Abstract
Virtually all bacterial pathogens require iron to infect vertebrates. The most abundant source of iron within vertebrates is in the form of heme as a cofactor of hemoproteins. Many bacterial pathogens have elegant systems dedicated to the acquisition of heme from host hemoproteins. Once internalized, heme is either degraded to release free iron or used intact as a cofactor in catalases, cytochromes, and other bacterial hemoproteins. Paradoxically, the high redox potential of heme makes it a liability, as heme is toxic at high concentrations. Although a variety of mechanisms have been proposed to explain heme toxicity, the mechanisms by which heme kills bacteria are not well understood. Nonetheless, bacteria employ various strategies to protect against and eliminate heme toxicity. Factors involved in heme acquisition and detoxification have been found to contribute to virulence, underscoring the physiological relevance of heme stress during pathogenesis. Herein we describe the current understanding of the mechanisms of heme toxicity and how bacterial pathogens overcome the heme paradox during infection.
Iron is an essential cofactor for many enzymes found within all kingdoms of life. Bacterial pathogens are no exception to this rule, and therefore, they must acquire iron from their hosts in order to cause disease. Iron is a transition metal that can cycle between redox states, making it a valuable cofactor for biological processes. Ferric iron is water insoluble, and as such, it requires specialized proteins to facilitate its mobilization and to maintain intracellular reservoirs. In mammalian species, lactoferrin and transferrin transport iron, while ferritin stores iron. The most abundant form of iron in vertebrates, however, is bound within a porphyrin ring as ferriprotoporphyrin IX (heme). Heme solubilizes iron and enhances its catalytic ability by 5 to 10 orders of magnitude (14, 111). This catalytic activity is harnessed by hemoproteins involved in oxygenation reactions, oxidative stress responses, electron transport, oxygen transport, oxygen sensing, and oxygen storage. While heme is a necessary prosthetic group for many proteins, it also has the potential to cause toxicity at high concentrations. This property of heme requires that the intracellular pool of heme be tightly regulated.
Intracellular heme concentrations within vertebrates are tightly controlled by balancing the rates of heme biosynthesis and catabolism (87). Free heme released into the plasma by the dissolution of hemoproteins from lysed erythrocytes is quickly scavenged by albumin, hemopexin, and the serum lipocalin α1-microglobulin (13, 23, 44, 75). Any hemoglobin released into the serum is tightly bound by haptoglobin and subsequently cleared by tissue macrophages (51). It is evident that the vital yet reactive nature of heme requires that its production, degradation, and availability be carefully controlled in metazoans. Meeting these demands reduces heme-mediated toxicity and minimizes surplus free heme. Most bacterial pathogens that infect vertebrate tissues have systems dedicated to the acquisition of heme for use as a nutrient iron source. However, the toxicity of heme presents a paradox for microorganisms that satisfy their nutrient iron requirement through heme acquisition. This heme paradox is resolved through tightly regulated systems dedicated to balancing the acquisition of heme with the prevention of heme-mediated toxicity.
HEME SOURCES EXPLOITED BY BACTERIAL PATHOGENS
Heme acquisition systems.
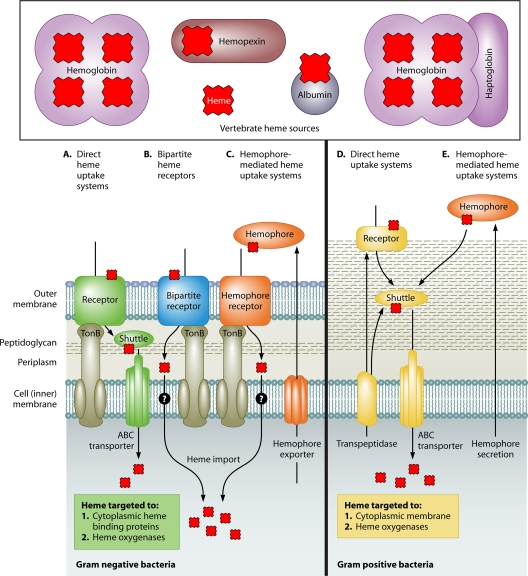
Bacterial pathogens can circumvent nearly every vertebrate form of heme sequestration. Bacterial heme acquisition systems that extract heme from hemopexin, heme-albumin, hemoglobin, and hemoglobin-haptoglobin have been identified in both Gram-negative and Gram-positive species (116) (Fig. 1). In Gram-negative bacteria there are three known classes of heme transport systems: direct heme uptake systems, bipartite heme receptors, and hemophore-mediated heme uptake systems (116).
FIG. 1.
Mechanisms of bacterial heme acquisition. Bacteria can utilize multiple heme sources found within the vertebrate host. Three types of heme acquisition systems have been identified for Gram-negative bacteria (left). These systems include direct heme uptake systems (A), bipartite heme receptors (B), and hemophore-mediated heme uptake systems (C). Most Gram-negative heme acquisition systems rely on an energy-transducing protein such as TonB to transport heme across the OM. Typically, Gram-positive bacteria (right) use direct heme uptake systems (D) to acquire heme. Recently, the first Gram-positive hemophore-mediated heme acquisition system (E) was identified in Bacillus anthracis. Heme is represented by the red polygons.
Direct heme uptake systems bind heme-containing proteins at the outer membrane (OM) of Gram-negative bacteria and transport heme into the periplasm in a TonB-dependent manner (116) (Fig. 1A). TonB is part of a cytoplasmic membrane complex that couples the proton motive force of the inner membrane to the outer membrane for the energy-dependent uptake of specific substrates, such as heme, into the periplasm (50, 100, 104). Once in the periplasm, heme is bound by a heme transport protein (HTP) (116). The heme-HTP complex shuttles heme to an ABC transporter in the inner membrane, through which heme is transported into the cytoplasm in an ATP-dependent process (116). A cytoplasmic protein is typically encoded within direct heme uptake operons, although the exact function of this family of proteins remains unclear. A specific example of a direct heme-binding uptake system is Pseudomonas aeruginosa phuR-phuSTUVW, where PhuR is the outer membrane receptor, PhuT is the HTP, PhuUVW is the ABC transporter, and PhuS is the cytoplasmic protein (53, 79, 117). Other pathogens encoding similar direct heme uptake systems include Bordetella pertussis, Yersinia pestis, Yersinia enterocolitica, Shigella dysenteriae, Vibrio cholerae, Campylobacter jejuni, Bartonella quintana, and Escherichia coli O157:H7 (69, 70, 84, 95, 112, 115, 118, 121).
Bipartite heme receptors have been identified only for Neisseria spp. These heme acquisition systems consist of a TonB-dependent outer membrane receptor, HpuB, and an outer membrane lipoprotein, HpuA (60) (Fig. 1B). The bipartite nature of HpuAB distinguishes it from the direct heme uptake systems, which have only a single-component TonB-dependent receptor (121). HpuA and HpuB form a functional complex, and both are required for the utilization of hemoglobin and hemoglobin-haptoglobin as nutrient iron sources (59, 61). Coincidentally, a spontaneous point mutation in Neisseria gonorrhoeae pilQ (pilQ1) suppresses the mutant hpuAB phenotype (16). PilQ forms a channel in the outer membrane and is required for pilus biogenesis, but its mutant form allows the entry of heme and various antimicrobial compounds with a TonB-independent, PilT-dependent mechanism (16). This suggests that PilQ or the pilus apparatus may regulate the free diffusion of heme into N. gonorrhoeae and highlights the concept that bacteria carefully regulate their intracellular pool of heme.
Hemophore-mediated heme uptake systems involve a secreted heme-binding protein called a hemophore (Fig. 1C). Holo-hemophores are recognized by an outer membrane receptor that mediates the import of heme. Two types of hemophore-mediated heme uptake systems have been described for Gram-negative bacteria: one uses the HasA hemophore, and the other uses the HxuA hemophore (116). The HasA hemophore-mediated heme uptake system has been identified in Serratia marcescens, P. aeruginosa, Pseudomonas fluorescens, and Y. pestis (4, 45, 56, 58, 96). The HasA system encodes an export complex (HasDEF), a hemophore receptor (HasR), and regulatory proteins (HasI and HasS) (7, 15, 33, 97, 116). Either TonB or the TonB homolog HasB provides the energy necessary for transporting heme from the surface-exposed HasR-HasA-heme complex into the periplasm (83). HasR can also bind free heme and hemoglobin-bound heme, but these processes are less efficient than HasA-mediated heme acquisition (33, 57). The HxuA system has been described only for Haemophilus influenzae type b (Hib) and consists of the hxuCBA gene cluster (19). HxuB is thought to export HxuA into the environment where HxuA binds the heme-hemopexin complex (19). The receptor for the heme-HxuA complex has not yet been determined, although HxuC is a likely candidate (19). While the HxuA system increases the diversity of utilizable heme substrates, the HasA system increases the efficiency of heme uptake (20).
Less diversity has been discovered for the heme uptake systems of Gram-positive bacteria. In general, Gram-positive heme uptake systems consist of surface-exposed receptors that shuttle heme through the cell wall to an ABC transporter for delivery into the cytoplasm (123) (Fig. 1D). The paradigm for Gram-positive heme uptake is represented by the Staphylococcus aureus iron-regulated surface determinant system (Isd). The Isd import machinery is encoded by 10 genes, including four cell wall-anchored proteins (IsdABCH), a transpeptidase (SrtB), a membrane transport system (IsdDEF), and two cytoplasmic heme oxygenases (IsdG and IsdI) (94). IsdB and IsdH are responsible for binding hemoglobin and hemoglobin-haptoglobin, respectively (24). IsdA extracts heme from IsdB or IsdH for passage to IsdC (64, 73, 94, 131). This efficient heme-scavenging system brings heme across the membrane into the cytoplasm through the IsdDEF ABC transporter, where it can either be degraded by the heme oxygenases IsdG and IsdI to release free iron or be trafficked intact to the cell membrane (68, 94). The function of exogenously acquired nondegraded heme in S. aureus remains to be elucidated. The Isd locus is present in numerous other Gram-positive pathogens, including Listeria monocytogenes, Bacillus anthracis, and Clostridium tetani (103, 116).
Other Gram-positive heme acquisition systems distinct from the Isd system have been identified for Corynebacterium spp. and Streptococcus spp. The Corynebacterium diphtheriae heme uptake system is encoded at a genomic locus containing htaC-htaA hmuTUV-htaB. HtaA and HtaB are membrane-associated and surface-exposed proteins thought to be heme receptors, while HmuUV is an ATP transporter that receives heme from HmuT and delivers it into the cytoplasm (3). The exact transport order of heme between HtaAB and HmuTUV has yet to be defined. In a manner similar to that of Corynebacterium diphtheriae Hta/Hmu, Streptococcus spp. encode surface-exposed heme-binding proteins (Shp and Shr) and an ABC transporter (HtaABC) (130). These systems are reminiscent of the Gram-negative direct heme transporters, except that the receptors and heme transport proteins are able to traffic heme through the thick cell wall of Gram-positive bacteria.
Only one hemophore-mediated heme uptake system has been described for Gram-positive bacteria (Fig. 1E). B. anthracis encodes an Isd heme uptake system that is both similar to and distinct from that of S. aureus (32, 67). Two proteins unique to B. anthracis are the secreted hemophores IsdX1 and IsdX2 (26, 67). IsdX1 is capable of removing heme from hemoglobin and passing heme to IsdC or IsdX2 (26). From IsdC, heme is transported into the cytoplasm through the Isd system. IsdX2 is both extracellular and associated with the cell surface, but the exact physiological role that IsdX2 serves in heme acquisition has yet to be defined (26).
Heme biosynthesis.
Most Gram-positive and Gram-negative pathogens have developed mechanisms for acquiring heme from their hosts, yet many of these organisms are also capable of synthesizing heme endogenously. Bacteria such as Streptococcus spp., Mycoplasma spp., H. influenzae, Enterococcus faecalis, Lactococcus lactis, Bartonella henselae, Borrelia burgdorferi, and Treponema pallidum do not have the complete machinery to make their own heme, and as such, they either do not require heme-iron or rely on heme acquired from the environment (12, 82, 88, 98). In comparison, the majority of bacteria with sequenced genomes contain the machinery for making heme, including bacteria from the Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Epsilonproteobacteria, Bacillales, Lactobacillales, Spirochaetales, and cyanobacteria (35, 43, 116).
The physiological relevance of both synthesizing and acquiring heme has remained elusive. Since the final step of heme biosynthesis requires iron to be inserted into the protoporphyrin IX (PPIX) ring, it is likely that bacteria synthesize heme only in environments where iron is available, as has been observed for Bradyrhizobium japonicum (90). In turn, when bacteria capable of both heme biosynthesis and acquisition are in environments that are iron poor, they may switch to utilizing exogenous heme as an iron source. In some bacteria such a response is controlled by the ferric uptake regulator Fur. Under iron-replete conditions, Fur represses the expression of heme uptake machinery in many species, including S. aureus, Y. pestis, and P. aeruginosa (68, 79, 115, 116). An alternative way of understanding the duality of heme acquisition and biosynthesis may lie in the energetic repercussions of heme biosynthesis compared to those of acquisition. Simply put, it may be less energetically expensive to acquire heme rather than synthesize it. While understandings of bacterial heme biosynthesis and acquisition processes individually have grown significantly, much has yet to be discovered about the interplay between the regulation and physiological uses of endogenous and exogenous heme.
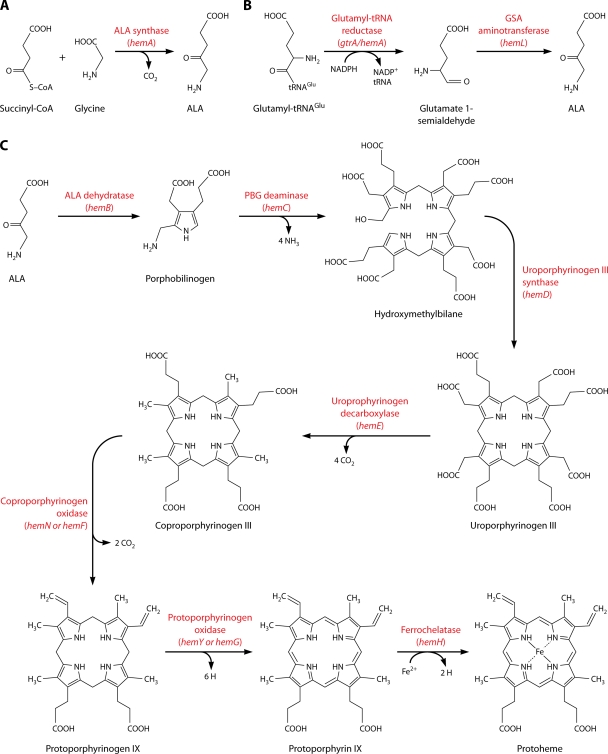
For those bacteria capable of heme biosynthesis, the process is fairly conserved. The first universal heme precursor is δ-aminolevulinic acid (ALA). ALA can be synthesized either from succinyl coenzyme A (CoA) and glycine by ALA synthase (hemA) (Fig. 2A) or from glutamyl-tRNAGlu by the C5 pathway using GtrA and HemL (Fig. 2B). Currently, no bacteria are known to use both routes to synthesize ALA, and hemA has been identified only in the Alphaproteobacteria (82). GtrA is the more ubiquitous enzyme used by bacteria to synthesize ALA. In some cases, the gtrA gene has been annotated as hemA, although this enzyme is not the same as ALA synthase and should be identified as gtrA to highlight its distinct function from that of HemA.
FIG. 2.
Schematic of heme biosynthesis in bacteria. The first universal heme precursor is δ-aminolevulinic acid (ALA). ALA can be synthesized by one of two routes, although no bacteria are known to use both ALA synthesis pathways. (A) Succinyl-CoA and glycine are converted into ALA by ALA synthase (hemA). HemA, however, has been identified only in members of the Alphaproteobacteria. (B) Most bacteria synthesize ALA from glutamyl-tRNAGlu by the C5 pathway using GtrA and HemL. (C) The rest of the heme biosynthesis pathway is highly conserved. ALA dehydratase (hemB) synthesizes porphobilinogen (PBG) from two ALA molecules. PBG deaminase (hemC) converts four PBG molecules into hydroxymethylbilane. Linear hydroxymethylbilane is fused into a ring by uroporphyrinogen III synthase (hemD) to make uroporphyrinogen III. Four carboxyl groups are removed by uroporphyrinogen decarboxylase (hemE) to make coproporphyrinogen III. Next, a coproporphyrinogen oxidase, either HemN (oxygen independent) or HemF (oxygen dependent), converts coproporphyrinogen III to protoporphyrinogen IX. Protoporphyrinogen IX is then oxidized to protoporphyrin IX by a protoporphyrinogen oxidase, either HemG (O2 independent) or HemY (O2 dependent) (8, 38). Finally, ferrochelatase (hemH) catalyzes the protoporphyrin IX chelation of ferrous iron to form protoheme. This protoheme can be utilized directly or modified further before being used as a prosthetic group in hemoproteins.
Once ALA has been synthesized, a series of seven reactions convert eight molecules of ALA into protoheme. This protoheme can be used directly or modified further before being employed as a prosthetic group in hemoproteins. The heme biosynthesis pathway is fully diagrammed in Fig. 2C. Two specific steps in heme biosynthesis of particular note are those that convert coproporphyrinogen III to protoporphyrinogen IX and then transform protoporphyrinogen IX into protoporphyrin IX. Each of these steps can be performed by two enzymes that are functionally redundant, but the distinguishing feature between each pair of enzymes is that one is oxygen dependent and the other is oxygen independent (82). The oxygen-dependent enzymes are found in eukaryotes and are less prevalent in prokaryotes (82). This may allow bacteria to be metabolically flexible and use alternative electron donors to synthesize heme in the absence of oxygen.
POTENTIAL MECHANISMS OF HEME TOXICITY
Whether through heme biosynthesis or through heme acquisition systems, most bacteria dedicate significant efforts to ensuring an adequate supply of heme. However, the utility of heme is inseparable from its toxic effects, and the degree of sensitivity to heme toxicity varies among bacteria. In general, Gram-positive bacteria are more sensitive to heme toxicity than are Gram-negative bacteria (78). Notable exceptions to this trend are the anaerobic Gram-positive Clostridium spp. and Gram-negative Porphyromonas spp., of which about 40% and 94% of tested strains are sensitive to heme toxicity, respectively (78). The variation in heme sensitivity observed across bacteria suggests one of two possibilities. First, bacteria less sensitive to heme may have more-robust mechanisms of heme tolerance. Alternatively, distinct bacterial genera may produce differing levels of toxic by-products upon heme exposure. Despite bacterial heme sensitivity being recognized for over 60 years, the mechanisms by which heme kills bacteria remain undefined.
One possible mechanism of heme toxicity may be mediated by free iron released during heme degradation by bacterial heme oxygenases (77, 102). Free iron causes intracellular damage mediated through the production of hydroxyl radicals via Fenton chemistry or through lipid peroxidation (25). The question remains, however, whether heme oxygenases release enough iron to exceed available iron-binding sites within the cell and cause cellular toxicity. The lack of in vivo data demonstrating that iron-mediated lipid peroxidation and Fenton chemistry are the causes of cellular damage after heme exposure support the idea that iron released from heme may not be the cause of heme toxicity. In addition, noniron metalloporphyrins are toxic to S. aureus, although they are not degraded by the staphylococcal heme oxygenases IsdG and IsdI (55, 113). This suggests that free metals, including iron, are not likely to be the main cause of metalloporphyrin toxicity. In this regard, it is likely that the offending agent is the heme itself (25).
Due to the lack of data regarding mechanisms of heme toxicity in bacteria, models of bacterial toxicity may be extrapolated from eukaryotes. In erythrocytes, free heme disrupts the cell membrane, resulting in hemolysis by a colloid-osmotic mechanism: the cell no longer maintains ion gradients, potassium leaks out, and water enters due to the osmotic gradient (18, 49, 99). Other eukaryotic cells do not undergo lysis in the presence of heme, although in vitro, endothelial cells are susceptible to heme toxicity by either the peroxidase-like or monooxygenase-like activities of heme (63). The monooxygenase-like reactivity of heme is the cause of heme-mediated DNA and protein damage in vitro and is most likely the mechanism of heme toxicity in metazoans (1, 2, 63).
The outer layers of the bacterial cell are notably different from eukaryotic membranes in lipid composition, cell wall, and structure. These outer layers act as an armament that protects the bacterial cell from a multitude of environmental insults. Therefore, it is possible that these eukaryotic mechanisms of heme toxicity do not translate to bacteria, as no reports of heme-induced bacterial lysis have been identified. Gram-negative species such as Y. pestis, Aeromonas salmonicida, Shigella flexneri, Prevotella spp., and Porphyromonas spp. accumulate heme in their outer surface (21, 34, 42, 62, 105). Rather than a source of toxicity, this accumulation of heme is thought to contribute to bacterial pathogenesis by increasing bacterial heme storage, utilization, or host invasion (21, 31, 34, 41, 105). Moreover, iron-replete S. aureus preferentially traffics exogenously acquired heme to the cell membrane, although the function of this process is still unknown (94). Of the species that accumulate heme in their membranes, S. aureus and Porphyromonas spp. are highly susceptible to heme toxicity, while Y. pestis and many Prevotella spp. are highly resistant to heme toxicity (48, 51, 60). This brief survey suggests that the accumulation of heme on the bacterial surface does not correlate with toxicity. Bacterial heme toxicity appears to depend more on species-specific properties. One piece of evidence to support this notion is that Gram-positive and not Gram-negative bacteria accumulate DNA damage when exposed to heme in vivo (76).
The mechanism of heme-mediated toxicity is multifaceted. While much is known about eukaryotic heme toxicity, the relevance of these findings to bacteria remains unclear. The main cause of bacterial heme toxicity is not due to the release of free iron by cellular monooxygenases or by the peroxidase-like activity of heme (55, 63, 111). Some heme toxicity may be due to its monooxygenase-like reactivity, but this has not been directly tested for bacteria (76). Based on what has been reported, a likely cause of bacterial heme toxicity is its ability to damage DNA; however, the two have not been directly correlated (63). Taking these facts into consideration, it is likely that many mechanisms of heme toxicity have yet to be discovered.
MECHANISMS OF BACTERIAL HEME TOLERANCE
While the mechanism of bacterial heme toxicity is not well defined, several means by which bacteria avoid heme toxicity have been characterized. The regulation of biosynthesis and the regulation of uptake are two ways by which bacteria control intracellular levels of heme. When the regulation of heme uptake and biosynthesis is not sufficient to prevent heme toxicity, other mechanisms that are utilized by bacteria include export, sequestration, and degradation. These are discussed in greater detail below and are summarized in Table 1.
TABLE 1.
Mechanisms of heme tolerance
| Mechanism | Organism(s) (reference[s]) |
|
|---|---|---|
| Characterizeda | Predicted | |
| Export | ||
| HrtAB | S. aureus (106), B. anthracis (107), S. agalactiae (27) | L. monocytogenes, L. inocua, B. thuringiensis, B. cereus, |
| PefAB-PefCD | S. agalactiae (27, 108) | L. lactis, S. epidermidis (22, 108) |
| MtrCDE | N. gonorrhoeae (37) | |
| Sequestration | ||
| HemS family | Y. enterocolitica (112), S. dysenteriae (128), P. aeruginosa (53), E. coli O157:H7 (114), Y. pestis (115) | A. tumefaciens |
| Degradation | ||
| HO family | N. gonorrhoeae (132), N. meningitidis (132), C. perfringens (39), C. tetani (11), L. interrogans (72), Helicobacter pylori (36), C. diphtheriae (126), Corynebacterium ulcerans (52), P. aeruginosa (125), C. jejuni (95) | D. radiodurans, cyanobacteria (30) |
| IsdG family | B. anthracis (101), S. aureus (102), B. japonicum (89), B. melitensis (89), M. tuberculosis (17) | Alphaproteobacteria, Agrobacterium, Streptomyces, Deinococcus-Thermus, Chloroflexi (89, 102) |
| Other | ||
| Ght | N. meningitidis (91) | E. coli O157:H7, V. cholerae, B. pertussis, P. multocida, H. influenzae, S. enterica serovar Typhimurium, C. burnetii (91) |
Bacterial species in which the listed proteins are required to alleviate heme toxicity are printed in boldface type. Those bacterial species that contain the listed protein but have not been shown to use the listed protein to alleviate heme toxicity are printed in lightface type.
Export.
S. aureus, one of the pathogens most sensitive to heme, has a heme-regulated transporter (HrtAB) that alleviates toxicity (119). A mutation of the hrtAB transporter genes results in a further increase in heme sensitivity (106). The mechanism by which HrtAB alleviates heme toxicity has yet to be elucidated; however, HrtAB is thought to pump out either heme directly or a toxic metabolite of heme accumulation. Orthologous HrtAB systems have been characterized for Streptococcus agalactiae, B. anthracis, and L. lactis (27, 85, 107). Mutations in either the B. anthracis or L. lactis Hrt systems also result in increased sensitivity to heme toxicity (85, 107). The S. agalactiae hrtAB mutant has not been generated, so the contribution of HrtAB to resisting heme toxicity in this organism has yet to be determined. Other Gram-positive pathogens and saprotrophs that encode putative Hrt systems include L. monocytogenes and Listeria inocua of the listeriae, Bacillus thuringiensis and Bacillus cereus of the bacilli, and Staphylococcus epidermidis of the staphylococci (22, 108). Notably absent from this list are the nonpathogenic, nonsaprotrophic bacilli B. subtilis and B. licheniformis. This observation suggests that the Hrt system may have evolved in bacteria that come into contact with vertebrate blood to protect them from heme toxicity. This point is underscored by the upregulation of B. anthracis hrtAB in an animal model of anthrax (107).
A dual-operon efflux system has recently been identified for S. agalactiae, comprised of pefAB and pefRCD (27). PefAB and PefCD are two putative heme and protoporphyrin IX (PPIX) efflux pumps, although the role of PefB may be as an accessory protein rather than an actual pump (27). When the pef operons are disrupted, the intracellular levels of heme and PPIX increase, causing enhanced sensitivity to heme toxicity (27). In addition to pefAB and pefRCD, S. agalactiae also encodes orthologs of hrtAB, which are transcribed at higher levels than pefAB and pefCD at high heme concentrations (27). This suggests that the pef transporters are utilized to fine-tune intracellular heme levels, while hrtAB is employed to protect S. agalactiae from heme toxicity in heme-rich environments (27). The differential activity of two heme-regulated transport systems highlights the delicate balance that S. agalactiae maintains to cope with the heme paradox.
Other transporters with broader substrate specificity also provide some protection from heme toxicity. The multiple-transferable-resistance (Mtr) efflux system provides resistance to hydrophobic agents in N. gonorrhoeae (37). The inactivation of mtrCDE causes increased susceptibility to heme, while the overexpression of this efflux pump results in increased tolerance to heme toxicity (9). It is possible that the ability of general efflux systems to provide some resistance to heme is a conserved detoxification strategy used by many bacteria.
Sequestration.
The best example of heme sequestration to avoid heme toxicity is in the eukaryotic parasite Plasmodium spp., the causative agents of malaria. During Plasmodium infection of erythrocytes, hemoglobin is digested into amino acids and heme (80). The amino acids are used as a nutrient, but the accumulation of heme is toxic. Plasmodium sequesters heme into a nontoxic, highly insoluble, dark brown substance called hemozoin (28). Hemozoin formation has been reported to be catalyzed by the heme detoxification protein (HDP) (47). Many bacteria utilize ferritin-like proteins to sequester free cellular iron, but heme sequestration tactics have not been well characterized for bacteria.
Some of the cytoplasmic heme-binding proteins associated with the direct heme uptake systems of Gram-negative bacteria have been proposed to function in heme sequestration and utilization. Proteins in this family include Y. enterocolitica HemS, E. coli O157:H7 ChuS, P. aeruginosa PhuS, S. dysenteriae ShuS, and Y. pestis HmuS. Since they were first described for Y. enterocolitica, we will refer to these proteins as the HemS family of proteins. Each of these cytoplasmic proteins is able to bind heme, but there is not a consensus on the function of this family of proteins. The deletion of Y. enterocolitica HemS is lethal, but hemS expression in E. coli prevents heme toxicity (112). It has been proposed that HemS degrades heme, but no biochemical data have been published to support this hypothesis. ChuS, the HemS homolog in E. coli O157:H7, however, has been shown to have heme oxygenase activity in the presence of ascorbate or an NADPH-dependent reductase (114). Whether ChuS degrades heme by an enzymatic or nonenzymatic process remains undefined. Another HemS family member, P. aeruginosa PhuS, degrades heme through a nonenzymatic process that occurs via free H2O2 oxidation of ferric PhuS (53). Rather than acting as a heme oxygenase itself, the most likely function of PhuS is to store intracellular heme and traffic heme to a distinct heme monooxygenase, PigA (53). S. dysenteriae ShuS is necessary for efficient heme utilization and protection from heme toxicity in S. dysenteriae, but data suggest that it is not likely to be a heme oxygenase (128). A potential mechanism by which ShuS could provide resistance to heme toxicity is through the DNA-binding properties of apo-ShuS (48). Y. pestis HmuS is also thought to function in heme utilization, but its mechanism remains ill defined (115). Members of the HemS family of cytoplasmic heme-binding proteins have been assigned diverse functions as heme monooxygenases, heme-trafficking proteins, heme-sequestering proteins, and DNA-binding proteins. Although the functions of these proteins may be diverse, it is clear that they are important for heme utilization and tolerance to heme toxicity.
Another heme-binding protein that contributes to resistance to heme toxicity is the Haemophilus ducreyi Cu,Zn superoxide dismutase sodC (74). SodC is unique in its ability to bind a heme molecule at its dimer interface (81). The mutation of sodC results in an increased sensitivity of H. ducreyi to heme toxicity (74). Intriguingly, the mechanism for its protection against heme toxicity seems twofold, as both its antioxidant function and heme-binding function were individually able to rescue the heme sensitivity of the sodC mutant (74). Other cytoplasmic proteins such as AhpC in S. agalactiae and HutZ in V. cholerae also bind heme (54, 127). AhpC is a 2-Cys peroxiredoxin family protein, but its peroxidase activity does not depend on its heme-binding status (54). The mutation of either of these proteins, however, does not result in increased heme sensitivity (54, 127). It is thought that instead, these proteins function to store heme and promote its utilization (54, 127). Whereas many cytoplasmic proteins may bind heme, the specific function that such binding serves may be diverse. Identification of the exact function of heme-binding proteins has proven to be difficult, and much has yet to be learned about the regulation of heme trafficking within the bacterial cytoplasm.
Degradation.
Reducing heme toxicity can also be accomplished by the heme oxygenase-mediated degradation of heme. Although these proteins bind heme, they are not considered hemoproteins, as their heme-binding capacity is for the purpose of catalytically degrading heme. The HO family of heme monooxygenases was first identified for mammals, where they function primarily to protect cells from heme toxicity (116). In bacterial pathogens most heme oxygenases are implicated primarily in iron acquisition, although some have been identified to protect against heme toxicity.
In the presence of an electron donor, the canonical HO proteins degrade heme to free iron, CO, and α-biliverdin (120). HO family heme monooxygenases have been characterized for many bacterial species, a list of which can be found in Table 1. The P. aeruginosa PigA (also referred to as pa-HO) is an exception in the HO family of heme monooxygenases, as it produces a mixture of all four biliverdin isomers (92). Other bacteria predicted to encode an HO family heme monooxygenase include Deinococcus radiodurans, Agrobacterium tumefaciens, and cyanobacteria, including Anabaena sp. PCC7120, Thermosynechococcus elongatus, Prochlorococcus marinus, and Nostoc punctiforme (30).
The IsdG family of heme oxygenases, first described for S. aureus, degrades heme to release free iron and form staphylobilin in the presence of a reducing agent (94). IsdG family heme oxygenases have been characterized for S. aureus, B. anthracis, B. japonicum, Mycobacterium tuberculosis, and Brucella melitensis and are predicted to be encoded in the Alphaproteobacteria, Streptomyces, Deinococcus-Thermus, and Chloroflexi groups (17, 89, 101, 102). S. aureus encodes two IsdG family heme oxygenases (102). While functionally redundant, in that they both degrade heme to staphylobilin, they are differentially regulated by heme (93). IsdG degradation is inhibited in the presence of heme, while IsdI abundance is not affected by heme concentrations (93). In this way, S. aureus increases the rate of heme degradation as intracellular heme levels rise. This is yet another example of how bacteria refine intracellular levels of heme.
The functions of the heme oxygenase products (biliverdin and staphylobilin) in bacteria remain unclear. In cyanobacteria, algae, and plants, biliverdin is a precursor for light-harvesting phytobilin pigments (30). A reaction specific to mammals is the conversion of biliverdin to the potent antioxidant bilirubin (66, 110). It is possible that bacterial biliverdin and staphylobilin are excreted as waste products or further metabolized to be used as carbon and nitrogen sources. The energetically economical nature of bacteria, however, suggests that it is unlikely that biliverdin and staphylobilin are simply refuse. In the context of iron and heme homeostasis, it is possible that biliverdin and staphylobilin might function as signaling molecules or somehow provide protection from heme toxicity.
Although the role of biliverdin or staphylobilin in heme homeostasis or metabolism remains undefined, some heme oxygenases have been directly implicated in alleviating heme toxicity. The inactivation of N. gonorrhoeae hemO causes a growth defect when the mutant is grown in liquid culture in which heme is the only iron source (133). It is unclear whether this means that the N. gonorrhoeae hemO mutant is simply unable to utilize heme or if it is also more sensitive to heme toxicity. The disruption of B. anthracis isdG causes growth inhibition across all concentrations of heme, when heme is the sole iron source (101). This suggests that IsdG is needed both for the utilization of iron from heme and for the prevention of heme toxicity. As discussed above, PhuS delivers heme to PigA in P. aeruginosa (53). Whereas the sensitivity of the phuS and pigA mutants to heme toxicity has not been tested, the PhuS homologs HemS and ShuS do provide protection against heme toxicity (112, 128). It is possible that the HemS family of proteins shuttle heme to a heme monooxygenase and that the controlled degradation of heme by a heme monooxygenase provides protection against heme toxicity (53). Taken together, the regulated degradation of heme by heme oxygenases is a prime example of how bacteria solve the heme paradox by reaping nutritional benefits from heme while simultaneously eliminating the associated toxicity of heme.
Alternative strategies for heme detoxification.
Besides exportation, sequestration, and degradation systems, other mechanisms of heme resistance are employed by bacteria. One example of this is the N. meningitidis gene of hydrophobic agent tolerance, ght. The mutation of ght causes increased susceptibility to heme and other hydrophobic agents (91). A broad spectrum of Gram-negative bacteria have ght homologs, including the pathogens E. coli O157:H7, V. cholerae, B. pertussis, Pasteurella multocida, H. influenzae, Salmonella enterica serovar Typhimurium, and Coxiella burnetii (91). The mechanism for the increased sensitivity of the ght mutant to heme and other hydrophobic agents has yet to be elucidated; nonetheless, it is separate from the Mtr efflux system and PilQ (91).
BACTERIAL HEME SENSING
Many of the systems involved in resistance to heme toxicity are not constitutively expressed. Rather, bacteria regulate their expression in response to heme toxicity signals. In S. aureus and B. anthracis, the heme sensor system (HssRS) activates the transcription of the hrtAB ABC transporter (107, 119). HssRS is a two-component system (TCS) composed of HssS as a membrane-bound sensor kinase and HssR as a cytoplasmic response regulator. The mutation of hssRS results in increased heme sensitivity. L. lactis encodes a heme-regulated hrtAB ortholog, ygfBA, but no hssRS ortholog (85, 119). Instead, the regulator of ygfBA is thought to be the neighboring ygfC gene, but this has yet to be confirmed.
The putative heme and PPIX efflux pumps PefAB and PefCD in S. agalactiae are regulated by the MarR superfamily repressor PefR. The PefR-dependent inhibition of the two operons is alleviated when it binds heme or PPIX (27). PefR relieves its inhibition in the presence of >0.3 μM heme and plateaus at between 1 and 10 μM heme (27). S. agalactiae also encodes putative HssRS and HrtAB systems. In comparison to the regulation of the pef operons, S. agalactiae hrtAB is activated in 1 μM heme and continues to be highly activated in 10 μM heme. The activation of these two distinct efflux systems at different concentrations of heme further highlights the intricate regulatory mechanisms that bacteria employ to control intracellular concentrations of heme.
Another heme-responsive TCS distinct from HssRS is ChrAS from Corynebacterium diphtheriae (46). ChrAS activates the transcription of the heme monooxygenase hmuO and inhibits the transcription of the heme biosynthesis gene hemA (gtrA) in the presence of heme (5, 6). The counterregulation of heme degradation and synthesis by ChrAS is subtly elegant. The disruption of hmuO does not cause heme sensitivity, but the mutation of either chrA or chrS results in growth inhibition and a loss of viability in the presence of high concentrations of heme (5). This indicates that ChrAS has other transcriptional targets and that these targets are involved in protecting C. diphtheriae from heme toxicity. The ChrAS-regulated factors that protect C. diphtheriae from heme toxicity have not yet been identified.
Like hmuO, the hemO heme monooxygenase in Clostridium perfringens is regulated to maintain a certain level of iron and heme in the cell. In the presence of heme, hemO is upregulated, but it is downregulated in the presence of iron (39). The factors responsible for the iron-dependent downregulation of hemO have not been identified. The VirSR TCS and the VirR-regulated RNA (VR-RNA) contribute to the positive regulation of hemO and other C. perfringens virulence genes; however, the stimulus of VirSR is still unknown (39, 65). It is likely that virulence factors are regulated in the presence of host factors, and it is possible that host heme might be such an environmental signal.
ROLE OF HEME IN PATHOGENESIS
Most bacteria require iron and heme for full virulence, as measured by bacterial growth in animal models of infection. For bacteria that do not synthesize heme, heme acquisition allows them to aerobically respire or activate catalases, which protect against the oxidative burst of host phagocytes (29, 129). In this way, heme acquisition provides an advantage during infection. This is exemplified by the inactivation of the S. agalactiae cytochrome bd quinol oxidase cydA, which prevents acquired heme from activating aerobic respiration reducing bacterial fitness in a murine model (129). The competitive advantage provided by heme is also likely a factor for bacteria that synthesize heme, as S. aureus heme auxotrophs manifest as slow-growing small-colony variants (122). Most important, however, is the observation that many heme uptake mutants are attenuated for virulence. The inactivation of genes involved in heme acquisition in B. pertussis, V. cholerae, Haemophilus spp., and S. aureus all result in reduced virulence in animal models (10, 40, 71, 86, 109). While heme provides a distinct advantage for bacteria during infection, the heme paradox requires that bacteria must carefully balance intracellular concentrations of this valuable nutrient source, as it can also be toxic at high concentrations.
The virulence contributions of some mechanisms that respond to and prevent heme toxicity have been assessed in animal models. The deletion of pefR in S. agalactiae results in decreased intracellular levels of heme and reduced virulence in a mouse model of infection (27). Conversely, the deletion of staphylococcal hssR results in an accumulation of heme toxicity and hypervirulence in the mouse liver (119). These data are consistent with the idea that reducing intracellular heme levels might starve pathogens, while increasing intracellular heme triggers hypervirulence. The N. gonorrhoeae MtrCDE efflux pump provides protection from heme and other hydrophobic agents (9, 37). Clinically relevant mutations that result in the overexpression of the MtrCDE efflux pump generally result in increased resistance to hydrophobic agents and, correspondingly, increased bacterial burden in vaginal lavage fluids of infected mice (124). Those authors proposed that the direct relationship between virulence and MtrCDE expression levels is due to increased resistance to innate immune effectors. An alternative possibility is that these mutants are also more resistant to heme toxicity and that this may provide an advantage in vivo. Taken together, these results demonstrate the variable impact that disrupting heme homeostasis can have on a bacterial pathogen. Moreover, this further highlights the importance of resolving the heme paradox in pathogenesis.
SUMMARY
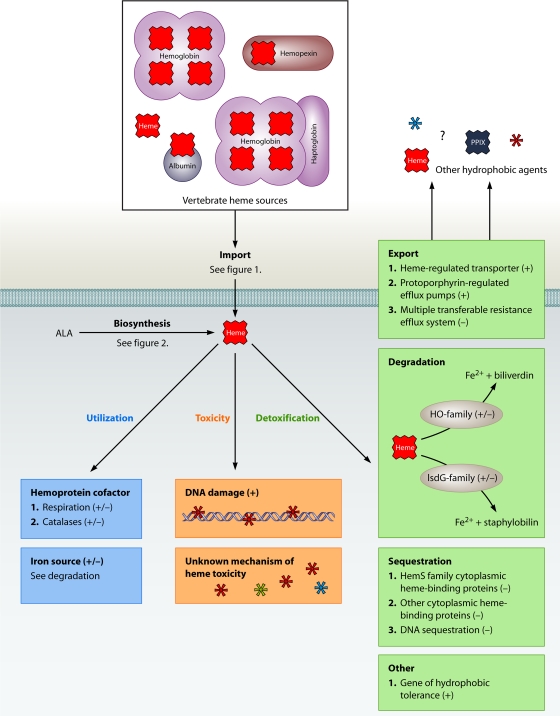
Heme is a required cofactor and a useful source of nutrient iron for most bacterial pathogens (Fig. 3). Bacteria may synthesize their own heme and/or acquire it from the host environment (Fig. 3). While we have discovered much about each of these processes independently, the interplay between heme biosynthesis and acquisition is an understudied area of infectious disease biology. For example, it is not known if heme is differentially segregated depending on whether it is acquired exogenously or synthesized endogenously. Moreover, the impact of heme acquisition on heme synthesis has not been evaluated. While it is known that the utility of heme as a redox cycling cofactor poses the risk of heme toxicity; how heme toxicity contributes to the outcome of host-pathogen interactions remains to be determined.
FIG. 3.
Overview of the heme paradox in bacteria. Most Gram-negative and Gram-positive bacteria are capable of acquiring or synthesizing heme. Heme can then be used for cellular processes (blue boxes). However, an accumulation of intracellular heme can cause toxicity to the cell (orange boxes). Bacteria utilize multiple mechanisms to eliminate heme toxicity (green boxes). Processes found for Gram-negative bacteria are marked with a “−” in parentheses, while those identified for Gram-positive bacteria are marked with a “+” in parentheses.
Bacterial heme toxicity is multifactorial, and although it is possible that the DNA-damaging activity of heme impacts its toxicity, other factors are likely to contribute to this process (Fig. 3). Despite our incomplete understanding of how heme is toxic to bacteria, we have made much progress in identifying the methods that bacteria employ to solve the heme paradox. These mechanisms include export, sequestration, and degradation strategies (Fig. 3). These heme tolerance mechanisms are often carefully regulated by TCSs and transcription factors that respond to changes in environmental and intracellular heme. Now that many of the heme-sensing systems have been identified, future work will focus on determining how these systems sense heme as a component of vertebrate blood. An understanding of the exact mechanisms of bacterial heme toxicity not only will provide basic scientific knowledge about how heme causes toxicity but also will provide novel targets for therapeutic intervention.
Acknowledgments
We thank members of the Skaar laboratory for their critical reading of the manuscript.
Work in the Skaar laboratory is supported by the Searle Scholars Program, NIH grant U54 AI057157-06 from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense, and grants AI069233 and AI073843 from the National Institute of Allergy and Infectious Diseases, NIH. L.L.A. is supported by grant 5 T32 HL069765 from the National Institute of Allergy and Infectious Diseases, NIH. E.P.S. is a Burroughs Wellcome Investigator in the pathogenesis of infectious diseases.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Biography
 Laura L. Anzaldi was born and raised in Pittsburgh, PA. In 2008, she received a B.S. in Chemistry with a concentration in Pharmacology from Duke University. Some of her academic distinctions include graduation summa cum laude and induction into the ΦΒΚ honors society. She completed her undergraduate honors thesis under the direction of Dr. Eric Toone. She also spent two summers at the University of Pittsburgh in the Research Experience for Undergraduates (REU) program. There she was trained in the laboratory of Dr. David Waldeck. After completing her undergraduate degree, she began training at Vanderbilt University in the laboratory of Dr. Eric Skaar, where she is currently working on her Ph.D. Her research focuses on determining the mechanisms by which Gram-positive pathogens tolerate heme toxicity. She is interested in the use of small molecules as molecular probes for studying bacterial pathogenesis and the identification of novel therapeutic targets.
Laura L. Anzaldi was born and raised in Pittsburgh, PA. In 2008, she received a B.S. in Chemistry with a concentration in Pharmacology from Duke University. Some of her academic distinctions include graduation summa cum laude and induction into the ΦΒΚ honors society. She completed her undergraduate honors thesis under the direction of Dr. Eric Toone. She also spent two summers at the University of Pittsburgh in the Research Experience for Undergraduates (REU) program. There she was trained in the laboratory of Dr. David Waldeck. After completing her undergraduate degree, she began training at Vanderbilt University in the laboratory of Dr. Eric Skaar, where she is currently working on her Ph.D. Her research focuses on determining the mechanisms by which Gram-positive pathogens tolerate heme toxicity. She is interested in the use of small molecules as molecular probes for studying bacterial pathogenesis and the identification of novel therapeutic targets.
 Eric P. Skaar received a B.S. in Bacteriology from the University of Wisconsin at Madison in 1996, where he trained in the laboratory of Dr. Timothy Donohue. Upon completion of his undergraduate degree, Dr. Skaar initiated graduate training at Northwestern University in the laboratory of Dr. Hank Seifert, where he received both his Ph.D. and M.P.H. degrees in 2002. Dr. Skaar pursued postdoctoral training with Dr. Olaf Schneewind at the University of Chicago, where his research focused on the contribution of metal metabolism to the pathogenesis of Gram-positive infections. Since 2005, he has been on the faculty of the Department of Microbiology and Immunology, where he is currently an Associate Professor. Research in the Skaar laboratory is focused on determining how bacteria acquire nutrients when they are inside their vertebrate hosts, with the long-term goal of being to inhibit these processes.
Eric P. Skaar received a B.S. in Bacteriology from the University of Wisconsin at Madison in 1996, where he trained in the laboratory of Dr. Timothy Donohue. Upon completion of his undergraduate degree, Dr. Skaar initiated graduate training at Northwestern University in the laboratory of Dr. Hank Seifert, where he received both his Ph.D. and M.P.H. degrees in 2002. Dr. Skaar pursued postdoctoral training with Dr. Olaf Schneewind at the University of Chicago, where his research focused on the contribution of metal metabolism to the pathogenesis of Gram-positive infections. Since 2005, he has been on the faculty of the Department of Microbiology and Immunology, where he is currently an Associate Professor. Research in the Skaar laboratory is focused on determining how bacteria acquire nutrients when they are inside their vertebrate hosts, with the long-term goal of being to inhibit these processes.
Editor: A. T. Maurelli
Footnotes
Published ahead of print on 2 August 2010.
REFERENCES
- 1.Aft, R. L., and G. C. Mueller. 1983. Hemin-mediated DNA strand scission. J. Biol. Chem. 258:12069-12072. [PubMed] [Google Scholar]
- 2.Aft, R. L., and G. C. Mueller. 1984. Hemin-mediated oxidative degradation of proteins. J. Biol. Chem. 259:301-305. [PubMed] [Google Scholar]
- 3.Allen, C. E., and M. P. Schmitt. 2009. HtaA is an iron-regulated hemin binding protein involved in the utilization of heme iron in Corynebacterium diphtheriae. J. Bacteriol. 191:2638-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnoux, P., R. Haser, N. Izadi-Pruneyre, A. Lecroisey, and M. Czjzek. 2000. Functional aspects of the heme bound hemophore HasA by structural analysis of various crystal forms. Proteins 41:202-210. [DOI] [PubMed] [Google Scholar]
- 5.Bibb, L. A., N. D. King, C. A. Kunkle, and M. P. Schmitt. 2005. Analysis of a heme-dependent signal transduction system in Corynebacterium diphtheriae: deletion of the chrAS genes results in heme sensitivity and diminished heme-dependent activation of the hmuO promoter. Infect. Immun. 73:7406-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibb, L. A., C. A. Kunkle, and M. P. Schmitt. 2007. The ChrA-ChrS and HrrA-HrrS signal transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae. Infect. Immun. 75:2421-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biville, F., H. Cwerman, S. Letoffe, M. S. Rossi, V. Drouet, J. M. Ghigo, and C. Wandersman. 2004. Haemophore-mediated signalling in Serratia marcescens: a new mode of regulation for an extra cytoplasmic function (ECF) sigma factor involved in haem acquisition. Mol. Microbiol. 53:1267-1277. [DOI] [PubMed] [Google Scholar]
- 8.Boynton, T. O., L. E. Daugherty, T. A. Dailey, and H. A. Dailey. 2009. Identification of Escherichia coli HemG as a novel, menadione-dependent flavodoxin with protoporphyrinogen oxidase activity. Biochemistry 48:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bozja, J., K. Yi, W. M. Shafer, and I. Stojiljkovic. 2004. Porphyrin-based compounds exert antibacterial action against the sexually transmitted pathogens Neisseria gonorrhoeae and Haemophilus ducreyi. Int. J. Antimicrob. Agents 24:578-584. [DOI] [PubMed] [Google Scholar]
- 10.Brickman, T. J., C. K. Vanderpool, and S. K. Armstrong. 2006. Heme transport contributes to in vivo fitness of Bordetella pertussis during primary infection in mice. Infect. Immun. 74:1741-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brüggemann, H., R. Bauer, S. Raffestin, and G. Gottschalk. 2004. Characterization of a heme oxygenase of Clostridium tetani and its possible role in oxygen tolerance. Arch. Microbiol. 182:259-263. [DOI] [PubMed] [Google Scholar]
- 12.Bryan-Jones, D. G., and R. Whittenbury. 1969. Haematin-dependent oxidative phosphorylation in Streptococcus faecalis. J. Gen. Microbiol. 58:247-260. [DOI] [PubMed] [Google Scholar]
- 13.Bullen, J. J., and E. Griffiths. 1999. Iron and infection: molecular, physiological and clinical aspects. John Wiley & Sons, New York, NY.
- 14.Calvin, M. 1961. Chemical evolution. Oregon State System of Higher Education, Eugene, OR.
- 15.Cescau, S., H. Cwerman, S. Létoffé, P. Delepelaire, C. Wandersman, and F. Biville. 2007. Heme acquisition by hemophores. Biometals 20:603-613. [DOI] [PubMed] [Google Scholar]
- 16.Chen, C. J., D. M. Tobiason, C. E. Thomas, W. M. Shafer, H. S. Seifert, and P. F. Sparling. 2004. A mutant form of the Neisseria gonorrhoeae pilus secretin protein PilQ allows increased entry of heme and antimicrobial compounds. J. Bacteriol. 186:730-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chim, N., A. Iniguez, T. Q. Nguyen, and C. W. Goulding. 2009. Unusual diheme conformation of the heme-degrading protein from Mycobacterium tuberculosis. J. Mol. Biol. 395:595-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou, A., and C. Fitch. 1981. Mechanism of hemolysis induced by ferriprotoporphyrin IX. J. Clin. Invest. 68:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cope, L., R. Yogev, U. Muller-Eberhard, and E. Hansen. 1995. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J. Bacteriol. 177:2644-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cope, L. D., S. E. Thomas, Z. Hrkal, and E. J. Hansen. 1998. Binding of heme-hemopexin complexes by soluble HxuA protein allows utilization of this complexed heme by Haemophilus influenzae. Infect. Immun. 66:4511-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daskaleros, P. A., and S. M. Payne. 1987. Congo red binding phenotype is associated with hemin binding and increased infectivity of Shigella flexneri in the HeLa cell model. Infect. Immun. 55:1393-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Been, M., M. J. Bart, T. Abee, R. J. Siezen, and C. Francke. 2008. The identification of response regulator-specific binding sites reveals new roles of two-component systems in Bacillus cereus and closely related low-GC Gram-positives. Environ. Microbiol. 10:2796-2809. [DOI] [PubMed] [Google Scholar]
- 23.Dockal, M., D. C. Carter, and F. Ruker. 1999. The three recombinant domains of human serum albumin. J. Biol. Chem. 274:29303-29310. [DOI] [PubMed] [Google Scholar]
- 24.Dryla, A., D. Gelbmann, A. Von Gabain, and E. Nagy. 2003. Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol. Microbiol. 49:37-53. [DOI] [PubMed] [Google Scholar]
- 25.Everse, J., and N. Hsia. 1997. The toxicities of native and modified hemoglobins. Free Radic. Biol. Med. 22:1075-1099. [DOI] [PubMed] [Google Scholar]
- 26.Fabian, M., E. Solomaha, J. S. Olson, and A. W. Maresso. 2009. Heme transfer to the bacterial cell envelope occurs via a secreted hemophore in the Gram-positive pathogen Bacillus anthracis. J. Biol. Chem. 284:32138-32146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez, A., D. Lechardeur, A. Derre-Bobillot, E. Couve, P. Gaudu, and A. Gruss. 2010. Two coregulated efflux transporters modulate intracellular heme and protoporphyrin IX availability in Streptococcus agalactiae. PLoS Pathog. 6:e1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fitch, C. D. 1998. Involvement of heme in the antimalarial action of chloroquine. Trans. Am. Clin. Climatol. Assoc. 109:97-106. [PMC free article] [PubMed] [Google Scholar]
- 29.Frankenberg, L., M. Brugna, and L. Hederstedt. 2002. Enterococcus faecalis heme-dependent catalase. J. Bacteriol. 184:6351-6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frankenberg-Dinkel, N. 2004. Bacterial heme oxygenases. Antioxid. Redox Signal. 6:825-834. [DOI] [PubMed] [Google Scholar]
- 31.Garduno, R. A., and W. W. Kay. 1992. Interaction of the fish pathogen Aeromonas salmonicida with rainbow trout macrophages. Infect. Immun. 60:4612-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gat, O., G. Zaide, I. Inbar, H. Grosfeld, T. Chitlaru, H. Levy, and A. Shafferman. 2008. Characterization of Bacillus anthracis iron-regulated surface determinant (Isd) proteins containing NEAT domains. Mol. Microbiol. 70:983-999. [DOI] [PubMed] [Google Scholar]
- 33.Ghigo, J., S. Letoffe, and C. Wandersman. 1997. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J. Bacteriol. 179:3572-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grenier, D. 1991. Hemin-binding property of Porphyromonas gingivalis outer membranes. FEMS Microbiol. Lett. 61:5. [DOI] [PubMed] [Google Scholar]
- 35.Guégan, R., J.-M. Camadro, I. S. Girons, and M. Picardeau. 2003. Leptospira spp. possess a complete haem biosynthetic pathway and are able to use exogenous haem sources. Mol. Microbiol. 49:745-754. [DOI] [PubMed] [Google Scholar]
- 36.Guo, Y., G. Guo, X. Mao, W. Zhang, J. Xiao, W. Tong, T. Liu, B. Xiao, X. Liu, Y. Feng, and Q. Zou. 2008. Functional identification of HugZ, a heme oxygenase from Helicobacter pylori. BMC Microbiol. 8:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagman, K. E., W. Pan, B. G. Spratt, J. T. Balthazar, R. C. Judd, and W. M. Shafer. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611-622. [DOI] [PubMed] [Google Scholar]
- 38.Hansson, M., and L. Hederstedt. 1992. Cloning and characterization of the Bacillus subtilis hemEHY gene cluster, which encodes protoheme IX biosynthetic enzymes. J. Bacteriol. 174:8081-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hassan, S., K. Ohtani, R. Wang, Y. Yuan, Y. Wang, Y. Yamaguchi, and T. Shimizu. 2010. Transcriptional regulation of hemO encoding heme oxygenase in Clostridium perfringens. J. Microbiol. 48:96-101. [DOI] [PubMed] [Google Scholar]
- 40.Henderson, D. P., and S. M. Payne. 1994. Vibrio cholerae iron transport systems: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect. Immun. 62:5120-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinnebusch, B., R. Perry, and T. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 42.Hirst, I. D., T. S. Hastings, and A. E. Ellis. 1994. Utilization of haem compounds by Aeromonas salmonicida. J. Fish Dis. 17:365-373. [Google Scholar]
- 43.Hopkinson, B. M., K. L. Roe, and K. A. Barbeau. 2008. Heme uptake by Microscilla marina and evidence for heme uptake systems in the genomes of diverse marine bacteria. Appl. Environ. Microbiol. 74:6263-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hrkal, Z., Z. Vodrázka, and I. Kalousek. 1974. Transfer of heme from ferrihemoglobin and ferrihemoglobin isolated chains to hemopexin. Eur. J. Biochem. 43:73-78. [DOI] [PubMed] [Google Scholar]
- 45.Idei, A., E. Kawai, H. Akatsuka, and K. Omori. 1999. Cloning and characterization of the Pseudomonas fluorescens ATP-binding cassette exporter, HasDEF, for the heme acquisition protein HasA. J. Bacteriol. 181:7545-7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito, Y., S. Nakagawa, A. Komagata, M. Ikeda-Saito, Y. Shiro, and H. Nakamura. 2009. Heme-dependent autophosphorylation of a heme sensor kinase, ChrS, from Corynebacterium diphtheriae reconstituted in proteoliposomes. FEBS Lett. 583:2244-2248. [DOI] [PubMed] [Google Scholar]
- 47.Jani, D., R. Nagarkatti, W. Beatty, R. Angel, C. Slebodnick, J. Andersen, S. Kumar, and D. Rathore. 2008. HDP—a novel heme detoxification protein from the malaria parasite. PLoS Pathog. 4:e1000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaur, A. P., and A. Wilks. 2007. Heme inhibits the DNA binding properties of the cytoplasmic heme binding protein of Shigella dysenteriae (ShuS). Biochemistry 46:2994-3000. [DOI] [PubMed] [Google Scholar]
- 49.Kirschner-Zilber, I., E. Rabizadeh, and N. Shaklai. 1982. The interaction of hemin and bilirubin with the human red cell membrane. Biochim. Biophys. Acta 690:10. [DOI] [PubMed] [Google Scholar]
- 50.Krewulak, K. D., and H. J. Vogel. 2008. Structural biology of bacterial iron uptake. Biochim. Biophys. Acta 1778:1781-1804. [DOI] [PubMed] [Google Scholar]
- 51.Kristiansen, M., J. H. Graversen, C. Jacobsen, O. Sonne, H. J. Hoffman, S. K. Law, and S. K. Moestrup. 2001. Identification of the haemoglobin scavenger receptor. Nature 409:198-201. [DOI] [PubMed] [Google Scholar]
- 52.Kunkle, C., and M. Schmitt. 2007. Comparative analysis of hmuO function and expression in Corynebacterium species. J. Bacteriol. 189:3650-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lansky, I. B., G. S. Lukat-Rodgers, D. Block, K. R. Rodgers, M. Ratliff, and A. Wilks. 2006. The cytoplasmic heme-binding protein (PhuS) from the heme uptake system of Pseudomonas aeruginosa is an intracellular heme-trafficking protein to the delta-regioselective heme oxygenase. J. Biol. Chem. 281:13652-13662. [DOI] [PubMed] [Google Scholar]
- 54.Lechardeur, D., A. Fernandez, B. Robert, P. Gaudu, P. Trieu-Cuot, G. Lamberet, and A. Gruss. 2010. The 2-Cys peroxiredoxin alkyl hydroperoxide reductase C binds heme and participates in its intracellular availability in Streptococcus agalactiae. J. Biol. Chem. 285:16032-16041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee, W. C., M. L. Reniere, E. P. Skaar, and M. E. Murphy. 2008. Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J. Biol. Chem. 283:30957-30963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Létoffé, S., J. M. Ghigo, and C. Wandersman. 1994. Secretion of the Serratia marcescens HasA protein by an ABC transporter. J. Bacteriol. 176:5372-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Létoffé, S., F. Nato, M. E. Goldberg, and C. Wandersman. 1999. Interactions of HasA, a bacterial haemophore, with haemoglobin and with its outer membrane receptor HasR. Mol. Microbiol. 33:546-555. [DOI] [PubMed] [Google Scholar]
- 58.Létoffé, S., V. Redeker, and C. Wandersman. 1998. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol. Microbiol. 28:1223-1234. [DOI] [PubMed] [Google Scholar]
- 59.Lewis, L. A., M. Gipson, K. Hartman, T. Ownbey, J. Vaughn, and D. W. Dyer. 1999. Phase variation of HpuAB and HmbR, two distinct haemoglobin receptors of Neisseria meningitidis DNM2. Mol. Microbiol. 32:977-989. [DOI] [PubMed] [Google Scholar]
- 60.Lewis, L. A., E. Gray, Y. P. Wang, B. A. Roe, and D. W. Dyer. 1997. Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis. Mol. Microbiol. 23:737-749. [DOI] [PubMed] [Google Scholar]
- 61.Lewis, L. A., M. H. Sung, M. Gipson, K. Hartman, and D. W. Dyer. 1998. Transport of intact porphyrin by HpuAB, the hemoglobin-haptoglobin utilization system of Neisseria meningitidis. J. Bacteriol. 180:6043-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lillard, J. J. W., J. D. Fetherston, L. Pedersen, M. L. Pendrak, and R. D. Perry. 1997. Sequence and genetic analysis of the hemin storage (hms) system of Yersinia pestis. Gene 193:13-21. [DOI] [PubMed] [Google Scholar]
- 63.Lin, H., and J. Everse. 1987. The cytotoxic activity of hematoheme: evidence for two different mechanisms. Anal. Biochem. 161:323-331. [DOI] [PubMed] [Google Scholar]
- 64.Liu, M., W. N. Tanaka, H. Zhu, G. Xie, D. M. Dooley, and B. Lei. 2008. Direct hemin transfer from IsdA to IsdC in the iron-regulated surface determinant (Isd) heme acquisition system of Staphylococcus aureus. J. Biol. Chem. 283:6668-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lyristis, M., A. E. Bryant, J. Sloan, M. M. Awad, I. T. Nisbet, D. L. Stevens, and J. I. Rood. 1994. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol. Microbiol. 12:761-777. [DOI] [PubMed] [Google Scholar]
- 66.Macias, R. I., J. J. Marin, and M. A. Serrano. 2009. Excretion of biliary compounds during intrauterine life. World J. Gastroenterol. 15:817-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maresso, A. W., T. J. Chapa, and O. Schneewind. 2006. Surface protein IsdC and sortase B are required for heme-iron scavenging of Bacillus anthracis. J. Bacteriol. 188:8145-8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mazmanian, S., E. Skaar, A. Gaspar, M. Humayun, P. Gornicki, J. Jelenska, A. Joachmiak, D. Missiakas, and O. Schneewind. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906-909. [DOI] [PubMed] [Google Scholar]
- 69.Mey, A. R., and S. M. Payne. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42:835-849. [DOI] [PubMed] [Google Scholar]
- 70.Mills, M., and S. M. Payne. 1997. Identification of shuA, the gene encoding the heme receptor of Shigella dysenteriae, and analysis of invasion and intracellular multiplication of a shuA mutant. Infect. Immun. 65:5358-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morton, D. J., T. W. Seale, L. L. Madore, T. M. VanWagoner, P. W. Whitby, and T. L. Stull. 2007. The haem-haemopexin utilization gene cluster (hxuCBA) as a virulence factor of Haemophilus influenzae. Microbiology 153:215-224. [DOI] [PubMed] [Google Scholar]
- 72.Murray, G. L., K. M. Ellis, M. Lo, and B. Adler. 2008. Leptospira interrogans requires a functional heme oxygenase to scavenge iron from hemoglobin. Microbes Infect. 10:791-797. [DOI] [PubMed] [Google Scholar]
- 73.Muryoi, N., M. T. Tiedemann, M. Pluym, J. Cheung, D. E. Heinrichs, and M. J. Stillman. 2008. Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J. Biol. Chem. 283:28125-28136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Negari, S., J. Sulpher, F. Pacello, K. Ingrey, A. Battistoni, and B. Lee. 2008. A role for Haemophilus ducreyi Cu,ZnSOD in resistance to heme toxicity. Biometals 21:249-258. [DOI] [PubMed] [Google Scholar]
- 75.Nielsen, M. J., H. J. Møller, and S. K. Moestrup. 2010. Hemoglobin and heme scavenger receptors. Antioxid. Redox Signal. 12:261-273. [DOI] [PubMed] [Google Scholar]
- 76.Nir, U., H. Ladan, Z. Malik, and Y. Nitzan. 1991. In vivo effects of porphyrins on bacterial DNA. J. Photochem. Photobiol. B Biol. 11:295-306. [DOI] [PubMed] [Google Scholar]
- 77.Nitzan, Y., H. Ladan, and Z. Malik. 1987. Growth-inhibitory effect of hemin on staphylococci. Curr. Microbiol. 14:279-284. [Google Scholar]
- 78.Nitzan, Y., H. M. Wexler, and S. M. Finegold. 1994. Inactivation of anaerobic bacteria by various photosensitized porphyrins or by hemin. Curr. Microbiol. 29:125-131. [DOI] [PubMed] [Google Scholar]
- 79.Ochsner, U. A., Z. Johnson, and M. L. Vasil. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146:185-198. [DOI] [PubMed] [Google Scholar]
- 80.Olliaro, P. L., and D. E. Goldberg. 1995. The Plasmodium digestive vacuole: metabolic headquarters and choice drug target. Parasitol. Today 11:294-297. [DOI] [PubMed] [Google Scholar]
- 81.Pacello, F., P. R. Langford, J. S. Kroll, C. Indiani, G. Smulevich, A. Desideri, G. Rotilio, and A. Battistoni. 2001. A novel heme protein, the Cu,Zn-superoxide dismutase from Haemophilus ducreyi. J. Biol. Chem. 276:30326-30334. [DOI] [PubMed] [Google Scholar]
- 82.Panek, H., and M. R. O'Brian. 2002. A whole genome view of prokaryotic haem biosynthesis. Microbiology 148:2273-2282. [DOI] [PubMed] [Google Scholar]
- 83.Paquelin, A., J. M. Ghigo, S. Bertin, and C. Wandersman. 2001. Characterization of HasB, a Serratia marcescens TonB-like protein specifically involved in the haemophore-dependent haem acquisition system. Mol. Microbiol. 42:995-1005. [DOI] [PubMed] [Google Scholar]
- 84.Parrow, N. L., J. Abbott, A. R. Lockwood, J. M. Battisti, and M. F. Minnick. 2009. Function, regulation, and transcriptional organization of the hemin utilization locus of Bartonella quintana. Infect. Immun. 77:307-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pedersen, M. B., C. Garrigues, K. Tuphile, C. Brun, K. Vido, M. Bennedsen, H. Mollgaard, P. Gaudu, and A. Gruss. 2008. Impact of aeration and heme-activated respiration on Lactococcus lactis gene expression: identification of a heme-responsive operon. J. Bacteriol. 190:4903-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pishchany, G., S. E. Dickey, and E. P. Skaar. 2009. Subcellular localization of the Staphylococcus aureus heme iron transport components IsdA and IsdB. Infect. Immun. 77:2624-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ponka, P. 1999. Cell biology of heme. Am. J. Med. Sci. 318:241. [DOI] [PubMed] [Google Scholar]
- 88.Posey, J. E., and F. C. Gherardini. 2000. Lack of a role for iron in the Lyme disease pathogen. Science 288:1651-1653. [DOI] [PubMed] [Google Scholar]
- 89.Puri, S., and M. O'Brian. 2006. The hmuQ and hmuD genes from Bradyrhizobium japonicum encode heme-degrading enzymes. J. Bacteriol. 188:6476-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qi, Z., I. Hamza, and M. R. O'Brian. 1999. Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc. Natl. Acad. Sci. U. S. A. 96:13056-13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rasmussen, A. W., H. L. Alexander, D. Perkins-Balding, W. M. Shafer, and I. Stojiljkovic. 2005. Resistance of Neisseria meningitidis to the toxic effects of heme iron and other hydrophobic agents requires expression of ght. J. Bacteriol. 187:5214-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ratliff, M., W. Zhu, R. Deshmukh, A. Wilks, and I. Stojiljkovic. 2001. Homologues of Neisserial heme oxygenase in Gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183:6394-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reniere, M. L., and E. P. Skaar. 2008. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol. Microbiol. 69:1304-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reniere, M. L., V. J. Torres, and E. P. Skaar. 2007. Intracellular metalloporphyrin metabolism in Staphylococcus aureus. Biometals 20:333-345. [DOI] [PubMed] [Google Scholar]
- 95.Ridley, K., J. Rock, Y. Li, and J. Ketley. 2006. Heme utilization in Campylobacter jejuni. J. Bacteriol. 188:7862-7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rossi, M. S., J. D. Fetherston, S. Letoffe, E. Carniel, R. D. Perry, and J. M. Ghigo. 2001. Identification and characterization of the hemophore-dependent heme acquisition system of Yersinia pestis. Infect. Immun. 69:6707-6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rossi, M. S., A. Paquelin, J. M. Ghigo, and C. Wandersman. 2003. Haemophore-mediated signal transduction across the bacterial cell envelope in Serratia marcescens: the inducer and the transported substrate are different molecules. Mol. Microbiol. 48:1467-1480. [DOI] [PubMed] [Google Scholar]
- 98.Sander, A., S. Kretzer, W. Bredt, K. Oberle, and S. Bereswill. 2000. Hemin-dependent growth and hemin binding of Bartonella henselae. FEMS Microbiol. Lett. 189:55-59. [DOI] [PubMed] [Google Scholar]
- 99.Schmitt, T. H., W. A. Frezzatti, and S. Schreier. 1993. Hemin-induced lipid membrane disorder and increased permeability: a molecular model for the mechanism of cell lysis. Arch. Biochem. Biophys. 307:96-103. [DOI] [PubMed] [Google Scholar]
- 100.Schöffler, H., and V. Braun. 1989. Transport across the outer membrane of Escherichia coli K12 via the FhuA receptor is regulated by the TonB protein of the cytoplasmic membrane. Mol. Gen. Genet. 217:378-383. [DOI] [PubMed] [Google Scholar]
- 101.Skaar, E. P., A. H. Gaspar, and O. Schneewind. 2006. Bacillus anthracis IsdG, a heme-degrading monooxygenase. J. Bacteriol. 188:1071-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Skaar, E. P., A. H. Gaspar, and O. Schneewind. 2004. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279:436-443. [DOI] [PubMed] [Google Scholar]
- 103.Skaar, E. P., and O. Schneewind. 2004. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 6:390-397. [DOI] [PubMed] [Google Scholar]
- 104.Skare, J. T., and K. Postle. 1991. Evidence for a TonB-dependent energy transduction complex in Escherichia coli. Mol. Microbiol. 5:2883-2890. [DOI] [PubMed] [Google Scholar]
- 105.Smalley, J. W., and A. J. Birss. 1999. Iron protoporphyrin IX-albumin complexing increases the capacity and avidity of its binding to the periodontopathogen Porphyromonas gingivalis. Microb. Pathog. 26:131-137. [DOI] [PubMed] [Google Scholar]
- 106.Stauff, D. L., D. Bagaley, V. J. Torres, R. Joyce, K. L. Anderson, L. Kuechenmeister, P. M. Dunman, and E. P. Skaar. 2008. Staphylococcus aureus HrtA is an ATPase required for protection against heme toxicity and prevention of a transcriptional heme stress response. J. Bacteriol. 190:3588-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stauff, D. L., and E. P. Skaar. 2009. Bacillus anthracis HssRS signalling to HrtAB regulates haem resistance during infection. Mol. Microbiol. 72:763-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stauff, D. L., and E. P. Skaar. 2009. The heme sensor system of Staphylococcus aureus. Contrib. Microbiol. 16:120-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stevens, M., S. Porcella, J. Klesney-Tait, S. Lumbley, S. Thomas, M. Norgard, J. Radolf, and E. Hansen. 1996. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect. Immun. 64:1724-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stocker, R., Y. Yamamoto, A. F. McDonagh, A. N. Glazer, and B. N. Ames. 1987. Bilirubin is an antioxidant of possible physiological importance. Science 235:1043-1046. [DOI] [PubMed] [Google Scholar]
- 111.Stojiljkovic, I., B. D. Evavold, and V. Kumar. 2001. Antimicrobial properties of porphyrins. Expert Opin. Invest. Drugs 10:309-320. [DOI] [PubMed] [Google Scholar]
- 112.Stojiljkovic, I., and K. Hantke. 1994. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13:719-732. [DOI] [PubMed] [Google Scholar]
- 113.Stojiljkovic, I., V. Kumar, and N. Srinivasan. 1999. Non-iron metalloporphyrins: potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol. Microbiol. 31:429-442. [DOI] [PubMed] [Google Scholar]
- 114.Suits, M., G. Pal, K. Nakatsu, A. Matte, M. Cygler, and Z. Jia. 2005. Identification of an Escherichia coli O157:H7 heme oxygenase with tandem functional repeats. Proc. Natl. Acad. Sci. U. S. A. 102:16955-16960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Thompson, J. M., H. A. Jones, and R. D. Perry. 1999. Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect. Immun. 67:3879-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tong, Y., and M. Guo. 2009. Bacterial heme-transport proteins and their heme-coordination modes. Arch. Biochem. Biophys. 481:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tong, Y., and M. Guo. 2007. Cloning and characterization of a novel periplasmic heme-transport protein from the human pathogen Pseudomonas aeruginosa. J. Biol. Inorg. Chem. 12:735-750. [DOI] [PubMed] [Google Scholar]
- 118.Torres, A. G., and S. M. Payne. 1997. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23:825-833. [DOI] [PubMed] [Google Scholar]
- 119.Torres, V. J., D. L. Stauff, G. Pishchany, J. S. Bezbradica, L. E. Gordy, J. Iturregui, K. L. Anderson, P. Dunman, S. Joyce, and E. P. Skaar. 2007. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1:109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Unno, M., T. Matsui, G. C. Chu, M. Couture, T. Yoshida, D. L. Rousseau, J. S. Olson, and M. Ikeda-Saito. 2004. Crystal structure of the dioxygen-bound heme oxygenase from Corynebacterium diphtheriae: implications for heme oxygenase function. J. Biol. Chem. 279:21055-21061. [DOI] [PubMed] [Google Scholar]
- 121.Vanderpool, C. K., and S. K. Armstrong. 2001. The Bordetella bhu locus is required for heme iron utilization. J. Bacteriol. 183:4278-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.von Eiff, C., C. Heilmann, R. A. Proctor, C. Woltz, G. Peters, and F. Gotz. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179:4706-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611-647. [DOI] [PubMed] [Google Scholar]
- 124.Warner, D. M., W. M. Shafer, and A. E. Jerse. 2008. Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol. Microbiol. 70:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wegele, R., R. Tasler, Y. Zeng, M. Rivera, and N. Frankenberg-Dinkel. 2004. The heme oxygenase(s)-phytochrome system of Pseudomonas aeruginosa. J. Biol. Chem. 279:45791-45802. [DOI] [PubMed] [Google Scholar]
- 126.Wilks, A., and M. Schmitt. 1998. Expression and characterization of a heme oxygenase (HmuO) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. J. Biol. Chem. 273:837-841. [DOI] [PubMed] [Google Scholar]
- 127.Wyckoff, E., M. Schmitt, A. Wilks, and S. Payne. 2004. HutZ is required for efficient heme utilization in Vibrio cholerae. J. Bacteriol. 186:4142-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wyckoff, E. E., G. F. Lopreato, K. A. Tipton, and S. M. Payne. 2005. Shigella dysenteriae ShuS promotes utilization of heme as an iron source and protects against heme toxicity. J. Bacteriol. 187:5658-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yamamoto, Y., C. Poyart, P. Trieu-Cuot, G. Lamberet, A. Gruss, and P. Gaudu. 2005. Respiration metabolism of group B Streptococcus is activated by environmental haem and quinone and contributes to virulence. Mol. Microbiol. 56:525-534. [DOI] [PubMed] [Google Scholar]
- 130.Zhu, H., M. Liu, and B. Lei. 2008. The surface protein Shr of Streptococcus pyogenes binds heme and transfers it to the streptococcal heme-binding protein Shp. BMC Microbiol. 8:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhu, H., G. Xie, M. Liu, J. S. Olson, M. Fabian, D. M. Dooley, and B. Lei. 2008. Pathway for heme uptake from human methemoglobin by the iron-regulated surface determinants system of Staphylococcus aureus. J. Biol. Chem. 283:18450-18460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhu, W., D. Hunt, A. Richardson, and I. Stojiljkovic. 2000. Use of heme compounds as iron sources by pathogenic neisseriae requires the product of the hemO gene. J. Bacteriol. 182:439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhu, W., A. Wilks, and I. Stojiljkovic. 2000. Degradation of heme in Gram-negative bacteria: the product of the hemO gene of neisseriae is a heme oxygenase. J. Bacteriol. 182:6783-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]