Abstract
CD44, the primary receptor for hyaluronic acid, plays an important role in tumor growth and metastasis. CD44-hyaluronic acid interactions can be exploited for targeted delivery of anti-cancer agents specifically to cancer cells. Although various splicing variants of CD44 are expressed on the plasma membrane of cancer cells, the hyaluronic acid binding domain (HABD) is highly conserved among the CD44 splicing variants. Using a novel two-step process, we have identified monothiophosphate-modified aptamers (thioaptamers) that specifically bind to the CD44’s HABD with high affinities. Binding affinities of the selected thioaptamers for the HABD were in the range of 180–295 nM, significantly higher affinity than that of hyaluronic acid (Kd > μM range). The selected thioaptamers bound to CD44 positive human ovarian cancer cell lines (SKOV3, IGROV, and A2780), but failed to bind CD44 negative NIH3T3 cell line. Our results indicated that thio substitution at specific positions of the DNA phosphate backbone result in specific and high affinity binding of thioaptamers to CD44. The selected thioaptamers will be of great interest for further development as a targeting or imaging agent to deliver therapeutic payloads for cancer tissues.
The CD44 proteoglycan family of transmembrane glycoproteins is ubiquitously expressed in physiological and pathological systems (1). Studies of tumorigenesis using CD44 antibodies and vaccines have shown the importance of CD44 in tumor growth and metastasis (2, 3). CD44 is the primary cell surface receptor for hyaluronic acid (HA)1, binding this ligand via a Link module, a lectin-like fold (4, 5). The HA binding domain (HABD) is stabilized by three disulfide bridges and is centered around the N-terminal Link module in the extra cellular domain of CD44, but extends beyond the Link involving additional basic residues. The Link module is highly conserved among CD44 family members and has only two disulfide bridges. The solution and crystal structures of the HABD are available (6–8). Although CD44 is coded by a single gene, numerous transcripts are formed by alternative splicing. Standard CD44 (CD44S) is comprised of the constant, non-variant exon products, whereas the variant isoforms arise by splicing of additional exon products into a single site within the membrane-proximal region of the ectodomain. Carcinomas typically produce several CD44 variants as well as decreased proportions of CD44S, and many studies have implicated CD44 variants rather than CD44S in tumor progression (9, 10). CD44 requires “activation” with respect to HA binding and consequent signaling, possibly through localization of CD44 within specialized plasma membrane lipid micro-domains or rafts, from which endocytosis of HA and CD44 occurs (11).
CD44 is increasingly recognized as a marker for subpopulations of tumor-initiating cells or cancer stem cells (CSCs), that are highly malignant and chemoresistant, likely because of increased anti-apoptotic pathway activity and enrichment of multidrug transporters (12). Epithelial/mesenchymal transition (EMT) has been linked to the properties of CD44+/CD24− CSCs, as well as to HA (13). There is increasing evidence for HA-dependent association of CD44 with receptor tyrosine kinases and transporters, important in drug resistance and malignancy (14). Recent studies of CD44+ CSC-like subpopulation of cells isolated from human patient epithelial ovarian carcinoma specimens and ascites revealed that the CSCs are enriched with ABC-family drug transporters, ABCG2/BCRP1 and MDR1, as well as activated TLRR4/MyD88 and NF-κB (15–17). These mechanisms may account for the drug-resistance of CSC, a critical aspect of their phenotype and importance in therapeutic response. HA-CD44 interactions can be exploited for delivery of chemotherapeutic drugs and other anticancer agents to cancer cells. Many investigators have shown increased efficacy in cell and animal tumor models by conjugating drugs to HA or anti-CD44 antibodies, as well as incorporating drugs or siRNAs into vehicles that have been decorated with HA or antibodies (18, 19). Further selective targeting to tumor-relevant and over-expressed variants of CD44 is a substantial goal. However, there is evidence that HA interactions with the CD44 splice variants are diminished compared to those with CD44S (20), contrary to this goal. Thus, the application of aptamer technology to develop a library of high-affinity and individually specific CD44S and CD44 splice-variant aptamers is a significant first step to achieving this level of refined targeting.
Aptamers are small, structurally distinct oligonucleotide molecules that exhibit specific and high binding affinities to proteins and other biological macromolecules. Aptamers are emerging as attractive alternatives over conventional ligand such as antibodies and peptides for diagnostic and therapeutic applications (21–24). Aptamers can be obtained through an in vitro selection process (25, 26) against virtually any kind of molecule whereas antibodies generally require biological systems that must be induced by an immune response. Compared to antibodies, the smaller size of aptamers makes them easier to synthesize in large quantities as well as to introduce a wide range of chemical modifications. Through chemical modifications, the kinetic parameters and binding affinities can be modified for the aptamers (27). Aptamers, in general, exhibit longer shelf life and they are easy to store.
Native oligonucleotides are susceptible to digestion by cellular nucleases but sugar-phosphate backbone modifications can render them more resistant to degradation (28, 29). Among the several modifications reported over the last two decades, the sulfur substitution of the phosphate backbone is the most commonly performed (29). Thioaptamers are backbone modified aptamers where one (monothio) or both (dithio) of the non-bridging phosphoryl oxygens are substituted with sulphur. Since sulfur substitution often increases the binding affinity of the oligonucleotide to the protein (30, 31), complete substitution of the phosphate backbone might lead to non-specific binding of the selected aptamer. A novel method developed in our laboratory (32, 33) allows us to optimize both the number and position of the thio substitutions in the DNA aptamer sequence.
Using combinatorial selection methods, a strategy for identifying nucleotide sequences with high affinity for the target protein, we have selected DNA thioaptamers that specifically bind to the HABD of CD44. In this paper, we report the selection, binding and cell-based assays of the thioaptamers selected against the HABD of CD44.
MATERIALS AND METHODS
Materials
Cell-free recombinant protein expression kits (RTS500HY) containing E. coli lysates were obtained from Roche Diagnostics (Indianapolis, IN). The CD44 HABD construct, cloned into the pET19b vector, was obtained from Genescript Inc, (Piscataway, NJ). Templates for the DNA synthesis were obtained from Midland Certified Reagents (Midland, TX). Taq polymerase and chirally pure Sp isomer of dATP(αS) were obtained from Axxora LLC (San Diego, CA). Magnetic (MPG) Streptavidin beads, used for the single-strand, biotinylated DNA isolation, were purchased from Pure Biotech, (Middlesex, NJ). Recombinant human CD44-Fc chimera, containing residues 21–220 of CD44 fused with human IgG (residues 100–330), was obtained from R&D Systems Inc (Minneapolis, MN). HABP was purchased from EMD Chemicals (Gibbstown, NJ). CD44 antibody was purchased from Abcam (Cambridge, MA). TOPO TA cloning kits were obtained from Invitrogen (Carlsbad, CA) and the plasmid isolation kits were obtained from Qiagen (Foster city, CA). Biotinylated HA was purchased from Sigma (St. Louis, MO).
Cell lines and culture
The human epithelial CD44+ ovarian cancer cell lines, SKOV3, IGROV, and A2780 were kind gifts from Dr. Anil K. Sood (MD Anderson Cancer Center). The cells were maintained in RPMI-1640 medium supplemented with 15% fetal bovine serum and 0.1% Gentamicin sulfate. CD44− NIH3T3 mouse fibroblast cell line was maintained in Dulbecco modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum. All experiments were performed with 70 to 80% confluent cultures with 5% CO2 at 37 °C.
Protein expression and purification
The CD44 HABD (20–178 aminoacid residues) was produced by in-vitro (cell-free) expression methods using the RTS500HY kit (E. coli based cell-free expression system). DNA encoding residues 20–178 was synthesized and cloned into the expression vector pET19b between the NdeI and BamH1 sites. A 3 mL reaction mixture was used for the cell-free expression, using the RTS500 HY kit, following procedures described previously (34). After 12 hours of incubation at 30°C with gentle agitation, the protein was recovered by centrifugation and refolded and purified following published procedures (35). The purity and concentration of the recovered protein were analyzed by analytical gel-electrophoresis and UV absorbance, respectively. HA binding to the isolated CD44-HABD was confirmed using biotinylated HA. The tertiary structure and the proper folding of the CD44 HABD were confirmed by testing with conformation-specific monoclonal antibody F10.44.2 and 15N-HSQC NMR spectroscopy (6, 7).
Thioaptamer library synthesis
The initial library was synthesized from a 73-mer DNA template containing a 30-nt random region flanked by 21-mer and 22-mer PCR primer regions. The library was annealed with the reverse primer, subjected to Klenow reaction for 5 hrs at 37 °C, and amplified by PCR using AmpliTaq DNA polymerase and a mixture of dATP(aS), dTTP, dCTP, and dGTP to give the thio-substituted library. Reaction conditions for the PCR amplifications were: oligonucleotide library (40 nM), dNTP mixture of dATP(aS), dTTP, dCTP, and dGTP (80 μM each), MgCl2 (2 mM), 5′-biotinylated forward primer and reverse primer (300 nM of each) and AmpliTaq DNA polymerase (1 U) in a total volume of 100 μL. PCR was run for 35 cycles of 94°C (2 min), 55°C (2 min) and 72°C (2 min). The resulting 73-mer library contained monothiophosphate substitutions (in the Sp configuration) on the 5′ side of every dA residue with the exception of the primer region on the non-template strand.
Single-stranded DNA (ss-DNA) was isolated from the double-stranded PCR products by the following procedure using the MPG Streptavidin beads. The 5′-biotinylated DNA strands were mixed with MPG Streptavidin beads in buffer containing 10mM Tris-HCl, 2M NaCl, and 1mM EDTA, (pH 7.4) and incubated for 30 minutes. After washing the magnetic beads bound oligonucleotides 3 times, the melting solution (0.1M NaOH) was added to remove the non-biotinylated strand. The solution of ss-DNA TA library was pooled, purified and denatured in binding buffer at 95°C for 5 min and quenched on ice. Fluorescein-labeled, single-stranded TA library was synthesized from the PCR reaction of the template with 5′-biotinylated forward primer and 5′-fluorescein-labeled reverse primer.
Combinatorial selection of thioaptamers
Purified CD44 HABD containing the histidine tag was bound to the Nickel-Nitro-triacetic acid (Ni-NTA) beads in buffer containing 50 mM Tris-HCl, 50 mM NaCl, pH 7.4. Prior to screening the TA library to select sequences binding the CD44-HABD, the sequences that could bind the Ni-NTA beads were excluded from the library by incubating the initial ssDNA library with Ni-NTA beads, in binding buffer (50 mM Tris-HCl, 20 mM MgCl2, pH 7.4). The reaction mixture was spun at 500 g for 5 minutes and the supernatant was carefully removed, and the unbound TA library in the supernatant was used in the selection experiments. This pre-screened TA library (200 pmoles) in binding buffer (50 mM Tris-HCl, 20 mM MgCl2, pH 7.4) was mixed with CD44-HABD protein bound to the Ni-NTA beads (400-100pmols) and incubated with gentle shaking for 30–60 minutes. The beads were spun, placed on a magnet and the supernatant liquid was carefully removed. The TA bound Ni-NTA beads were washed 4 times with the binding buffer to remove unbound sequences, and the bound TAs were subsequently eluted with 500 mM imidazole in binding buffer. The eluted TA library was PCR amplified and taken to the next cycle of the iteration. At each iterative cycle, the stringency of binding was increased by gradually decreasing, both the amount of protein and incubation time and increasing the number of washes of the TA-protein complex on the beads. All incubations were done at room temperature.
Cloning procedure and structural analysis
The selected TA libraries from the 5th and 10th rounds were amplified with a dNTP mixture and unmodified primers and cloned into pCT2.1 TOPO vector and transformed into E. coli TOPO 10F′ cells. The E. coli colonies containing plasmid DNA with single sequences were picked, the plasmids were isolated using the miniprep kit (Qiagen), and sequenced. A subset of sequences from the initial library, the 5th and 10th selection rounds were determined. The primary sequences obtained from the 10th cycle were analyzed using the ClustalW program (36) (www.ebi.ac.uk/clustalw) and were grouped into six groups based on the convergence of the primary sequence.
Filter binding assays
A filter binding assay was used to measure the equilibrium binding constants of selected TAs to both the CD44-Fc chimera and the recombinant CD44-HABD. Biotinylated TAs (15 nM) were incubated with varying concentrations of the CD44 proteins (20 to 1500 nM) in 3.5 μL of buffer-T (10 mM Tris-HCl, pH 7.4) for 30 minutes. The dot-blot apparatus (BioRad) was set up with filter pads at the bottom, nylon membrane in the middle (to trap the uncomplexed TAs) and nitrocellulose membrane on top (to trap the CD44-TA complex) and all the membranes were wetted with buffer-T. After incubation samples were diluted to 30 μL with buffer-T and transferred to the dot-blot apparatus and filtered under vacuum. The membranes were washed with buffer-T (100 μL) three times to wash away any unbound TAs from the nitrocellulose membrane down to the nylon membrane. After the washes, the nitrocellulose membrane was vacuum-baked and the nylon membrane was UV cross-linked to immobilize the retained aptamers. The membranes were processed for chemiluminescent detection using Pierce biotinylated nucleic acid detection kit following the manufacturer’s instructions. The chemiluminescent signals were detected and imaged on a Chemimager (Alpha Innotech). Image analysis and quantification of spot intensities were carried out using the ImageJ program (ver.1.43) (37). For binding analysis on nitrocellulose, background spot intensity due to buffer effect was subtracted from all data points and saturation binding curves were generated by curve fits assuming a single binding site. In the case of HABD, data from protein dependent TA depletion on the nylon membrane was considered for analysis. The nylon spot intensity in the absence of protein was considered as total TA present. The depleted spot intensities due to protein binding for each protein concentration were calculated by subtracting from the intensity corresponding to total TA present. To confirm the specificity of the selected TAs to CD44 HABD, the selected TAs were tested for binding to another HA binding protein, HABP. This protein has the conserved Link module involving the HA binding site, and shares 32.3% sequence identity with CD44 (38). Failure of the TAs to bind to HABP shows that the TA binding to CD44 HABD is not due to non-specific electrostatic interactions.
CD44 immunostaining
The cells were grown on glass chamber slides and fixed with 4% para-formaldehyde for 10 minutes and then washed with PBS buffer. The slides were incubated with protein block (10% normal goat serum in PBS) for 30 minutes and incubated with primary rat anti-mouse CD44 antibody (1:800 dilution, Abcam) at room temperature for 1 hour. The same amount of affinity-purified normal IgG from the corresponding species was used as a negative control. The slides were washed and incubated with secondary goat anti-rat Alexa 568 IgG (1:400 dilution) for 30 min at room temperature in the dark. The slides were then washed with PBS and rinsed with deionized water. Nuclear counterstaining was performed with Hoechst 33342 (Sigma) for 5 min at room temperature. The images were acquired under the same condition using fluorescent microscopy (TE2000-E, Nikon) and processed by using Nikon Elements. Five randomly selected high power fields at x60 magnification were shown.
Cell binding assay
CD44 positive ovarian cancer cell lines (SKOV3, IGROV, and A2780) and the CD44 negative NIH3T3 cell line were plated onto 8-well glass chamber slides to allow them to attach for 24 hours. The cells were incubated with 50 nM of fluorescein-labeled TAs for 10 minutes at 37°C. The slides were immediately washed with ice cold PBS and nuclei were counterstained with Hoechst 33342 (Sigma) for 5 min at room temperature. Series of images were obtained under the same conditions when fluorescence intensities needed to be compared.
RESULTS
Sequences of selected thioaptamers
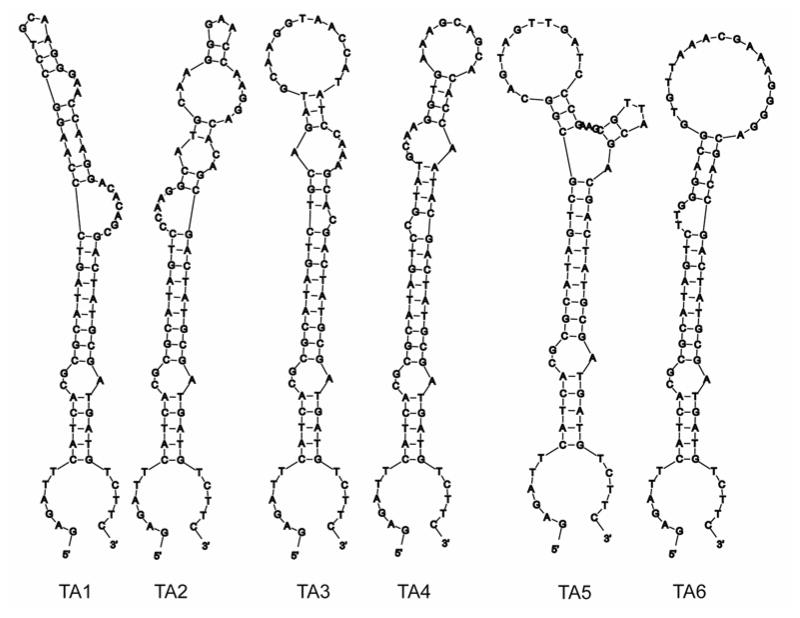
After 10 rounds of selection using a combinatorial library containing potentially 1014 different sequences, the TA library converged to several sequence families. Based on the primary sequence alignments determined by the ClustalW program (36), the TA sequences were categorized into six groups (TA1-TA6). The variable region of the selected sequences is shown in Figure 1. The -TGCAAGG(G/T)AACCA- motif was found to occur in four out of the six groups. Primary sequences of TA1 and TA2 differ by only one base. Since only dATP (αS) was used in the polymerization steps, all adenosines in the selected sequences beyond position 22, have a 5′-monothiophosphate substitution. The MFOLD (39) (mfold.bioinfo.rpi.edu) predicted secondary structures indicated that all 6 selected TA sequences can form hairpin loop structures with the random region forming the loop and the primer regions making up the stem regions (Figure 2).
Figure 1.
Primary sequences of the thioaptamers selected after the 10th round of selection. Only the sequences of the variable, random region are shown. Sequences were aligned using the ClustalW program (36).
Figure 2.
Secondary structures of the six thioaptamers. Secondary structures were predicted with the MFold program (39). The random regions are shown with a box. (A) TA1. (B) TA2. (C) TA3. (D) TA4. (E) TA5. (F) TA6.
Specific binding of selected thioaptamers to CD44+ cells
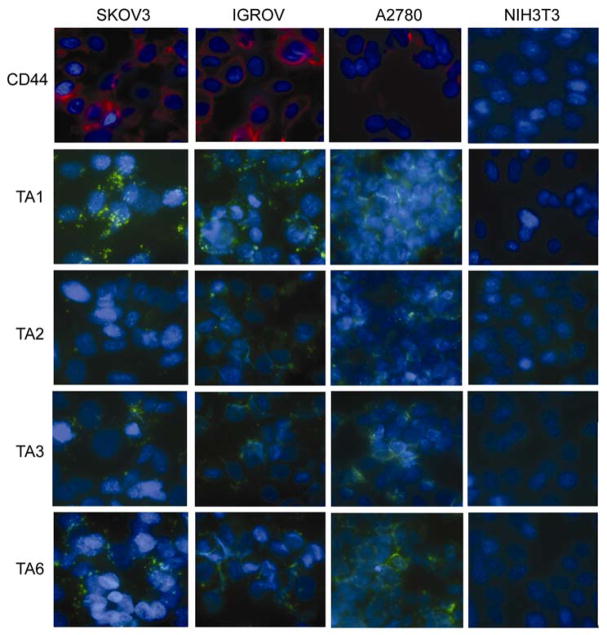
To test the selective binding of TAs to CD44, fluorescently tagged TAs (50 nM) were incubated with both CD44+ ovarian cancer cell lines (SKOV3, IGROV, and A2780) (39) and the CD44− NIH3T3 cell line for 10 min at 37°C. The fluorescent intensities and localization were compared using a fluorescent microscope. TA1 and TA6 demonstrated rapid binding to the cell membranes of all CD44+ ovarian cancer cell lines at a concentration of 50 nM (Figure 3), whereas TA2 and TA3 showed weaker binding to the CD44+ cells. TA4 and TA5 did not show any binding to the CD44+ cells. In sharp contrast to the TA binding to CD44+ cells, none of TAs bound to CD44− NIH3T3 cells (Figure 3). Among 5 TA sequences, TA1 showed the highest binding to the CD44+ ovarian cancer cells tested. This is supported by the high number of occurrences of TA1 sequences within the set of cloned sequences in the final selection round (data not shown). TAs were found in punctate structures at the membrane, which coincides with known CD44 surface expression patterns (40).
Figure 3.
TA binding to CD44+ cells. CD44+ ovarian cancer cell lines (SKOV3, IGROV, and A2780) and the CD44 negative NIH3T3 cell line were incubated with 50 nM of fluorescein-labeled TAs for 10 minutes at 37°C. The slides were counterstained with Hoechst 33342. green, fluorescein labeled CD44 TA; blue, nuclear counter staining; red, CD44.
Binding of selected thioaptamers to CD44 HABD and CD44-Fc
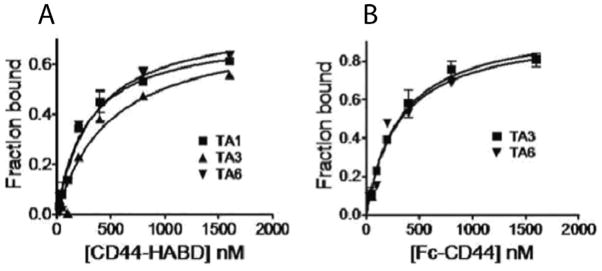
Filter-binding methods were used to quantitatively measure the binding of the selected TAs to the CD44. We used two constructs for this purpose, the HABD and CD44-Fc proteins. Filter-binding assays were performed with the three TAs that showed binding to the CD44+ cells. All three TAs, −TA1, TA3 and T6, showed binding to CD44-HABD. Only TA3 and TA6 showed binding to CD44-Fc. TA1 did not show binding to the CD44-Fc. The saturation binding curves are shown in Figure 4. The equilibrium dissociation constants, Kd, were derived from these curves and are listed in Table 1. All TAs tested showed nanomolar range of binding constants to the CD44 proteins (180–285 nM). The binding affinities (nM) of the CD44 TAs are significantly better than that of HA (Kd = 61 μM (6)). TA6 bound to the CD44 HABD and CD44-Fc with same affinity, while TA3 showed higher binding affinity towards CD44-Fc. Both TA3 and TA6 showed similar binding constants with CD44-Fc. TA1 did not show any detectable binding to CD44-Fc. To confirm the specificity of our TAs to CD44, we tested the binding of a random, 73-mer monothio-substituted oligonucleotide to CD44. This random oligonucleotide failed to bind to both the recombinant CD44-HABD and CD44-Fc. All three TAs failed to bind to another HA binding protein, HABP. Both these results confirm that the selected TAs are specific to the CD44-HABD.
Figure 4.
Saturation binding curves of thioaptamers to CD44. Biotinylated thioaptamers were used in the assays. (A) Binding curves for CD44-Fc. The chemilumniscent spot intensities on the nitrocellulose membrane, due to retention of thioaptamers, are plotted as a function of total CD44-Fc concentration. (B) Binding curves for CD44-HABD. The chemilumniscent spot intensities on the nylon membrane, due to depletion of thioaptamers, are plotted as a function of total CD44-HABD concentrations.
Table 1.
Equilibrium dissociation constants of selected thioaptamers towards CD44-Fc and CD44-HABD. The dissociation constants were derived from saturation binding curves shown in Figure 4. Binding experiments were performed at room temperature and pH 7.4.
| Thioaptamer |
CD44-HABD |
CD44-Fc |
|---|---|---|
| TA1 | 190.6 ± 25.4 nM | |
| TA3 | 285.4 ± 83.1 nM | 188.1 ± 26.8 nM |
| TA6 | 187.0 ± 30.6 nM | 179.4 ± 60.8 nM |
DISCUSSION
In this study, we have selected TAs that specifically bind to the CD44’s HABD with the goal of using these TAs for targeted delivery of therapeutic drugs to cancer cells expressing CD44. Targeted delivery of anti-cancer drugs specifically to the cancer cells will greatly enhance the efficacy and reduce adverse effects. Since CD44 is a membrane protein that is over-expressed on the plasma membrane of cancer cells and internalized into the cells upon ligand binding, CD44 targeted delivery presents a powerful strategy for selective cancer cell targeting. Since the HABD is highly conserved among all the splicing variants and all splicing variants contain the HABD in their extracellular domain, TAs specific to the HABD are valuable tools to recognize cancer cells that express CD44 splicing variants.
Previous studies have reported using HA for selective targeting of cancer cells expressing CD44. HA-bound liposomes were used to deliver encapsulated doxorubicin to tumor cells expressing high levels of CD44 (41). However, there are several limitations in using HA for targeted delivery. HA is naturally present in the human body and can bind to several proteins besides CD44 (42), leading to non-specific binding to other proteins. Our TAs showed specific binding to CD44, and failed to bind to another HA binding protein, HABP. HA is a large polymer and difficult to synthesize compared to oligonucleotides. Furthermore, several studies have reported that HA interactions with the CD44 splice variants are diminished compared to those with CD44S (20). The binding constants for the selected TAs reported in this study, are significantly lower compared to the binding constants for HA to CD44. Since TAs show higher affinity and specificity than the HA, TAs would be a better ligand of HABD to achieve higher level of refined targeting.
HA is known to regulate tumor progression through different mechanisms. Several studies have shown that HA binding activates CD44 and stimulates tumor progression (11, 43, 44). HA, in its very large, high molecular weight form is part of the extra-cellular matrix and is reported to promote cancer initiation and progression by activating signaling pathways and host-tumor interactions (45, 46). Since the TAs are selected against the HABD of CD44, binding of the TAs would disrupt any further binding of HA to CD44 and could potentially interfere with tumor progression that depends on the HA binding mediated activation of CD44. While the high molecular weight forms of HA have been shown to promote tumor progression, the small, low molecular weight forms of HA have been shown to suppress tumor progression (47). The TAs are similar to the low molecular weight forms of the HA, and therefore, might not trigger the same signaling pathways as the high-molecular weight HAs, and promote tumor progression. Furthermore, binding of small HA oligomers to the surface of CD44 appears to enhance the HA uptake (48). The selected TAs, being small oligonuelcotides, might increase the internalization upon binding to CD44. TAs have several advantages over unmodified oligonucleotide aptamers. Thio-modified oligonucleotides exhibit higher level of resistance to digestion by cellular nucleases (28, 29). TAs also show tighter binding towards target proteins compared to unmodified aptamers (30, 49). It appears that the thiophosphates do not bind sodium ions as effectively as the unmodified phosphates, and thus the thio-substituted phosphate esters act as nearly bare anions (50). Since less energy is required to strip the cations from the backbone of thio-substituted oligonucleotides, these agents can in principle bind more tightly to proteins. However, complete thio-substitution of oligonucleotides could lead to non-specific binding to other cellular proteins (31). Therefore, to achieve selective, high-affinity binding, optimization of the number of thio substitutions is critical. Our combinatorial selection methods allow us to select TAs with hybrid backbones where the number and the position of the sulfur substitutions are optimized. This approach allows us to simultaneously select for both the sequence and the best monothiophosphate substitutions. In contrast, almost all modified aptamers are first selected by SELEX and then post-selection modified. As we have shown previously (49–51), monothiothiophosphate substitutions can perturb both the structure and interaction with the target and therefore, it is better to select both sequence and modifications simultaneously. While the TAs selected showed specific binding to CD44, unmodified oligonucleotides containing the same primary sequences failed to bind to the CD44 proteins or to the CD44+ cells (data not shown). This observation shows that sulfur substitution at specific positions of the oligonucleotides led to specificity and higher affinity binding to the CD44.
We have used recombinant CD44-HABD for the iterative selection cycles. For membrane binding assays, we have used both the CD44-Fc chimera and CD44-HABD recombinant protein. It has been shown that the CD44 HABD (20–178) and CD44-Fc are structurally similar and share a common tertiary fold. Based on the similar reactivity of CD44-HABD and CD44-Fc towards the conformation sensitive mAbs BRIC-235 and F10.44.2, Teriete et al (52) showed these two constructs are structurally similar. The concentration-dependent binding of HA to CD44-Fc is very similar to that of CD44-HABD. Our data demonstrated that TAs selected against CD44-HABD bound to both CD44-HABD and CD44-Fc with similar dissociation constants, supporting the notion of bioequivalence between these two protein constructs.
All TAs showed rapid and selective binding to cultured CD44+ cells, but not to the CD44− cells. Although pre-incubation of the CD44+ cells with HA (1 mM, 10 kD) failed to block the binding of CD44 TA (data not shown), it does not preclude the HABD selective binding of CD44 TA since there are substantial difference in the binding affinities between TA and HA to CD44. We identified TAs that bind to the CD44 HABD at nanomolar affinity, significantly higher binding than its natural ligand HA, and will be of great interest for further development as a targeting or imaging agent to deliver therapeutic payloads for cancer tissues.
Acknowledgments
We thank Dr. Anil Sood of the University of Texas MD Anderson Cancer Center for the gift of CD44 positive cancer cell lines used in this study, and Dr. David Volk of the University of Texas Health Science Center for helpful discussions.
Footnotes
This work was supported by National Institutes of Health (GM084552, N01-HV28184, CC987654321, R01CA128797, 2U54CA096300-06A1), Department of Defense (W81XWH-07-2-0101, W81XWH-09-1-0212), Welch Foundation (H-1296), and the Alliance for Nano Health.
Abbreviations: HA, hyaluronic acid; HABD, hyaluronic acid binding domain; HABP, hyaluronic acid binding protein; IgG, immunoglobulin G; dNTP, deoxy nucleotide triphosphate; TA, thioaptamer;
References
- 1.Naor D, Sionov RV, Ish-Shalom D. CD44: structure, function, and association with the malignant process. Adv Cancer Res. 1997;71:241–319. doi: 10.1016/s0065-230x(08)60101-3. [DOI] [PubMed] [Google Scholar]
- 2.Strobel T, Swanson L, Cannistra SA. In vivo inhibition of CD44 limits intra-abdominal spread of a human ovarian cancer xenograft in nude mice: a novel role for CD44 in the process of peritoneal implantation. Cancer Res. 1997;57:1228–1232. [PubMed] [Google Scholar]
- 3.Wallach-Dayan SB, Rubinstein AM, Hand C, Breuer R, Naor D. DNA vaccination with CD44 variant isoform reduces mammary tumor local growth and lung metastasis. Mol Cancer Ther. 2008;7:1615–1623. doi: 10.1158/1535-7163.MCT-07-2383. [DOI] [PubMed] [Google Scholar]
- 4.Kohda D, Morton CJ, Parkar AA, Hatanaka H, Inagaki FM, Campbell ID, Day AJ. Solution structure of the Link module: a hyaluronan-binding domain involved in extracellular matrix stability and cell migration. Cell. 1996;86:767–775. doi: 10.1016/s0092-8674(00)80151-8. [DOI] [PubMed] [Google Scholar]
- 5.Banerji S, Ni J, Wang SX, Clasper S, Su J, Tammi R, Jones M, Jackson DG. LYVE-1, a new homologue of the CD44 glycoprotein, is a lymph-specific receptor for hyaluronan. J Cell Biol. 1999;144:789–801. doi: 10.1083/jcb.144.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerji S, Wright AJ, Noble M, Mahoney DJ, Campbell ID, Day AJ, Jackson DG. Structures of the CD44-hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat Struct Mol Biol. 2007;14:234–239. doi: 10.1038/nsmb1201. [DOI] [PubMed] [Google Scholar]
- 7.Takeda M, Ogino S, Umemoto R, Sakakura M, Kajiwara H, Sugahara KN, Hayasaka H, Miyasaka M, Shimada I. Ligand-induced structural changes of the CD44 hyaluronan-binding domain revealed by NMR. J Biol Chem. 2006;281:40089–40095. doi: 10.1074/jbc.M608425200. [DOI] [PubMed] [Google Scholar]
- 8.Ogino S, Nishida N, Umemoto R, Suzuki M, Takeda M, Terasawa H, Kitayama J, Matsumoto M, Hayasaka H, Miyasaka M, Shimada I. Two-state conformations in the hyaluronan-binding domain regulate CD44 adhesiveness under flow condition. Structure. 2010;18:649–656. doi: 10.1016/j.str.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Skotheim RI, Nees M. Alternative splicing in cancer: noise, functional, or systematic? Int J Biochem Cell Biol. 2007;39:1432–1449. doi: 10.1016/j.biocel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 10.Afify A, Pang L, Howell L. Diagnostic utility of CD44 standard, CD44v6, and CD44v3–10 expression in adenocarcinomas presenting in serous fluids. Appl Immunohistochem Mol Morphol. 2007;15:446–450. doi: 10.1097/01.pai.0000213154.49063.22. [DOI] [PubMed] [Google Scholar]
- 11.Bourguignon LYW, Singleton P, Zhu H, Zhou B. Hyaluronan (HA) promotes signaling interaction between CD44 and the TGF-RI receptor in metastatic breast tumor cells. J Biol Chem. 2002;277:39703–39712. doi: 10.1074/jbc.M204320200. [DOI] [PubMed] [Google Scholar]
- 12.An Y, Ongkeko WM. ABCG2: the key to chemoresistance in cancer stem cells? Expert Opin Drug Metab Toxicol. 2009;5:1529–1542. doi: 10.1517/17425250903228834. [DOI] [PubMed] [Google Scholar]
- 13.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toole BP, Slomiany MG. Hyaluronan, CD44 and Emmprin: partners in cancer cell chemoresistance. Drug Resist Update. 2008;11:110–121. doi: 10.1016/j.drup.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvero AB, Chen R, Fu HH, Montagna M, Schwartz PE, Rutherford T, Silasi DA, Steffensen KD, Waldstrom M, Visintin I, Mor G. Molecular phenotyping of human ovarian cancer stem cells unravels the mechanisms for repair and chemoresistance. Cell Cycle. 2009;8:158–166. doi: 10.4161/cc.8.1.7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu L, McArthur C, Jaffe RB. Ovarian cancer stem-like side-population cells are tumourigenic and chemoresistant. British J Cancer. 2010;13:1276–1283. doi: 10.1038/sj.bjc.6605626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fong MY, Kakar SS. The role of cancer stem cells and the side population in epithelial ovarian cancer. Histol Histopathol. 2010;25:113–120. doi: 10.14670/HH-25.113. [DOI] [PubMed] [Google Scholar]
- 18.Peer D, Margalit R. Tumor-targeted hyaluronan nanoliposomes increase the antitumor activity of liposomal Doxorubicin in syngeneic and human xenograft mouse tumor models. Neoplasia. 2004;6:343–353. doi: 10.1593/neo.03460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auzenne E, Ghosh SC, Khodadadian M, Rivera B, Farquhar D, Price RE, Ravoori M, Kundra V, Freedman RS, Klostergaard J. Hyaluronic acid-paclitaxel: antitumor efficacy against CD44(+) human ovarian carcinoma xenografts. Neoplasia. 2007;9:479–486. doi: 10.1593/neo.07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson DG, Bell JI, Dickinson R, Timans J, Whittle JN. Proteoglycan forms of the lymphocyte homing receptor CD44 are alternatively spliced variants containing the v3 exon. J Cell Biol. 1995;128:673–685. doi: 10.1083/jcb.128.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gold L. Oligonucleotides as research, diagnostic and therapeutic agents. J Biol Chem. 1995;270:13581–13584. doi: 10.1074/jbc.270.23.13581. [DOI] [PubMed] [Google Scholar]
- 22.Thiviyanathan V, Somasunderam AD, Gorenstein DG. Combinatorial selection and delivery of thioaptamers. Biochem Soc Trans. 2007;35:50–52. doi: 10.1042/BST0350050. [DOI] [PubMed] [Google Scholar]
- 23.Cho EJ, Lee JW, Ellington AD. Applications of aptamers as sensors. Annu Rev Anal Chem. 2009;2:241–264. doi: 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- 24.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nature Rev. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands tobacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 26.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 27.Luzi E, Minunni S, Tombelli S, Mascini M. New trends in affinity sensing: aptamers for ligand binding. Trends in Anal Chem. 2003;22:810–818. [Google Scholar]
- 28.Jhaveri S, Olwin B, Ellington AD. In vitro selection of phosphorothiolated aptamers. Bioorg Med Chem Lett. 1998;8:2285–2290. doi: 10.1016/s0960-894x(98)00414-4. [DOI] [PubMed] [Google Scholar]
- 29.Micklefield J. Backbone modification of nucleic acids; synthesis, structure and theraputic applications. Curr Med Chem. 2001;8:1157–1179. doi: 10.2174/0929867013372391. [DOI] [PubMed] [Google Scholar]
- 30.Milligan JF, Uhlenbeck OC. Determination of RNA-protein contacts using thiophosphate substitutions. Biochemistry. 1989;28:2849–2855. doi: 10.1021/bi00433a016. [DOI] [PubMed] [Google Scholar]
- 31.King DJ, Bassett SE, Li X, Fennewald SA, Herzog NK, Luxon BA, Shope R, Gorenstein DG. Combinatorial selection and binding of phosphorothioate aptamers targeting human NF-kappaB RelA(p65) and p50. Biochemistry. 2002;41:9696–9706. doi: 10.1021/bi020220k. [DOI] [PubMed] [Google Scholar]
- 32.King DJ, Ventura DA, Brasier AR, Gorenstein DG. Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry. 1998;37:16489–16493. doi: 10.1021/bi981780f. [DOI] [PubMed] [Google Scholar]
- 33.Somasunderam A, Ferguson MR, Rojo DR, Thiviyanathan V, Li X, O’Brien WA, Gorenstein DG. Combinatorial selection, inhibition and antiviral activity of DNA thioapatamers targeting the RNase H domain of HIV-1 reverse transcriptase. Biochemistry. 2005;44:10388–10395. doi: 10.1021/bi0507074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neerathilingam M, Greene LH, Colebrooke SA, Campbell ID, Staunton D. Quantitation of protein expression in a cell-free system: efficient detection of yields and 19F NMR to identify folded protein. J Biomol NMR. 2005;31:11–19. doi: 10.1007/s10858-004-5357-6. [DOI] [PubMed] [Google Scholar]
- 35.Banerji S, Day AJ, Kahmann JD, Jackson DG. Characterization of a functional hyaluronan-binding domain from the human CD44 molecule expressed in Escherichia coli. Protein Expr Purif. 1998;14:371–381. doi: 10.1006/prep.1998.0971. [DOI] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nuc Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Intl. 2004;11:36–42. [Google Scholar]
- 38.Kohda D, Morton CJ, Parker AA, Inagaki FM, Campbell ID, Day AJ. Solution Structure of the Link Module: A Hyaluronan-binding domain involved in extracellular matrix stability and cell migration. Cell. 1996;86:767–775. doi: 10.1016/s0092-8674(00)80151-8. [DOI] [PubMed] [Google Scholar]
- 39.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nuc Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H, Hao J, Wang L, Li Y. Co-expression of invasive markers (uPA, CD44) and multiple drug-resistance proteins (MDR1, MRP2) is correlated with epithelial ovarian cancer progression. British J Cancer. 2009;101:432–440. doi: 10.1038/sj.bjc.6605185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eliaz RE, Szoka FC. Liposome-encapsulated Doxyrubicin targeted to CD44. Cancer Res. 2001;61:2592–2601. [PubMed] [Google Scholar]
- 42.Yokoo M, Miyahayashi Y, Naganuma T, Kimura N, Sasada H, Sato E. Identification of hyaluronic acid-binding proteins and their expressions in porcine cumulusoocyte complexes during in vitro maturation. Biol Reproduction. 2002;67:1165–1171. doi: 10.1095/biolreprod67.4.1165. [DOI] [PubMed] [Google Scholar]
- 43.Heldin P, Karousou E, Bernert B, Porsch H, Nishitsuka K, Skandalia SS. Importance of Hyaluronan-CD44 interactions in inflammation and tumorigenesis. Connective Tissue Res. 2008;49:215–218. doi: 10.1080/03008200802143323. [DOI] [PubMed] [Google Scholar]
- 44.Turley EA, Nobel PW, Bourguignon LYW. Signaling properties of hyaluronan receptors. J Biol Chem. 2002;277:4589–4592. doi: 10.1074/jbc.R100038200. [DOI] [PubMed] [Google Scholar]
- 45.Itano N, Zhuo L, Kimata K. Impact of the hyaluronan-rich tumor microenvironment on cancer initiation and progression. Cancer Sci. 2008;99:1720–1725. doi: 10.1111/j.1349-7006.2008.00885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Misra S, Hascall VC, Berger FG, Markwalk RR, Ghatak S. Hyaluronan, CD44 and cyclooxygenase-2 in colon cancer. Connect Tissue Res. 2008;49:219–224. doi: 10.1080/03008200802143356. [DOI] [PubMed] [Google Scholar]
- 47.Alaniz L, Rizzo M, Malvicini M, Jaunarena J, Avella D, Atorrasagasti C, Aquino JB, Garcia M, Matar P, Silva M, Mazzolini G. Low molecular weight hyaluronan inhibits colorectal carcinoma growth by decreasing tumor cell proliferation and stimulating immune response. Cancer Lett. 2009;278:9–16. doi: 10.1016/j.canlet.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 48.Slomiany MG, Dai L, Bomer PA, Knackstedt TJ, Kranc DA, Tolliver L, Maria BL, Toole BP. Abrogating drug resistance in malignant peripheral nerve sheath tumors by disrupting hyaluronan-CD44 interactions with small hyaluronan oligosaccharides. Cancer Res. 2009;69:4992–4998. doi: 10.1158/0008-5472.CAN-09-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X, Gorenstein DG. Progress in thioaptamer development. Curr Drug Targets. 2004;5:705–715. doi: 10.2174/1389450043345074. [DOI] [PubMed] [Google Scholar]
- 50.Volk DE, Power TD, Gorenstein DG, Luxon BA. An ab initio study of phosphorothioate and phosphorodithioate interactions with sodium cation. Tett Lett. 2002;43:4443–4447. [Google Scholar]
- 51.Volk DE, Yang X, Fennewald SM, King DJ, Bassett SE, Venkitachalam S, Herzog N, Luxon BA, Gorenstein DG. Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB. Bioorg Chem. 2002;30:396–419. doi: 10.1016/s0045-2068(02)00510-2. [DOI] [PubMed] [Google Scholar]
- 52.Teriete P, Banerji S, Noble M, Blundell CD, Wright AJ, Pickford AR, Lowe E, Mahoney DJ, Tammi MI, Kahmann JD, Campbell ID, Day AJ, Jackson DG. Structure of the regulatory hyaluronan binding domain in the inflammatory leukocyte homing receptor CD44. Mol Cell. 2004;13:483–496. doi: 10.1016/s1097-2765(04)00080-2. [DOI] [PubMed] [Google Scholar]