Abstract
The most intensely studied of the Vibrio vulnificus virulence factors is the capsular polysaccharide (CPS). All virulent strains produce copious amounts of CPS. Acapsular strains are avirulent. The structure of the CPS from the clinical isolate ATCC 27562 is unusual. It is serine modified and contains, surprisingly, N-acetylmuramic acid. We identified the complete 25-kb CPS biosynthesis locus from ATCC 27562. It contained 21 open reading frames and was allelic to O-antigen biosynthesis loci. Two of the genes, murACPS and murBCPS, were paralogs of the murAPG and murBPG genes of the peptidoglycan biosynthesis pathway; only a single copy of these genes is present in the strain CMCP6 and YJ016 genomes. Although MurACPS and MurBCPS were functional when expressed in Escherichia coli, lesions in either gene had no effect on CPS production, virulence, or growth in V. vulnificus; disruption of 8 other genes within the locus resulted in an acapsular phenotype and attenuated virulence. Thus, murACPS and murBCPS were functional but redundant. Comparative genomic analysis revealed that while completely different CPS biosynthesis loci were found in the same chromosomal region in other V. vulnificus strains, most of the CPS locus of ATCC 27562 was conserved in another marine bacterium, Shewanella putrefaciens strain 200. However, the average GC content of the CPS locus was significantly lower than the average GC content of either genome. Furthermore, several of the encoded proteins appeared to be of Gram-positive and archaebacterial origin. These data indicate that the horizontal transfer of intact and partial CPS loci drives CPS diversity in marine bacteria.
The capsular polysaccharide (CPS) often constitutes the outermost layer of the bacterial cell. As such, it can mediate direct interactions between the bacterium and its environment and is an important virulence factor for many animal and plant pathogens (11). CPS, like O antigen, is composed of monosaccharides joined by glycosidic linkages, and an incredibly diverse range of branched and modified CPS molecules is possible. The CPS is synthesized and assembled by a series of proteins that are encoded by genes clustered in specific biosynthesis loci. CPS biosynthesis has been divided into 4 main groups that are distinguished based on their mechanisms of synthesis and assembly and genomic organization (47). Group I and IV capsule biosynthesis (G1C and G4C, respectively) parallels O-antigen biosynthesis.
Vibrio vulnificus is a Gram-negative pathogen that is responsible for more than 95% of all seafood-related deaths in the United States (31). These bacteria are found inhabiting estuarine waters in subtropical climates where they associate with zooplankton and are found colonizing filter feeders such as shellfish. Infection may occur through two routes: consumption of contaminated seafood or wound exposure to contaminated material. In susceptible individuals, the consumption of contaminated food can lead to septicemia with mortality rates greater than 50%. The mortality rates associated with wound infection, though lower, are still significant at 20 to 30%. The individuals most at risk are those with a compromised immune system, impaired liver function, and/or increased serum iron levels.
The V. vulnificus CPS is an essential virulence factor. V. vulnificus produces multiple capsular types; of 120 V. vulnificus isolates examined, 94 different CPS carbotypes were detected by using high-performance anion-exchange chromatography (8). The most prevalent monosaccharides identified in the CPS of V. vulnificus strains are rhamnose, galactosamine uronate, galactose, glucosamine, and galactosamine. The CPS of the V. vulnificus type strain ATCC 27562 (hereinafter referred to as strain 27562) is unusual (25). It contains a serine modification and is composed of N-acetylglucosamine (GlcNAc), galacturonic acid (GalA), rhamnose (Rha), and surprisingly, N-acetylmuramic acid (MurNAc). MurNAc is unique to bacteria and is an essential component of bacterial peptidoglycan. MurNAc is rarely found in other bacterial polysaccharides, such as lipopolysaccharide (LPS) or CPS. Indeed, there are only 3 known examples of muramic acid incorporation outside bacterial peptidoglycan: in the LPS of Yersinia ruckerii (5) and in the CPS of V. vulnificus strains 27562 and UMCP/67b (8, 26). Amino acid modification of CPS is also uncommon.
The importance of the CPS as a virulence factor and the incredible diversity of V. vulnificus CPS carbotypes suggest that the elaboration of CPS diversity in V. vulnificus is an active process, yet little is known about the biochemical or genetic mechanisms that drive this diversity. We previously identified a polysaccharide polymerase (Wzy), a tyrosine autokinase (Wzc), and an epimerase (RmlC) that were required for CPS biosynthesis in strain 27562 (39). Here, we show that these genes are part of an unusual 25-kb CPS biosynthesis locus containing 21 open reading frames (ORFs). Interestingly, genes coding for a UDP-N-acetylglucosamine 1-carboxyvinyl transferase (murA) and a UDP-N-acetylmuramate dehydrogenase (murB) were among the ORFs identified in the CPS locus. MurA and MurB synthesize UDP-N-acetylmuramic acid, an important constituent of peptidoglycan and a component of the CPS. Strain 27562 in fact harbored two copies of the murA and murB genes. We show that the murA and murB genes of the CPS locus (murACPS and murBCPS, respectively) are expressed in strain 27562 and are functional when expressed in Escherichia coli. However, murACPS and murBCPS were dispensable for CPS production, virulence, and growth in rich and nutrient-limited media. Comparative genomic analysis revealed that while completely different CPS biosynthesis loci were found in the same chromosomal region in V. vulnificus strains MO6-24/O, CMCP6, and YJ016, most of the CPS locus of strain 27562 was conserved in another marine bacterium, Shewanella putrefaciens strain 200. The average GC content of the CPS locus was significantly lower than the average GC content of the 27562 and S. putrefaciens 200 genomes. Furthermore, the closest homologs of five of the encoded proteins were of Gram-positive and archaebacterial origin. These observations suggest that horizontal gene transfer (HGT) may be a prominent mechanism for the exchange and evolution of CPS loci in marine bacteria and provide an explanation for the redundancy of the murACPS and murBCPS genes.
MATERIALS AND METHODS
Bacterial strains, media, and PCR.
E. coli DH5α λpir was used in transformation experiments. E. coli S17.1 λpir was used as the donor strain in conjugation experiments. A rifampin-resistant derivative of the encapsulated V. vulnificus type strain ATCC 27562 (referred to hereinafter as 27562) was obtained from the Collection de l'Institut Pasteur and used as the recipient. Both strains were grown in LB medium (Sigma). Antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg/ml; rifampin (Rf), 100 μg/ml; kanamycin (Km), 160 μg/ml for V. vulnificus and 10 μg/ml for E. coli; and chloramphenicol (Cm), 1 μg/ml for V. vulnificus and 6 μg/ml for E. coli. Isopropyl-β-d-thiogalactopyranoside (IPTG) was used at a concentration of 1 mM. Deoxyribosylthymine (dT) was used at a final concentration of 0.3 mM. Mueller-Hinton (MH) medium was used for antibiotic susceptibility testing. PCRs were performed in 50-μl volumes using Taq DNA polymerase (BioTools) or Platinum Pfx DNA polymerase (Invitrogen) following the manufacturer's instructions. When necessary, PCR products were cloned into the pCR2.1 vector (Invitrogen) and sequenced by using the M13F and M13R primers at The Centre for Applied Genomics (the Hospital for Sick Children, Toronto).
Identification and sequencing of the CPS locus of strain 27562.
The pNKTXI-SceI plasmid (39), which carries a Kmr mini-Tn10 transposon (Tn), was conjugated from S17.1 λpir to strain 27562, and mutants with Tn insertions were selected on LB Rf Km plates. Acapsular mutants appeared translucent on solid medium, and these colonies were selected for further analysis. Translucent colonies were grown overnight on LB Km plates. Bacteria were scraped from the plate, and genomic DNA (gDNA) was extracted using DNAZOL reagent (Invitrogen). The gDNA was digested with BglII or EcoRI and self-ligated. Ligations were transformed into E. coli DH5α λpir and selected on LB plates containing Km. Plasmid DNA was prepared using a Sigma plasmid miniprep kit (Sigma). Inserts were sequenced with primers miniTn10-km4 (GCAGACAGTTTTATTGTTCATGATG) and pirseq (ACACTTAACGGCTGACATGG) to obtain sequence adjacent to the insertion point. Primers used for sequencing the CPS locus by primer walking are listed in Table 1 . Strains with Tn insertions in the CPS are listed in Table 2.
TABLE 1.
Primers used in this study
| Primer and purpose | Sequence |
|---|---|
| Generation of targeted deletions | |
| ΔmurAx-F | GGGGTACCCTTTACCAGGAGGTTGTGCC |
| ΔmurAx-R | GGGGTACCCCCCATCTGCAATACAGTTC |
| murBxΔ600F | GGGGGGTACCCATTCCTGTTGTTATTCAGTTAAA |
| murBxΔ600R | GGGGGGTACCGCACTAACTTTATACCCAGAG |
| Probes and RT-PCR | |
| murA5′ | ATGTGCGCTGCTTTATTAGTAG |
| murA3′ | GAGGCTCCTACCGCTGCTA |
| murAF | ATGGAAAAGTTCCGTGTAATTG |
| murAR | TTAGTTTGACTCACGAAAACG |
| murBF | CAAATTAAGCAAAATATCTCTT |
| murBR | TCATGACTCTACTCTTAGC |
| murBXF | ATTAAATGGGATGACGAAACTT |
| murBXR | TTCTCCGACTTCTACGCC |
| RTpgmurBF | GTATCCTGCTGGTGAACAGA |
| RTpgmurBR | GCAAATAAGGCTGGCCAACT |
| RTmurBxF | AAGAAGAAACCACGTCTGTAGC |
| RTmurBxR | TTCTCCGACTTCTACGCCGTA |
| RTmurAxF | CCAGGAGGTTGTGCCATTGG |
| RTmurAxR | GCCAAAGTAGCGGCCAGTAT |
| cBspHImurBx5′ | TCATGATTGAAAAATTTAATGTTCAAAT |
| cBHImurBx3′ | GGATCCTTACCAACAAGCAGGTTCTAC |
| NcoImurAxaltF | CCATGGATGTGGAAAAGTTAGAAATAATT |
| cBHIMurA3′ | GGATCCTCATTTAACTCTCTTCACAGAG |
| Sequencing the CPS locus | |
| WzaR-OUT | ACGATATCGTCTATGTCACCG |
| WzcF | CTTCTGCTAGCGATGCTAGC |
| 2wzcF | CTAGTGATCTAGAAAACTGCTC |
| Wzcinv | GGTGTTGTCATTGTCTCGTTCAT |
| Wzcinv2 | AGCTAATGCCGCCCGAGC |
| WzcRk/o | CTTCTGCTAGCGATGCTAGC |
| RmlBseq1 | CATGCCATCGCCATATACAG |
| RmlBseq2 | CATAAATTAATCACTCTCGGTC |
| 1395murbx6 | TCTCAATGATGGCTCGCTGAA |
| RmlAseq | TTCAGCGAGCCATCATTGAG |
| 1395murbx7 | GCTTTCTTTTGCCATTAACCG |
| 1395murbx8 | CAATATTTCAAGCGGCAGAAC |
| RmlCBspHI | ATGAAAGTTATTGATACAAACATAC |
| RmlC R | CTATCCTAAAAGATATTTACATT |
| MurBseq | GCAAAGGCAGGTGAGAATTG |
| 1395murbx2 | GTTTTAAGGAAATTTGATGTGGA |
| 1395murbx3 | ATAGCTGATAGGATAGAGGCT |
| 1475F5R | GCTCCTACCGCTGCTAGC |
| 1475F4R | CTAGGCTTTTGTTTACTTTGACT |
| 1395murbx4 | GTGAGTCTCCTGTGCTTGAA |
| 1395murbx5 | GAACTTCCTGTTGTATTGAATG |
| 1475F3R | CTTCCTATCAAATCTAAACTCG |
| 1475FX | GCTATTTGAACTATTAGGCAGC |
| BspH1wzy | GGGTTTTCATGATTACATACAACGTTAGATTAA |
| 1407F | GAATTCTTTACGATCCAACAAG |
| b4wzy | CCAATGGCCAGGATATTAAAG |
| 1407FF | TGGGTCTGTCCTTTCTCCTGT |
| 1407FFC | ACAGGAGAAAGGACAGACCCA |
| Wzcseq | GAGGGCTTTGGAGGAGATA |
| WzyRIseq | GAGGGCTTTGGAGGAGATA |
| Wzcseq2 | ACGTGAGTGATTTGTCAAGAG |
| 1475R1 | TACCAGAGAGGATTAAGAGGTT |
| 1475R2 | CCCAGAGCTAAATTATCAACC |
| 1475R3 | CAGGCTAATGGTATACAAAGTG |
| 1475 R3next | GAGGGATATTGCGATCAGGC |
| 1475FX6 | TCCTTTGCCTTTCGAGCGGT |
| 1475-FX5 | GCCAGACAGAGAACACCTC |
| 1475FX3 | TGGTGTGAGCCAAGTAGAAAG |
| 1475FX4 | TGGTGTGAGCCAAGTAGAAAG |
| 1475F2 | CCACGTTACAAACAATATTGCC |
| 1475F1 | TTATCGTATAACGCCTTACCTT |
| EPI-REV | GTTATAAACCGAATATGGAGCA |
TABLE 2.
V. vulnificus CPS− strains isolated in this study
| Strain or genotype | Description | Reference or source |
|---|---|---|
| ATCC 27562 | Wild type; Rfr | CIPa |
| Δwzc | Targeted disruption of wzc with pSW23T; Cmr | 38 |
| ΔrmlC | Tn10 insertion in rmlC; Kmr | 38 |
| ΔmurBCPS | Targeted disruption of murBCPS with pSW23T; Cmr | This study |
| ΔmurACPS | Targeted disruption of murACPS with pSW23T; Cmr | This study |
| ΔcppA | Tn10 insertion in cppA; Kmr | This study |
| Δwzx | Tn10 insertion in wzx; Kmr | This study |
| Δwzy | Tn10 insertion in wzy; Kmr | 39 |
| ΔwcvD | Targeted disruption of wcvD with pSW23T; Cmr | This study |
| ΔwcvE | Targeted disruption of wcvE with pSW23T; Cmr | This study |
| ΔwcvF | Tn10 insertion in wcvF; Kmr | This study |
CIP, Collection de l'Institut Pasteur.
Bioinformatic analysis.
The Artemis Comparison Tool (ACT) (9) and Mauve (13) were used for genomic comparisons. Geneious (Biomatters Ltd.) was used to annotate the 27562 CPS locus and to determine its GC content and nucleotide and amino acid identities. Homology searches were conducted using BLAST analysis (2), and protein parameters and domain predictions were performed with ProtParam (22) and the PROSITE (29) research tool from the Expert Protein analysis system.
Constructing the ΔmurACPS, ΔmurBCPS, ΔwcvD, and ΔwcvE strains.
The primer pairs ΔmurAx-F/ΔmurAx-R and murBxΔ600F/murBxΔ600R (Table 1) were designed to amplify and clone 620-bp and 614-bp internal fragments of murACPS and murBCPS, respectively, into the KpnI site of the pir-dependent suicide vector, pSW23T (14). Primers ΔwcvDBHI/ΔwcvDMfe and ΔwcvEBHI/ΔwcvEMfe were used to amplify and clone 616-bp and 631-bp internal fragments of wcvD and wcvE, respectively, into the BamHI and EcoRI sites of pSW23T. The resulting plasmids were transformed into S17.1 λpir and conjugated to V. vulnificus. Correct integration was verified by PCR using a primer targeting the cat gene of pSW23T and a second primer that annealed to chromosomal DNA flanking the respective target genes.
Complementation of mutant strains.
The wild-type wcvD, wcvE, and cppA genes were amplified from V. vulnificus strain 27562 genomic DNA with primer pairs wcvDMfeI/wcvDSalI, wcvEMfeI/wcvESalI, and cppAEcoRI/cppASalI. The products were cut with corresponding restriction enzymes and cloned into the EcoRI and SalI sites of pBAD24T (38). Plasmids were transformed into S17.1 λpir and then conjugated to the respective mutant. Transconjugants were selected on LB Rf Ap plates containing Km or Cm. Expression of the wild-type genes was induced with 0.2% l-arabinose.
Southern analysis.
gDNA was extracted from V. vulnificus 27562 using DNAZOL reagent following the manufacturer's protocol (Invitrogen). gDNA was digested with selected restriction enzymes and separated on a 0.7% (final) agarose gel overnight at 50 V/cm. The gel was then treated and the fragments transferred to Hybond N+ nylon membranes (Amersham Pharmacia) according to the manufacturer's instructions. The membrane was probed with murACPS or murBCPS that was amplified with the murA5′/murA3′ or murBXF/murBXR primer pair, respectively. The murAPG and murBPG genes were amplified with the murAF/murAR and murBF/murBR primer pairs and also used as probes. The primers used to amplify probes are summarized in Table 1. Probes were labeled using the Gene Images AlkPhos direct labeling system (Amersham Pharmacia) according to the protocol provided by the manufacturer. Prehybridization, hybridization, and subsequent washes were performed at 55°C. The signal was detected using BioMax MS film (Kodak).
RNA isolation and RT-PCR.
An overnight culture of strain 27562 was diluted 1:100 into fresh LB supplemented with appropriate antibiotics and grown to an optical density at 600 nm (OD600) of 0.6. RNA was isolated using a Qiagen RNeasy mini kit. RNA was then treated with RNase-free DNase (Qiagen) for 30 min and cleaned using an RNeasy mini kit. Total RNA was then quantified using a Biomate 3 spectrophotometer (Thermo Spectronic). An Invitrogen SuperScript reverse transcriptase III (RT)-PCR kit was used to obtain cDNA. PCR was then performed using Taq polymerase (Biotools). Primers used for RT-PCR are summarized in Table 1.
Fosfomycin susceptibility tests for MurACPS activity.
The murACPS gene was amplified with primers NcoImurAxaltF and cBHIMurA3′ and cloned into pTRC99A cut with NcoI and BamHI. KAM32 (28), a drug-hypersensitive E. coli strain, was transformed with pTRC99A::murACPS or the empty vector. The strains were grown with and without IPTG induction to an OD600 of 1.0. Aliquots of 100 μl were spread on MH agar plates containing Ap with and without IPTG. A fosfomycin susceptibility disk (Oxoid) was positioned in the middle of each plate, and the plates were incubated at 37°C overnight. Zones of inhibition were measured the following day. The assay was performed 6 times for each strain.
Complementation of an E. coli murB conditional mutant.
The murBCPS gene was amplified from strain 27562 using the primers cBspH1MurBx5′ and cBH1MurBx3′ and cloned into the NcoI and BamHI sites of pTRC99AT (39). The construct was transformed into the conditional E. coli ST5 murB mutant (37) and grown at 37°C. The resulting colonies were restreaked on LB agar plates containing Ap and dT and grown at 37°C or 42°C. The same strain carrying the empty pTRC99AT vector served as the control. Primer sequences are summarized in Table 1.
Antiserum production and slide agglutination tests.
Rabbit antiserum to formalin-killed wild-type V. vulnificus 27562 cells was prepared as previously described (39). Overnight cultures of V. vulnificus wild-type and mutant cells were diluted to an OD600 of 1.0. Duplicate 20-μl aliquots of each sample were applied onto separate microscope slides. One aliquot was mixed with an equal volume of the rabbit antiserum. The other aliquot was mixed with an equal volume of serum obtained from the baseline bleed and served as the negative control. Agglutination results were scored after incubation of the mixtures at room temperature for 5 min. A distinct and immediate agglutination was registered as positive, while weak or no agglutination after 5 min was considered a negative test. Samples were tested in triplicate.
Growth assays in LB medium.
Overnight cultures of the parental 27562 and the ΔmurACPS and ΔmurBCPS strains were grown at 37°C in LB containing the appropriate antibiotics and diluted to an OD600 of 0.05 in fresh medium on the following day. Optical densities were determined at 1-h intervals, and the results were plotted. The assay was repeated in triplicate.
Growth assays in PP3 medium.
Proteose peptone 3 (PP3) is an animal tissue digest used to generate an environment supportive of bacterial virulence maintenance, polymer production, and phase variation in Vibrio species (1). Cultures of the parental 27562 and the ΔmurACPS and ΔmurBCPS strains were incubated overnight at 37°C in LB. The following day, the cells were washed three times in PP3 medium supplemented to 1% (wt/vol) NaCl. Cells were then inoculated into the same medium supplemented with appropriate antibiotics to an OD600 of 0.02 and incubated for 7 days. Each day, OD measurements were taken, and dilutions were spread on LB agar plates supplemented with appropriate antibiotics. The percentage of opaque versus translucent colony types was calculated as a function of the total number of CFU. The test was repeated in triplicate.
Growth assays in minimal medium.
M9 at 5× strength (Na2HPO4, 0.3 M; KH2PO4, 0.1 M; NaCl, 0.04 M; and NH4Cl, 0.09 M) was diluted to 1× and supplemented with MgSO4 (2 mM), glucose (0.2% [wt/vol]), CaCl2 (0.1 mM), Casamino Acids (0.2% wt/vol), yeast extract (0.0006% [wt/vol]), and NaCl (1% [wt/vol]). Cultures of the parental 27562 and the ΔmurACPS and ΔmurBCPS strains were grown overnight at 37°C in LB, washed in the supplemented M9 medium, diluted to an OD600 of 0.01 in the same medium with antibiotics, and incubated for 7 days. Each day, OD600 measurements were taken, and dilutions were plated to determine the number of CFU. The assay was repeated in triplicate.
CPS isolation and LD50 determinations.
CPS isolation and 50% lethal dose (LD50) determinations for the various strains were performed as previously described (39).
Nucleotide sequence accession number.
All nucleic acid sequences have been deposited at the National Center for Biotechnology Information (NCBI) GenBank database under accession number HM099886.
RESULTS
Identification of an unusual CPS biosynthesis locus in V. vulnificus 27562.
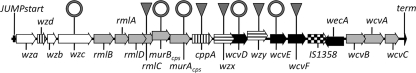
We previously identified a polysaccharide polymerase (wzy), a dTDP-4-dehydrorhamnose 3,5-epimerase (rmlC), and a tyrosine autokinase (wzc) required for CPS production in strain 27562 (38, 39). We have now determined that these genes map to a single 25-kb CPS biosynthesis locus containing 21 genes (Fig. 1 and Table 3). All of the ORFs, with the exception of wecA, encoding the initiating glycosyltransferase WecA, are transcribed in the same orientation. Nine ORFs encode enzymes that synthesize 3 of the sugars needed for completion of the polysaccharide repeat unit. The region also included a polysaccharide transport system, a polysaccharide polymerase, a flippase, and 4 glycosyltransferases.
FIG. 1.
Genetic organization of the CPS locus of V. vulnificus ATCC 27562. Arrows represent the locations and direction of transcription of the respective genes. Black, white, checkered, horizontally striped, light gray, and vertically striped arrows denote transferase, transport, insertion sequence, processing, nucleotide sugar biosynthesis, and unknown genes. The dark gray triangles and rings above the locus mark genes disrupted by Tn or suicide plasmid insertions, respectively. The JUMPstart and terminator sequences are also indicated.
TABLE 3.
Putative functions of the V. vulnificus ATCC 27562 CPS proteins
| Gene | %GC | Putative function | Organism | % Similarity | GenBank sequence accession no. | % Similarity in S. putrefaciens CPS locus | S. putrefaciens GenBank sequence accession no. |
|---|---|---|---|---|---|---|---|
| wza | 48.8 | Periplasmic protein involved in capsular polysaccharide export | Vibrio vulnificus | 98 | NP_759763 | ||
| wzb | 45.6 | Cytoplasmic phosphatase | V. vulnificus | 99 | ABD38618 | ||
| wzc | 45.4 | Tyrosine protein kinase | E. coli | 99 | YP_853169 | ||
| wzd | 41.9 | Unknown | V. vulnificus | 98 | NP_933131 | ||
| rmlB | 47.2 | dTDP-d-glucose-4,6-dehydratase | V. cholerae | 76 | ABI85357 | 69 | ZP_01705605 |
| rmlA | 43.2 | Glucose-1-phosphate thymidylyltransferase | V. cholerae | 96 | ZP_01949587 | 72 | ZP_01705606 |
| rmlD | 43.6 | dTDP-4-keto-l-rhamnose reductase | V. vulnificus | 90 | AAM34818 | 63 | ZP_01705607 |
| rmlC | 36.8 | dTDP-4-dehydrorhamnose 3,5-epimerase | V. cholerae | 88 | EDN15660 | 78 | ZP_01705608 |
| murBCPS | 32.6 | UDP-N-acetylpyruvoylglucosamine reductase | Neisseria gonorrhoeae | 47 | YP_207550 | 69 | ZP_01705609 |
| murACPS | 39.1 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | N. meningitidis | 52 | NP_283098 | 72 | ZP_01705610 |
| cppA | 31.5 | Unknown | Methanosarcina barkeri | 34 | YP_303597 | 56 | ZP_01705611 |
| wzx | 30.8 | Polysaccharide translocase | Bacillus thuringiensis | 21 | NZ_AAOX01000001 | 29 | ZP_01705640 |
| wcvD | 32.8 | Glycosyltransferase | Streptococcus pneumoniae | 45 | CAI34411 | 69 | ZP_01705641 |
| wzy | 31.2 | CPS polymerase | Bacillus cereus | 49 | NP_981681 | 68 | ZP_01705642 |
| wcvE | 32.5 | Glycosyltransferase | Methanosarcina mazei | 22 | CAI34411 | 67 | ZP_01705613 |
| wcvF | 38.3 | Rhamnosyltransferase | Shewanella frigidimarina | 99 | ZP_00638805 | 66 | ZP_01705614 |
| IS1358 | 43.0 | Transposase | V. cholerae | 99 | AAC82490.1 | ||
| wecA | 41.4 | Undecaprenylphosphate N-acetylglucosamine 1-phosphate transferase | V. vulnificus | 99 | AAO32663.1 | 60 | ZP_01705621 |
| wcvB | 41.7 | UDP-glucose 6-dehydrogenase | V. vulnificus | 099 | AAO32664.1 | 71 | ZP_01705620 |
| wcvA | 42.0 | UDP galacturonate 4-epimerase | V. vulnificus | 99 | AAO32665 | 71 | ZP_01705619 |
| wcvC | 42.9 | UTP-glucose-1-phosphate uridylyltransferase | V. vulnificus | 99 | NP_933159 |
Sugar biosynthesis proteins.
Genes encoding a UDP-glucose-6-dehydrogenase (wcvB), a UDP-glucuronic acid epimerase (wcvA), and a UTP-glucose-1-phosphate uridylyltransferase (wcvC) were found at the distal end of the cluster. Collectively, these enzymes catalyze the conversion of d-glucose-1-phosphate to UDP-d-galacturonic acid (UDP-GalA) (45). GalA is the central sugar moiety of the CPS of strain 27562, validating the CPS− phenotype of the mutant. The rmlBADC genes encode enzymes that convert glucose-1-phosphate into dTDP-l-rhamnose (36), and an rmlC mutant is acapsular and translucent. Complementation with the wild-type gene restored CPS production (38). The synthesis of MurNAc from UDP-GlcNAc is governed by MurA (a UDP-N-acetylglucosamine 1-carboxyvinyl transferase) and MurB (a UDP-N-acetylmuramate dehydrogenase), which are part of the highly conserved murABCDEF pathway for peptidoglycan biosynthesis. Two genes coding for homologs of MurA and MurB were identified 3′ of the rml cluster and designated murACPS and murBCPS, respectively. BLASTP analysis of murACPS and murBCPS indicated that they were most similar to the mur genes of the betaproteobacteria Neisseria meningitidis and Neisseria gonorrhoeae. The similarity of murACPS and murBCPS to orthologs from species outside the gammaproteobacteria was unexpected and prompted us to examine whether strain 27562 carried more than one paralog of the murA and murB genes. Primers were designed based on the murA and murB genes of peptidoglycan (PG) biosynthesis from strain CMCP6 and used to amplify products from strain 27562. The PCR products were cloned and sequenced. Protein sequence comparisons revealed that MurAPG and MurBPG were identical between strains 27562, YJ016, and CMCP6. However, MurACPS of strain 27562 shared only 54.5% identity with MurAPG from the same strain, and MurBCPS was only 29.9% identical to MurBPG. These results were verified by Southern blot analysis (data not shown). Thus, two copies of the murA and murB genes were present in the genome of strain 27562, whereas only a single copy of each of these genes was present in the genomes of CMCP6 and YJ016.
Glycosyltransferases.
The CPS repeat of strain 27562 contains four sugars, and four glycosyltransferases were identified in the cluster. A WecA initiating glycosyltransferase, which transfers GlcNAc to the undecaprenyl phosphate (UP) acceptor, and a rhamnosyltransferase (WcvF) account for the transfer of two of the four sugars. The wcvE and wcvD genes likely encode the glycosyltransferases responsible for the transfer of the two remaining sugars. A lesion in wcvD gave rise to an acapsular phenotype that could be complemented with the wild-type gene. Similar results were obtained with a wcvE lesion (data not shown).
Polymerization proteins.
Wzy and Wzx are transmembrane proteins characteristic of group I capsule (G1C) and group IV capsule (G4C) biosynthesis loci. We previously demonstrated the requirement of Wzy in CPS production and virulence in strain 27562 (39). The Tn insertion in wzx also yielded a CPS− phenotype (data not shown).
Transport proteins.
The wza, wzb, and wzc genes encode translocation proteins necessary for surface expression of the CPS in G1C and G4C. A targeted wzc knockout strain was translucent and acapsular, supporting the involvement of this gene in CPS production in strain 27562. Complementation with the wild-type gene restored CPS production (38).
Other features.
A cppA::Tn10 mutant was acapsular. Complementation with the wild-type gene restored opacity (data not shown). BLAST analysis did not provide a clear functional assignment for CppA. The highest similarities were to hypothetical proteins from S. putrefaciens (9e−83) and Methanosarcina barkeri (2e−46). The CPS of strain 27562 is serine modified, and all functions that may be inferred from the known CPS structure are accounted for, with the exception of the serine modification. We propose that cppA encodes the serine transferase. Additionally, the only other CPS serine transferase described in the literature shared no homology with any known proteins (3). The function of wzd is unknown.
The presence of a wecA gene supports a G4C designation for the locus. However, the wza-wzb-wzc genes, a defining feature of G1C loci, share the same locus as wecA. Hence, strain 27562 contains a G4C locus with an overall genetic organization that mirrors that of G1C loci. A JUMPstart (just upstream of many polysaccharides) element was identified 282 bp upstream of wza. Within this element is a potential uptake signal sequence (USS) and an ops. The USS (19) sequences are required for efficient uptake of DNA in some naturally transformable bacteria. The ops element is believed to recruit the transcription elongation regulator RfaH, which acts to decrease operon polarity in large transcriptional units (4).
The CPS locus is required for virulence.
The parental 27562 strain and derivatives that carried lesions in various genes of the CPS locus (wzc, rmlC, cppA, wzx, wzy, wcvD, wcvE, and wcvF) (Fig. 1) were tested for changes in virulence using a septicemic mouse model, and the results are presented in Table 4. The LD50 of the wild-type strain was 2.5 × 104. Disruption of each gene caused an increase of at least 3 orders of magnitude in the LD50. These results suggested that the CPS locus was required for virulence in V. vulnificus 27562.
TABLE 4.
LD50 values of the wild-type and mutant V. vulnificus strains in the iron-overloaded mouse model
| Strain or genotype | LD50 |
|---|---|
| 27562 | 2.5 × 104 |
| Δwzc | 1.9 × 107 |
| ΔrmlC | 2.9 × 107 |
| ΔmurBCPS | 4.2 × 105 |
| ΔmurACPS | 4.3 × 105 |
| ΔcppA | 1.3 × 107 |
| Δwzx | 2.2 × 107 |
| Δwzy | 3.8 × 107 |
| ΔwcvD | 3.1 × 107 |
| ΔwcvE | 1.6 × 107 |
| ΔwcvF | 3.2 × 107 |
murACPS and murBCPS are expressed and functional.
The presence of two copies of the murA and murB genes in the genome of strain 27562 was unexpected. To determine if murACPS and murBCPS were being expressed, we monitored their transcription by RT-PCR. The expression levels of the murBPG and rplT (ribosomal L20) genes were used as positive controls. No signal was obtained by PCR on DNase-treated RNA, confirming that the RNA was free of DNA contamination. RT-PCR on the same sample yielded products of the expected sizes for murACPS and murBCPS, indicating that murACPS and murBCPS were transcribed in vivo.
MurA is the target of the antimicrobial agent fosfomycin. Fosfomycin is a structural analogue of the MurA substrate phosphoenolpyruvate and inactivates the enzyme by forming a covalent bond with a cysteine residue in the active site (17). Overexpression of MurA leads to increased fosfomycin resistance (33). The functionality of MurACPS was determined by cloning the gene into the expression vector pTRC99A and monitoring the level of fosfomycin resistance in the antibiotic-hypersensitive E. coli strain KAM32 following overexpression of murACPS. The inhibition zones, with and without induction, were measured and compared with those of wild-type bacteria carrying the empty vector. The zone of inhibition surrounding the wild-type 27562 strain was 2.6 ± 0.16 cm (mean ± standard deviation). The introduction of empty pTRC99A vector had no significant effect on the zone of inhibition regardless of whether IPTG was present (2.6 ± 0.19 cm) or not (2.6 ± 0.15 cm). In cells carrying pTRC99A::murACPS, no change in the zone of inhibition was observed in the absence of induction (2.6 ± 0.06 cm). However, overproduction of MurACPS via IPTG induction resulted in a statistically significant (P < 0.005) decrease of 9 mm in the zone of inhibition (1.7 ± 0.11 cm). Hence, the overexpression of MurACPS in E. coli KAM32 led to decreased fosfomycin sensitivity, suggesting that MurACPS is a functional UDP-N-acetylglucosamine enolpyruvyltransferase.
To ascertain if MurBCPS was functional, we utilized the temperature-sensitive E. coli murB mutant, ST5. This mutant fails to grow at 42°C, but this growth defect can be complemented in trans with a functional murB gene. The murBCPS gene was cloned into pTRC99A and transformed into the ST5 strain. Complementation of the growth defect at 42°C was observed when the ST5 strain containing pTRC99A::murBCPS was grown under inducing conditions. No growth was visible for bacteria carrying the empty vector. These results suggested that MurBCPS is a functional UDP-N-acetylmuramate dehydrogenase.
MurACPS and MurBCPS are functionally redundant.
Mutations in wzc, rmlC, cppA, wzx, wzy, wcvD, wcvE, and wcvF resulted in an acapsular phenotype, demonstrating that these genes participated in CPS production. Although murACPS and murBCPS coded for functional enzymes, their physiological impact was unclear since the genome of strain 27562 also codes for murA and murB genes that are part of the peptidoglycan biosynthesis pathway. Targeted mutations were introduced into murACPS and murBCPS by using the suicide vector pSW23T. The ΔmurACPS and ΔmurBCPS mutant strains remained opaque and encapsulated. Moreover, slide agglutination assays revealed that antisera against strain 27562 agglutinated the ΔmurACPS and ΔmurBCPS strains, suggesting that the capsular polysaccharide was still being produced. The growth rates of the ΔmurACPS and ΔmurBCPS strains in different media (LB, M9, and PP3) were similar to those of wild-type cells, and phase variation rates were also unaffected (data not shown). Finally, the degrees of virulence of the wild-type and the ΔmurACPS and ΔmurBCPS strains were determined in vivo. Disruption of murACPS and murBCPS did not reduce the LD50 by three orders of magnitude as observed with lesions in other genes within the CPS locus. The LD50 for the wild-type strain was 2.5 × 104, while the LD50 for the ΔmurACPS and ΔmurBCPS mutant strains was 4.2 × 105. The modest 16-fold difference in virulence suggested that the ΔmurACPS and ΔmurBCPS lesions did not alter the capsule enough to cause a substantial decrease in the LD50. This suggested that murAPG and murBPG provide enough MurNAc to support both peptidoglycan and CPS biosynthesis and that murACPS and murBCPS are functionally redundant.
Genomic evidence for the exchange of CPS loci in the Vibrionaceae.
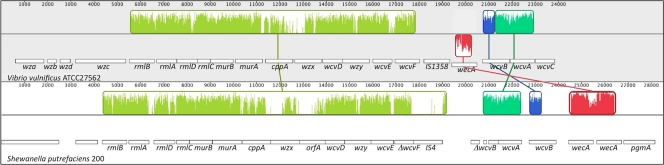
The CPS regions of V. vulnificus strain 27562 and S. putrefaciens strain 200 were compared using Mauve (Fig. 2). The loci were highly conserved over most of their length, as indicated by the yellow locally colinear block (LCB). The average GC content of the genes within the CPS locus (39.7%) was significantly lower than the 46.7% and 44.5% average GC content of the V. vulnificus and S. putrefaciens 200 genomes, respectively. In addition, the closest homologs of several of the proteins were enzymes from betaproteobacterial, Gram-positive, and archaebacterial species (Table 3). The conservation of gene synteny, amino acid identity, and GC content between the two clusters and the similarity of several of the encoded proteins to orthologs from species outside the gammaproteobacteria support the notion that this CPS locus was acquired by HGT in both V. vulnificus and S. putrefaciens 200. Despite these shared characteristics, several differences between the loci were of note. The orientation of the wecA gene was reversed in V. vulnificus with respect to its orientation in S. putrefaciens (Fig. 2, compare red LCBs), and the latter locus had two copies of wecA, which shared only 57% nucleotide identity. The wcvB nucleotide sugar dehydrogenase was duplicated in S. putrefaciens, but one copy had degenerated (Fig. 2, green and blue LCBs). IS1358 in the V. vulnificus locus was replaced by IS4 in S. putrefaciens, and IS4 disrupted wcvF. Finally, a gene that is not present in the V. vulnificus CPS locus follows the wzx gene in the CPS locus of S. putrefaciens 200.
FIG. 2.
The CPS locus of V. vulnificus ATCC 27562 is conserved in S. putrefaciens 200. The CPS loci of the two strains were aligned using Mauve. The alignment display is organized into one horizontal panel per input sequence. Each panel contains the strain designation, the organization of the genes within the locus (white boxes; genes in the forward orientation are shown on a single level while those in the reverse orientation are offset below), a scale showing the sequence coordinates in base pairs, and a single horizontal centerline. Connected colored segments (locally colinear blocks [LCBs]) indicate regions of a sequence that align with that of another sequence, and these regions are considered homologous. Within each colored segment is a similarity profile of the sequence. The height of the similarity profile corresponds to the average level of conservation in that region of the sequence. Areas within the colored segments that are completely white and regions outside the LCBs lack detectable homology among the input sequences and were not aligned; they contain sequence elements specific to a particular locus. Blocks lying above the centerline indicate regions that align in the forward orientation, and those below align in the reverse orientation. The relevant genes of each locus are labeled. The insertion of the two different IS elements (IS1358 in V. vulnificus and IS4 in S. putrefaciens, which disrupts wcvF), two copies of wecA, and an additional gene between wzx and wcvD in S. putrefaciens can be clearly seen.
DISCUSSION
The most intensely studied of the V. vulnificus virulence factors is the CPS, and virulence is absolutely dependent upon its production. The CPS mediates resistance of bacteria to complement-mediated bacteriolysis and phagocytosis. All virulent strains of V. vulnificus produce copious amounts of CPS, whereas acapsulated strains produce little or no CPS and are attenuated. We previously identified a cluster of 9 genes in V. vulnificus strain 27562 that were required for the production of a Wzy-dependent CPS. In the present study, we show that these genes are part of a novel 25-kb CPS locus containing 21 open reading frames. Lesions in 8 of these genes (wzc, rmlC, cppA, wzx, wcvD, wzy, wcvE, and wcvF) resulted in a translucent phenotype and attenuated virulence, supporting the involvement of this locus in CPS production. The locus includes a wecA, encoding the initiating glycosyltransferase WecA (a characteristic of G4C loci), and is also genetically linked to the wza-wzb-wzc translocation genes (a characteristic of G1C loci). Hence, the CPS cluster of strain 27562 shares characteristics with both G1C and G4C loci. This parallels the organization of the CPS locus in V. cholerae NRT36S, where wecA is also linked to the wza-wzb-wzc transport genes (10).
The CPS of strain 27562 contains repeats of d-GlcNAc, MurNAc, d-GalA, and l-Rha. A rhamnosyltransferase, WcvF, a WecA initiating glycosyltransferase, and two additional glycosyltransferases (WcvD and WcvE) were identified in the cluster. WcvD and WcvE likely participate in the transfer of muramic acid and glucuronic acid to the undecaprenyl phosphate carrier. Genes participating in the biosynthesis of the sugar rhamnose (rmlBADC) and d-GalA (wcvA, wcvB, and wcvC) were identified in the CPS cluster. Moreover, murACPS and murBCPS, which are paralogs of genes in the highly conserved murABCDEF operon that governs the synthesis of UDP-MurNAc for peptidoglycan biosynthesis (46), were identified in the CPS locus. This suggested that they participated in the production of muramic acid, which is found in the capsule. Since the intracellular pool of MurNAc must be shared between peptidoglycan and CPS biosynthesis in strain 27562, it was conceivable that MurACPS and MurBCPS acted to augment the intracellular MurNAc pool to sustain both processes and that disruption of either of these genes might result in slowed growth or attenuated virulence. However, lesions in murACPS and murBCPS had no effect on growth, CPS production, or virulence. The MurNAc sugar is not part of the linear chain in the CPS. It is thus conceivable that the CPS may continue to be polymerized and react with antisera despite lacking muramic acid. However, there are indications that the repeats must be complete for proper polymerization and transport of the CPS to the cell surface (3). Although we cannot discount the possibility that muramic acid content in the capsule is decreased in the ΔmurACPS and ΔmurBCPS strains, murACPS and murBCPS appear to be functionally redundant. While two murA genes are present in the genomes of several Gram-positive bacteria, such as Bacillus subtilis, Streptococcus pneumoniae, Streptococcus pyogenes, Staphylococcus aureus, and Clostridium acetobutylicum (6, 15, 32), only single copies of the murA and murB genes have ever been identified in the genomes of Gram-negative bacteria. To our knowledge, this is the first report of murA and murB paralogs in Gram-negative species.
V. vulnificus produces hundreds of different CPS carbotypes (8, 26). This diversity is driven in part by the plethora of different biosynthetic enzymes participating in CPS production in this species. The mechanism by which these different enzyme activities are acquired is unknown. HGT may be inferred from a variety of features, including an abnormally high sequence similarity to genes from unrelated organisms, GC content and codon bias differences, and a tendency of the recently acquired trait to be limited to the recipient strain and absent from closely related taxa. Most of the CPS locus of strain 27562 (from rmlB to wcvF) was conserved in S. putrefaciens strain 200. As a consequence, the S. putrefaciens 200 genome also harbors paralogs of murA and murB. The average GC content of the CPS locus (39.7%) was significantly lower than the 46.7% and 44.5% average GC contents of the V. vulnificus and S. putrefaciens 200 genomes, respectively. This supported an exogenous origin for the CPS cluster in both of these species, and the redundancy of the murACPS and murACPS genes is consistent with this notion. Direct transfer of CPS loci between these species has not been reported, but the two organisms have been isolated together from seawater and oysters, providing natural conditions under which gene transfer could occur (42). One of the most notable differences between the loci was the additional gene downstream of wzx in the S. putrefaciens CPS locus that was absent from the 27562 CPS locus. This additional gene codes for a poly-gamma-glutamate capsule biosynthesis enzyme. Poly-gamma-glutamate is a natural polymer that is required for virulence in Bacillus anthracis (18). Of particular interest was the observation that this gene was precisely inserted between the wzx and wcvD genes. This suggests that the same mechanism governing the transfer of complete CPS loci may also contribute to their evolution via the transfer of partial loci or individual genes to create novel CPS biosynthesis clusters.
HGT is an important source of genetic variation in many bacterial species. In one of the best-known examples, the epidemic Vibrio cholerae O139 serovar emerged from the pandemic biotype El Tor O1 serovar through the replacement of a 22-kb O-antigen region by a 40-kb O139-specific O-antigen DNA fragment (12). The CPS locus of strain 27562 contained IS1358. This IS element is also associated with CPS loci in V. cholerae, V. vulnificus, and Vibrio anguillarum (30, 44). IS1358 was shown to be active for transposition (16), making it possible to envision IS1358-mediated genetic exchange between Vibrio species that leads to the genesis of new CPS regions. However, only a single copy of IS1358 is present in the CPS locus of strain 27562, and in the S. putrefaciens CPS locus, IS1358 is replaced by IS4. The prevalence of IS elements may simply be due to the intermixing of CPS regions, with the IS elements providing targets for homologous recombination rather than playing a role via transposition. At present, the role, if any, of IS elements in the transfer of polysaccharide loci in the Vibrionaceae remains unclear.
Natural genetic transformation may be a powerful mechanism of horizontal gene transfer in bacterial populations. Many bacteria are naturally competent, including a number of Vibrio and other marine species (20, 21, 34, 35, 40, 41, 43). Chitin was recently demonstrated to induce natural competence in V. cholerae (34), and the conserved regions flanking the O-antigen biosynthesis loci were targets for homologous recombination during serogroup conversion of V. cholerae from O1 to O139 (7). Chitin also induces natural competence in V. vulnificus (24), and our genomic comparison of the CPS regions from V. vulnificus strains with different CPS carbotypes revealed that the genetically variable CPS loci were flanked by conserved chromosomal regions (Fig. 3). These regions, anchored by wza-wzb-wzc on one side and wcvB on the other, may serve as targets for the homologous recombination of intact exogenous CPS loci following their uptake by chitin-induced horizontal gene transfer. A JUMPstart element, first identified in Escherichia, Salmonella, Yersinia, and Vibrio strains (27), was identified upstream of wza. Within this element is a 9-bp sequence (AAGGGCGGT) that is almost identical to the uptake signal sequence (USS) of Haemophilus influenzae (23). The USS sequences are required for the efficient uptake of DNA in some naturally transformable bacteria (19), making it tempting to speculate that the HGT of polysaccharide loci in the Vibrionaceae may involve the preferential uptake of USS-tagged DNA by chitin-induced natural transformation.
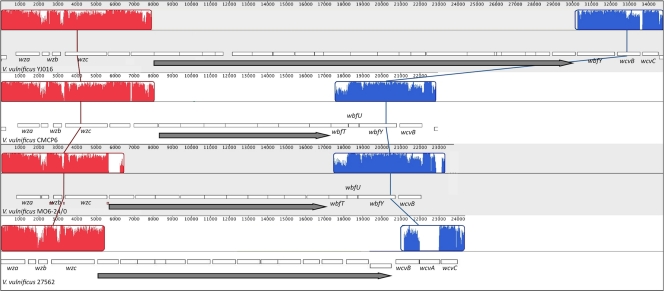
FIG. 3.
Comparative analysis of the CPS loci of V. vulnificus strains. The nucleotide sequences of the CPS loci of V. vulnificus strains YJ016, CMCP6, MO6-24/O, and ATCC 27562 were aligned using Mauve. The alignment display is organized as described in the Fig. 2 legend. Strain designations appear below the organization of the genes within the locus (white boxes). The long dark-gray arrows highlight carbotype-specific genes of the respective loci.
We previously showed that mutations in genes within the CPS locus also affected the production of LPS in strain 27562 (39). Furthermore, comparative genomics suggests that the putative LPS biosynthesis locus is adjacent to the CPS locus in the genomes of CMCP6, YJ016, and 27562. This organization is reminiscent of the situation in V. cholerae NRT36S, where both CPS and LPS genes occupy the same locus (10). It was proposed that the embedding of CPS and LPS genes within the same locus could provide a mechanism for the rapid emergence of new pathogenic and bacteriophage-resistant strains via the generation of novel capsular and O antigens by horizontal transfer. The exchange of LPS and CPS loci by chitin-induced natural transformation in V. vulnificus would provide a mechanistic basis for generating the large number of CPS types observed, and this phenomenon may simultaneously promote the emergence of new pathogenic strains through the acquisition of chromosomally linked LPS biosynthesis genes.
Acknowledgments
This work was supported by funding from the Canadian Institutes of Health Research (CIHR) to D.A.R.-M.
Editor: A. Camilli
Footnotes
Published ahead of print on 4 October 2010.
REFERENCES
- 1.Ali, A., M. H. Rashid, and D. K. Karaolis. 2002. High-frequency rugose exopolysaccharide production by Vibrio cholerae. Appl. Environ. Microbiol. 68:5773-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amor, P. A., J. A. Yethon, M. A. Monteiro, and C. Whitfield. 1999. Assembly of the K40 antigen in Escherichia coli: identification of a novel enzyme responsible for addition of L-serine residues to the glycan backbone and its requirement for K40 polymerization. J. Bacteriol. 181:772-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey, M. J., C. Hughes, and V. Koronakis. 1997. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26:845-851. [DOI] [PubMed] [Google Scholar]
- 5.Banoub, J. H., D. H. Shaw, H. Pang, J. J. Krepinsky, N. A. Nakhla, and T. Patel. 1990. Structural elucidation of the O-specific antigen of Yersinia ruckerii by fast atom bombardment mass spectrometry (FAB-MS). Biomed. Environ. Mass Spectrom. 19:787-790. [DOI] [PubMed] [Google Scholar]
- 6.Blake, K. L., A. J. O'Neill, D. Mengin-Lecreulx, P. J. Henderson, J. M. Bostock, C. J. Dunsmore, K. J. Simmons, C. W. Fishwick, J. A. Leeds, and I. Chopra. 2009. The nature of Staphylococcus aureus MurA and MurZ and approaches for detection of peptidoglycan biosynthesis inhibitors. Mol. Microbiol. 72:335-343. [DOI] [PubMed] [Google Scholar]
- 7.Blokesch, M., and G. K. Schoolnik. 2007. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 3:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bush, C. A., P. Patel, S. Gunawardena, J. Powell, A. Joseph, J. A. Johnson, and J. G. Morris. 1997. Classification of Vibrio vulnificus strains by the carbohydrate composition of their capsular polysaccharides. Anal. Biochem. 250:186-195. [DOI] [PubMed] [Google Scholar]
- 9.Carver, T. J., K. M. Rutherford, M. Berriman, M.-A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422-3423. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y., P. Bystricky, J. Adeyeye, P. Panigrahi, A. Ali, J. A. Johnson, C. A. Bush, J. G. Morris, Jr., and O. C. Stine. 2007. The capsule polysaccharide structure and biogenesis for non-O1 Vibrio cholerae NRT36S: genes are embedded in the LPS region. BMC Microbiol. 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comstock, L. E., and D. L. Kasper. 2006. Bacterial glycans: key mediators of diverse host immune responses. Cell 126:847-850. [DOI] [PubMed] [Google Scholar]
- 12.Comstock, L. E., D. Maneval, Jr., P. Panigrahi, A. Joseph, M. M. Levine, J. B. Kaper, J. G. Morris, Jr., and J. A. Johnson. 1995. The capsule and O antigen in Vibrio cholerae O139 Bengal are associated with a genetic region not present in Vibrio cholerae O1. Infect. Immun. 63:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darling, A. E., T. J. Treangen, X. Messeguer, and N. T. Perna. 2007. Analyzing patterns of microbial evolution using the Mauve genome alignment system. Methods Mol. Biol. 396:135-152. [DOI] [PubMed] [Google Scholar]
- 14.Demarre, G., A. M. Guerout, C. Matsumoto-Mashimo, D. A. Rowe-Magnus, P. Marliere, and D. Mazel. 2005. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 156:245-255. [DOI] [PubMed] [Google Scholar]
- 15.Du, W., J. R. Brown, D. R. Sylvester, J. Huang, A. F. Chalker, C. Y. So, D. J. Holmes, D. J. Payne, and N. G. Wallis. 2000. Two active forms of UDP-N-acetylglucosamine enolpyruvyl transferase in gram-positive bacteria. J. Bacteriol. 182:4146-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumontier, S., P. Trieu-Cuot, and P. Berche. 1998. Structural and functional characterization of IS1358 from Vibrio cholerae. J. Bacteriol. 180:6101-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Zoeiby, A., F. Sanschagrin, and R. C. Levesque. 2003. Structure and function of the Mur enzymes: development of novel inhibitors. Mol. Microbiol. 47:1-12. [DOI] [PubMed] [Google Scholar]
- 18.Ezzell, J. W., and S. L. Welkos. 1999. The capsule of Bacillus anthracis, a review. J. Appl. Microbiol. 87:250. [DOI] [PubMed] [Google Scholar]
- 19.Findlay, W. A., and R. J. Redfield. 2009. Coevolution of DNA uptake sequences and bacterial proteomes. Genome Biol. Evol. 2009:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frischer, M. E., J. M. Thurmond, and J. H. Paul. 1990. Natural plasmid transformation in a high-frequency-of-transformation marine Vibrio strain. Appl. Environ. Microbiol. 56:3439-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frischer, M. E., H. G. Williams, B. Bennison, G. R. Drake, D. L. Balkwill, and J. H. Paul. 1996. The naturally transformable marine bacterium WJT-1C formally identified as “Vibrio” is a pseudomonad. Curr. Microbiol. 33:287-291. [DOI] [PubMed] [Google Scholar]
- 22.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ.
- 23.Gonzalez-Fraga, S., M. Pichel, N. Binsztein, J. A. Johnson, J. G. Morris, Jr., and O. C. Stine. 2008. Lateral gene transfer of O1 serogroup encoding genes of Vibrio cholerae. FEMS Microbiol. Lett. 286:32-38. [DOI] [PubMed] [Google Scholar]
- 24.Gulig, P. A., M. S. Tucker, P. C. Thiaville, J. L. Joseph, and R. N. Brown. 2009. USER friendly cloning coupled with chitin-based natural transformation enables rapid mutagenesis of Vibrio vulnificus. Appl. Environ. Microbiol. 75:4936-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunawardena, S., G. P. Reddy, Y. Wang, V. S. Kolli, R. Orlando, J. G. Morris, and C. A. Bush. 1998. Structure of a muramic acid containing capsular polysaccharide from the pathogenic strain of Vibrio vulnificus ATCC 27562. Carbohydr. Res. 309:65-76. [DOI] [PubMed] [Google Scholar]
- 26.Hayat, U., G. P. Reddy, C. A. Bush, J. A. Johnson, A. C. Wright, and J. G. Morris, Jr. 1993. Capsular types of Vibrio vulnificus: an analysis of strains from clinical and environmental sources. J. Infect. Dis. 168:758-762. [DOI] [PubMed] [Google Scholar]
- 27.Hobbs, M., and P. R. Reeves. 1994. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Mol. Microbiol. 12:855-856. [DOI] [PubMed] [Google Scholar]
- 28.Huda, N., E. W. Lee, J. Chen, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. Molecular cloning and characterization of an ABC multidrug efflux pump, VcaM, in non-O1 Vibrio cholerae. Antimicrob. Agents Chemother. 47:2413-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hulo, N., A. Bairoch, V. Bulliard, L. Cerutti, E. De Castro, P. S. Langendijk-Genevaux, M. Pagni, and C. J. A. Sigrist. 2006. The PROSITE database. Nucleic Acids Res. 34:D227-D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jedani, K. E., U. H. Stroeher, and P. A. Manning. 2000. Distribution of IS1358 and linkage to rfb-related genes in Vibrio anguillarum. Microbiology 146(pt. 2):323-331. [DOI] [PubMed] [Google Scholar]
- 31.Jones, M. K., and J. D. Oliver. 2009. Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77:1723-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kock, H., U. Gerth, and M. Hecker. 2004. MurAA, catalysing the first committed step in peptidoglycan biosynthesis, is a target of Clp-dependent proteolysis in Bacillus subtilis. Mol. Microbiol. 51:1087-1102. [DOI] [PubMed] [Google Scholar]
- 33.Marquardt, J. L., E. D. Brown, W. S. Lane, T. M. Haley, Y. Ichikawa, C. H. Wong, and C. T. Walsh. 1994. Kinetics, stoichiometry, and identification of the reactive thiolate in the inactivation of UDP-GlcNAc enolpyruvoyl transferase by the antibiotic fosfomycin. Biochemistry 33:10646-10651. [DOI] [PubMed] [Google Scholar]
- 34.Meibom, K. L., M. Blokesch, N. A. Dolganov, C. Y. Wu, and G. K. Schoolnik. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824-1827. [DOI] [PubMed] [Google Scholar]
- 35.Miller, M. C., D. P. Keymer, A. Avelar, A. B. Boehm, and G. K. Schoolnik. 2007. Detection and transformation of genome segments that differ within a coastal population of Vibrio cholerae strains. Appl. Environ. Microbiol. 73:3695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchison, M., D. Bulach, T. Vinh, K. Rajakumar, S. Faine, and B. Adler. 1997. Identification and characterization of the dTDP-rhamnose biosynthesis and transfer genes of the lipopolysaccharide-related rfb locus in Leptospira interrogans serovar Copenhageni. J. Bacteriol. 179:1262-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyakawa, T., H. Matsuzawa, M. Matsuhashi, and Y. Sugino. 1972. Cell wall peptidoglycan mutants of Escherichia coli K-12: existence of two clusters of genes, mra and mrb, for cell wall peptidoglycan biosynthesis. J. Bacteriol. 112:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakhamchik, A., C. Wilde, and D. A. Rowe-Magnus. 2008. Cyclic-di-GMP regulates extracellular polysaccharide production, biofilm formation, and rugose colony development by Vibrio vulnificus. Appl. Environ. Microbiol. 74:4199-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakhamchik, A., C. Wilde, and D. A. Rowe-Magnus. 2007. Identification of a Wzy polymerase required for group IV capsular polysaccharide and lipopolysaccharide biosynthesis in Vibrio vulnificus. Infect. Immun. 75:5550-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paul, J. H., M. E. Frischer, and J. M. Thurmond. 1991. Gene transfer in marine water column and sediment microcosms by natural plasmid transformation. Appl. Environ. Microbiol. 57:1509-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul, J. H., J. M. Thurmond, M. E. Frischer, and J. P. Cannon. 1992. Intergeneric natural plasmid transformation between E. coli and a marine Vibrio species. Mol. Ecol. 1:37-46. [DOI] [PubMed] [Google Scholar]
- 42.Richards, G. P., M. A. Watson, and S. Parveen. 2005. Development of a simple and rapid fluorogenic procedure for identification of Vibrionaceae family members. Appl. Environ. Microbiol. 71:3524-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart, G. J., and C. D. Sinigalliano. 1990. Detection of horizontal gene transfer by natural transformation in native and introduced species of bacteria in marine and synthetic sediments. Appl. Environ. Microbiol. 56:1818-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stroeher, U. H., K. E. Jedani, and P. A. Manning. 1998. Genetic organization of the regions associated with surface polysaccharide synthesis in Vibrio cholerae O1, O139 and Vibrio anguillarum O1 and O2: a review. Gene 223:269-282. [DOI] [PubMed] [Google Scholar]
- 45.Sutherland, I. W. 1972. Bacterial exopolysaccharides. Adv. Microb. Physiol. 8:143-213. [DOI] [PubMed] [Google Scholar]
- 46.van Heijenoort, J. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25R-36R. [DOI] [PubMed] [Google Scholar]
- 47.Whitfield, C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75:39-68. [DOI] [PubMed] [Google Scholar]