade, and P. Kraiczy. 2002. Host association of Borrelia burgdorferi sensu lato—the key role of host complement. Trends Microbiol. 10:74-79. [DOI] [PubMed] [Google Scholar]
- 29.Lathem, W. W., P. A. Price, V. L. Miller, and W. E. Goldman. 2007. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science 315:509-513. [DOI] [PubMed] [Google Scholar]
- 30.Lawrenz, M. B., J. D. Lenz, and V. L. Miller. 2009. A novel autotransporter adhesin is required for efficient colonization during bubonic plague. Infect. Immun. 77:317-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Y., Y. Cui, Y. Hauck, M. E. Platonov, E. Dai, Y. Song, Z. Guo, C. Pourcel, S. V. Dentovskaya, A. P. Anisimov, R. Yang, and G. Vergnaud. 2009. Genotyping and phylogenetic analysis of Yersinia pestis by MLVA: insights into the worldwide expansion of Central Asia plague foci. PLoS One 4:e6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, F., H. Chen, E. M. Galvan, M. A. Lasaro, and D. M. Schifferli. 2006. Effects of Psa and F1 on the adhesive and invasive interactions of Yersinia pestis with human respiratory tract epithelial cells. Infect. Immun. 74:5636-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marani, P., S. Wagner, L. Baars, P. Genevaux, J. W. de Gier, I. Nilsson, R. Casadio, and G. von Heijne. 2006. New Escherichia coli outer membrane proteins identified through prediction and experimental verification. Protein Sci. 15:884-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcus, S., D. W. Esplin, and D. M. Donaldson. 1954. Lack of bactericidal effect of mouse serum on a number of common microorganisms. Science 119:877. [DOI] [PubMed] [Google Scholar]
- 35.Mecsas, J., R. Welch, J. W. Erickson, and C. A. Gross. 1995. Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J. Bacteriol. 177:799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montminy, S. W., N. Khan, S. McGrath, M. J. Walkowicz, F. Sharp, J. E. Conlon, K. Fukase, S. Kusumoto, C. Sweet, K. Miyake, S. Akira, R. J. Cotter, J. D. Goguen, and E. Lien. 2006. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat. Immunol. 7:1066-1073. [DOI] [PubMed] [Google Scholar]
- 37.Myers-Morales, T., C. Cowan, M. E. Gray, C. R. Wulff, C. E. Parker, C. H. Borchers, and S. C. Straley. 2007. A surface-focused biotinylation procedure identifies the Yersinia pestis catalase KatY as a membrane-associated but non-surface-located protein. Appl. Environ. Microbiol. 73:5750-5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ngampasutadol, J., S. Ram, S. Gulati, S. Agarwal, C. Li, A. Visintin, B. Monks, G. Madico, and P. A. Rice. 2008. Human factor H interacts selectively with Neisseria gonorrhoeae and results in species-specific complement evasion. J. Immunol. 180:3426-3435. [DOI] [PubMed] [Google Scholar]
- 39.Ngampasutadol, J., C. Tran, S. Gulati, A. M. Blom, E. A. Jerse, S. Ram, and P. A. Rice. 2008. Species-specificity of Neisseria gonorrhoeae infection: do human complement regulators contribute? Vaccine 26(Suppl. 8):I62-I66. [DOI] [PubMed] [Google Scholar]
- 40.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pier, G. B., M. Grout, T. S. Zaidi, J. C. Olsen, L. G. Johnson, J. R. Yankaskas, and J. B. Goldberg. 1996. Role of mutant CFTR in hypersusceptibility of cystic fibrosis patients to lung infections. Science 271:64-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prior, J. L., P. G. Hitchen, D. E. Williamson, A. J. Reason, H. R. Morris, A. Dell, B. W. Wren, and R. W. Titball. 2001. Characterization of the lipopolysaccharide of Yersinia pestis. Microb. Pathog. 30:49-57. [DOI] [PubMed] [Google Scholar]
- 43.Prior, J. L., J. Parkhill, P. G. Hitchen, K. L. Mungall, K. Stevens, H. R. Morris, A. J. Reason, P. C. Oyston, A. Dell, B. W. Wren, and R. W. Titball. 2001. The failure of different strains of Yersinia pestis to produce lipopolysaccharide O-antigen under different growth conditions is due to mutations in the O-antigen gene cluster. FEMS Microbiol. Lett. 197:229-233. [DOI] [PubMed] [Google Scholar]
- 44.Prokhorenko, I. R., S. V. Zubova, A. Y. Ivanov, and S. V. Grachev. 2009. Interaction of Gram-negative bacteria with cationic proteins: dependence on the surface characteristics of the bacterial cell. Int. J. Gen. Med. 2:33-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puente, X. S., L. M. Sanchez, A. Gutierrez-Fernandez, G. Velasco, and C. Lopez-Otin. 2005. A genomic view of the complexity of mammalian proteolytic systems. Biochem. Soc. Trans. 33:331-334. [DOI] [PubMed] [Google Scholar]
- 46.Razatos, A., Y. L. Ong, M. M. Sharma, and G. Georgiou. 1998. Molecular determinants of bacterial adhesion monitored by atomic force microscopy. Proc. Natl. Acad. Sci. U. S. A. 95:11059-11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rebeil, R., R. K. Ernst, B. B. Gowen, S. I. Miller, and B. J. Hinnebusch. 2004. Variation in lipid A structure in the pathogenic yersiniae. Mol. Microbiol. 52:1363-1373. [DOI] [PubMed] [Google Scholar]
- 48.Rietschel, E. T., H. Brade, O. Holst, L. Brade, S. Muller-Loennies, U. Mamat, U. Zahringer, F. Beckmann, U. Seydel, K. Brandenburg, A. J. Ulmer, T. Mattern, H. Heine, J. Schletter, H. Loppnow, U. Schonbeck, H. D. Flad, S. Hauschildt, U. F. Schade, F. Di Padova, S. Kusumoto, and R. R. Schumann. 1996. Bacterial endotoxin: chemical constitution, biological recognition, host response, and immunological detoxification. Curr. Top. Microbiol. Immunol. 216:39-81. [DOI] [PubMed] [Google Scholar]
- 49.Sebbane, F., D. Gardner, D. Long, B. B. Gowen, and B. J. Hinnebusch. 2005. Kinetics of disease progression and host response in a rat model of bubonic plague. Am. J. Pathol. 166:1427-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sebbane, F., C. O. Jarrett, D. Gardner, D. Long, and B. J. Hinnebusch. 2006. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc. Natl. Acad. Sci. U. S. A. 103:5526-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1:784-791. [Google Scholar]
- 52.Smith, M. J. 2000. Genetic regulation of type III secretion systems in Yersinia enterocolitica. PhD thesis. University of Idaho, Moscow, Idaho.
- 53.Stevenson, B., N. El-Hage, M. A. Hines, J. C. Miller, and K. Babb. 2002. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 70:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stoorvogel, J., M. J. van Bussel, J. Tommassen, and J. A. van de Klundert. 1991. Molecular characterization of an Enterobacter cloacae outer membrane protein (OmpX). J. Bacteriol. 173:156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stoorvogel, J., M. J. van Bussel, and J. A. van de Klundert. 1991. Biological characterization of an Enterobacter cloacae outer membrane protein (OmpX). J. Bacteriol. 173:161-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suomalainen, M., L. A. Lobo, K. Brandenburg, B. Lindner, R. Virkola, Y. A. Knirel, A. P. Anisimov, O. Holst, and T. K. Korhonen. 2010. Temperature-induced changes in the lipopolysaccharide of Yersinia pestis affect plasminogen activation by the Pla surface protease. Infect. Immun. 78:2644-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wachtel, M. R., and V. L. Miller. 1995. In vitro and in vivo characterization of an ail mutant of Yersinia enterocolitica. Infect. Immun. 63:2541-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Younger, J. G., S. Shankar-Sinha, M. Mickiewicz, A. S. Brinkman, G. A. Valencia, J. V. Sarma, E. M. Younkin, T. J. Standiford, F. S. Zetoune, and P. A. Ward. 2003. Murine complement interactions with Pseudomonas aeruginosa and their consequences during pneumonia. Am. J. Respir. Cell Mol. Biol. 29:432-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu, F., and S. Mizushima. 1982. Roles of lipopolysaccharide and outer membrane protein OmpC of Escherichia coli K-12 in the receptor function for bacteriophage T4. J. Bacteriol. 151:718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaidi, T. S., S. M. Fleiszig, M. J. Preston, J. B. Goldberg, and G. B. Pier. 1996. Lipopolysaccharide outer core is a ligand for corneal cell binding and ingestion of Pseudomonas aeruginosa. Invest. Ophthalmol. Vis. Sci. 37:976-986. [PubMed] [Google Scholar]